Abstract
The partner gene of MLL was identified in a patient with treatment-related acute myeloid leukemia in which the karyotype suggested t(3;11)(q25;q23). Prior therapy included the DNA topoisomerase II inhibitors, teniposide and doxorubicin. Southern blot analysis indicated that the MLL gene was involved in the translocation. cDNA panhandle polymerase chain reaction (PCR) was used, which does not require partner gene-specific primers, to identify the chimeric transcript. Reverse-transcription of first-strand cDNAs with oligonucleotides containing known MLL sequence at the 5′ ends and random hexamers at the 3′ ends generated templates with an intra-strand loop for PCR. In-frame fusions of either MLLexon 7 or exon 8 with the GMPS (GUANOSINE 5′-MONOPHOSPHATE SYNTHETASE) gene from chromosome band 3q24 were detected. The fusion transcript was alternatively spliced. Guanosine monophosphate synthetase is essential for de novo purine synthesis. GMPS is the first partner gene ofMLL on chromosome 3q and the first gene of this type in leukemia-associated translocations.
Introduction
Translocations of the MLL gene at chromosome band 11q23 are prevalent in patients with leukemia after treatment with epipodophyllotoxins and other DNA topoisomerase II inhibitors.1,2 The MLL gene fuses with one of many different partner genes, most of which remain unknown.1 Identification of the partner genes by genomic approaches can be difficult if the translocation breakpoints are within large, uncharacterized introns.3
The complex karyotype in a patient with treatment-related acute myeloid leukemia (t-AML) suggested a t(3;11)(q25;q23).4,5 Although the EAP/MDS1/EVI1 gene cluster on chromosome band 3q26 is involved in the t(3;21) in treatment-related myelodysplasia,6-8 no partner genes of MLL have been described on chromosome 3q. We used cDNA panhandle polymerase chain reaction (PCR) to determine the unknown partner gene in the t(3;11). This led to the identification of a novel MLLtranslocation partner.
Study design
A 3½-year-old boy previously designated patient 164 or 65 was diagnosed with metastatic neuroblastoma. The primary tumor was in the posterior mediastinum. There was bone marrow involvement and widespread bony metastases. He was administered cyclophosphamide (CPM), doxorubicin (ADR), teniposide (VM-26), and vincristine (VCR). At age 8 years, neuroblastoma recurred in the posterior mediastinum and the marrow. He received cisplatin, CPM, ADR, VM-26, and VCR. At age 12 years, neuroblastoma again recurred in the posterior mediastinum and the marrow, at which time the mediastinal mass was resected. He underwent local radiation to the primary tumor bed and then autologous bone marrow transplantation (ABMT) after conditioning with melphalan, VM-26, and total body irradiation. Four months after ABMT, he had a third mediastinal relapse, but there was no evidence of neuroblastoma in the marrow. The marrow karyotype was 46, XY, t(1;7)(q32;q32-34), inv2(p21q37), t(3;11)(q25;q23), add t(7)(q22) in all 29 cells examined. Two months later, at age 13 years, the white blood cell (WBC) count was 4.3 × 109/L with 7% circulating blasts. French-American-British (FAB) M4 AML was diagnosed. Three weeks later, when the WBC count increased to 8.3 × 109/L with 32% circulating blasts, the peripheral blood karyotype in 12 cells was the same as above. The patient received no further therapy. He died of leukemia within 3 months.
Detection of MLL gene rearrangement by Southern blot analysis
Genomic DNA and total RNA were isolated from cryopreserved peripheral blood mononuclear cells containing 32% leukemic blasts.MLL gene rearrangement was examined using the B859 fragment of ALL-1 cDNA and has been reported.5 The Institutional Review Board at The Children's Hospital of Philadelphia approved this research.
Fluorescence in situ hybridization analysis
The MLL probe (Oncor, Gaithersburg, MD) was labeled with digoxigenin and was detected with anti-digoxigenin labeled with rhodamine. For chromosome 3, a probe derived from the P1 clone 868-2, which hybridized to MDS1 cDNA, was used (G. Nucifora and J.D.R., unpublished data, 1995). The probe was labeled with biotin and detected with avidin fluorescein isothiocyanate.
cDNA panhandle PCR analysis of der11transcripts
First-strand cDNAs were synthesized from 1.4 μg total RNA using oligonucleotides containing MLL exon 5 sequence at the 5′ ends and random hexamers at the 3′ ends.9 Conditions and reagents for second-strand cDNA synthesis, formation of stem-loop templates, and PCR with MLL-specific primers have all been described.9 The cDNA panhandle PCR products were subcloned by recombination PCR.9 The subclones were screened by PCR and sequenced.9
Confirmation of der11 transcripts
Two microliters of the same first-strand cDNAs were amplified with MLL exon 6 sense primer 5′-CGCCCAAGTATCCCTGTAAA-3′ orMLL exon 7 sense primer 5′-GCAGATGGAGTCCACAGGAT-3′ and antisense primer 5′-TAGCACGGAATCCTTGTTCC-3′, corresponding to position 336 to 317 of the GMPS (GUANOSINE 5′-MONOPHOSPHATE SYNTHETASE) cDNA (GenBank accession no. NM_003875). Two microliters of the products were used as the template in second rounds of PCR with the same primer combinations. The products were gel-purified and sequenced.
Results and discussion
Because the karyotype suggested involvement of band 11q23 in the t(3;11), we examined peripheral blood mononuclear cells from the time of leukemia diagnosis for MLL gene rearrangement by Southern blot analysis.5 The probe detected 2 rearrangements, consistent with a translocation involvingMLL.5
Fluorescence in situ hybridization (FISH) analysis of 9 metaphase cells with the MLL probe showed hybridization with the normal chromosome 11 and split signals on the der(11) and der(3) chromosomes. This confirmed MLL involvement in the t(3:11) (data not shown). FISH analysis with the MDS1 probe showed that the candidate gene MDS1 and the more distal EVI1 gene were translocated to chromosome 11 but were not fused with MLL(data not shown).
We used cDNA panhandle PCR9 to identify the partner gene in the der(11) transcript. A population of products of various sizes was obtained (Figure 1A). Recombination PCR generated the subclones shown in Figure 1B-C, which demonstrate how cDNA panhandle PCR can identify several differentMLL-containing transcripts in the same reaction. Three subclones contained a fusion of MLL exon 7 to position 150 of the full-length 2212-base pair (bp) GMPScDNA10 (GenBank accession no. NM_003875) (Figure 1B-C, top). Two subclones contained a fusion of MLL exon 8 to the same position of GMPS (Figure 1B-C, top). Ten subclones contained the MLL sequence only. All 10 contained theMLL intron 7 sequence, and 2 contained the MLLintron 8 sequence, suggesting incompletely processed transcripts (Figure 1B-C, bottom).
Analysis of t-AML by cDNA panhandle PCR.
(A) cDNA panhandle PCR analysis of total RNA from peripheral blood mononuclear cells at t-AML diagnosis. As shown by the smear in the second lane of gel (t-AML), a population of products of various sizes was obtained from reverse transcribing first-strand cDNAs with 5′-MLL-random hexamer-3′ oligonucleotides, generating second-strands by MLL primer extension and forming stem-loop templates and 2 sequential PCRs with primers, all fromMLL.9 dH2O control reactions with and without reverse transcriptase (RT) are in lanes 3 and 4. (B) Recombination PCR-generated subclones were screened with the sameMLL primers used in nested PCR.9 The 15 subclones contained inserts that ranged in size from 727 bp to 1653 bp. Subclone numbers above the lanes correspond with the sequences in panel C. Control lane is reagent control. (C) Three subclones contained an in-frame fusion of MLL exon 7 to GMPS at position 150 of the 2212-bp full-length cDNA (GenBank accession no. NM_003875)(top). Two subclones contained a fusion of MLLexon 8 to the same position of GMPS (top). Ten subclones suggested incompletely processed MLL-containing transcripts (bottom). (D) MLL exon 6/GMPS gene-specific primer set yielded 472-bp and 358-bp products, confirming transcripts fusing MLL exon 8 or MLL exon 7, respectively, toGMPS. Lane 2 shows purified product with MLL exon 8-GMPS fusion. Lane 3 shows purified product withMLL exon 7-GMPS fusion. Lane 4 shows a 223-bp product obtained with MLL exon 7/GMPSgene-specific primer set that confirmed MLL exon 7-GMPS fusion.
Analysis of t-AML by cDNA panhandle PCR.
(A) cDNA panhandle PCR analysis of total RNA from peripheral blood mononuclear cells at t-AML diagnosis. As shown by the smear in the second lane of gel (t-AML), a population of products of various sizes was obtained from reverse transcribing first-strand cDNAs with 5′-MLL-random hexamer-3′ oligonucleotides, generating second-strands by MLL primer extension and forming stem-loop templates and 2 sequential PCRs with primers, all fromMLL.9 dH2O control reactions with and without reverse transcriptase (RT) are in lanes 3 and 4. (B) Recombination PCR-generated subclones were screened with the sameMLL primers used in nested PCR.9 The 15 subclones contained inserts that ranged in size from 727 bp to 1653 bp. Subclone numbers above the lanes correspond with the sequences in panel C. Control lane is reagent control. (C) Three subclones contained an in-frame fusion of MLL exon 7 to GMPS at position 150 of the 2212-bp full-length cDNA (GenBank accession no. NM_003875)(top). Two subclones contained a fusion of MLLexon 8 to the same position of GMPS (top). Ten subclones suggested incompletely processed MLL-containing transcripts (bottom). (D) MLL exon 6/GMPS gene-specific primer set yielded 472-bp and 358-bp products, confirming transcripts fusing MLL exon 8 or MLL exon 7, respectively, toGMPS. Lane 2 shows purified product with MLL exon 8-GMPS fusion. Lane 3 shows purified product withMLL exon 7-GMPS fusion. Lane 4 shows a 223-bp product obtained with MLL exon 7/GMPSgene-specific primer set that confirmed MLL exon 7-GMPS fusion.
Amplification of the same first-strand cDNAs with MLL exon 6- and GMPS-specific primers and sequencing of the 358-bp and 472-bp products confirmed both fusion transcripts (Figure 1D). Amplification of a 223-bp product with MLL exon 7- andGMPS-specific primers gave additional verification of theMLL exon 7–GMPS fusion. It has been shown that the MLL gene can be alternatively spliced.11These results indicate that the MLL-GMPS fusion transcript was alternatively spliced.
The GMPS gene, which has been mapped to chromosome band 3q24,12 is a new partner gene of MLL. No other cloned partner genes of MLL are located on chromosome 3q. The locus is distinct from and centromeric to theEAP/MDS1/EVI1 genes at chromosome band 3q26, which are involved in treatment-related myelodysplasia.6-8 Recently, the AF3p21 gene, which encodes a protein with a Src homology 3 domain,13 was identified as the only other partner gene of MLL on chromosome 3.
GMPS is the first gene of this type identified in leukemia-associated translocations. The human GMPS cDNA was cloned after purification of the protein.10 TheGMPS gene produces a single 2.4-kb mRNA.10 The 2079-bp open reading frame encodes a 693-amino acid protein10 with 2 post-translational modification variants.14 The protein is an amidotransferase and is essential for de novo purine synthesis.14,15 It catalyzes the formation of guanosine monophosphate (GMP) by incorporating an amide (NH2) group into xanthosine 5′-monophosphate.16 The amide donor can be either glutamine or ammonia.16 The transfer of the amide group requires hydrolysis of adenosine triphosphate (ATP) to adenosine monophosphate and inorganic pyrophosphate.16 Therefore, GMP synthetase contains a glutamine amide transfer (GAT) domain for glutamine hydrolysis and a synthetase domain for the hydrolysis of ATP (Figure 2).16 There is greater GMP synthetase mRNA and protein expression in leukemic cell lines such as HL60 and U937 and in transformed, lymphoblastoid lines and neoplastic tissues than in non-transformed, non-proliferating cells.10
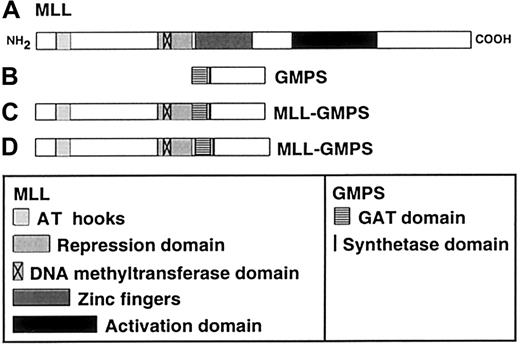
Schematic of putative MLL-GMPS chimeric oncoproteins.
(A) The 3969-amino acid (aa) MLL protein contains an AT hook DNA-binding motif (aa 170-310), a repression domain (aa 1101-1400), a DNA methyltransferase domain (aa 1153-1219), zinc fingers (aa 1434-1917), and an activation domain (aa 2340-3123).21,22(B) The 693-aa GMPS protein contains a GAT domain (aa 1-215) and a synthetase domain (aa 244-250)10 14 (GenBank accession no.P49915). (C) Predicted 2090-aa fusion protein resulting fromMLL exon 7-GMPS-fusion transcript. (D) Predicted 2128-aa fusion protein resulting from MLL exon 8-GMPS-fusion transcript. The MLL AT hook motif, repression domain and DNA methyltransferase domain, and the GMPS GAT and synthetase domains are retained in the chimeric oncoproteins; the zinc fingers and activation domain of MLL would not be retained.
Schematic of putative MLL-GMPS chimeric oncoproteins.
(A) The 3969-amino acid (aa) MLL protein contains an AT hook DNA-binding motif (aa 170-310), a repression domain (aa 1101-1400), a DNA methyltransferase domain (aa 1153-1219), zinc fingers (aa 1434-1917), and an activation domain (aa 2340-3123).21,22(B) The 693-aa GMPS protein contains a GAT domain (aa 1-215) and a synthetase domain (aa 244-250)10 14 (GenBank accession no.P49915). (C) Predicted 2090-aa fusion protein resulting fromMLL exon 7-GMPS-fusion transcript. (D) Predicted 2128-aa fusion protein resulting from MLL exon 8-GMPS-fusion transcript. The MLL AT hook motif, repression domain and DNA methyltransferase domain, and the GMPS GAT and synthetase domains are retained in the chimeric oncoproteins; the zinc fingers and activation domain of MLL would not be retained.
MLL gene translocations are thought to be leukemogenic by producing chimeric oncoproteins.17 Figure 2 shows the structural domains of putative chimeric oncoproteins that would be produced by the MLL-GMPS fusion. The effects of the MLL-GMPS fusion protein on MLL and GMPS function are unknown. TheMLL-GMPS transcripts contain all but the first 27 bases of the GMPS open-reading frame, suggesting that the GAT and synthetase domains both are intact.
We identified GMPS as a novel partner gene of MLLin a patient with FAB M4 AML diagnosed 9½ years from the start of protracted, multimodality therapy for primary and then multiply-relapsed metastatic neuroblastoma. This therapy included teniposide and doxorubicin, both DNA topoisomerase II inhibitors associated with leukemia as a treatment complication.18,19Epipodophyllotoxin-related leukemias with MLL gene translocations usually present as FAB M4 or FAB M5 AML; in this regard, the leukemia was typical.5,18 The mean latency from drug exposure to onset of leukemia is approximately 2 years.18 20 In the patient whom we studied, it cannot be determined when in the course of treatment the translocation first occurred nor which agent(s) contributed.
C.A.F. is supported by National Institutes of Health grants CA66140, CA77683, CA80175, and CA85469 and a Leukemia and Lymphoma Society Scholar Award.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Carolyn A. Felix, Division of Oncology, Abramson Research Center, Room 902B, The Children's Hospital of Philadelphia, 3516 Civic Center Blvd, Philadelphia, PA 19104-4318; e-mail:felix@emailchop.edu.