Abstract
This study reports the first use of gene array technology for the identification of a tumor-specific marker in lymphoid neoplasms. The differential gene expression of 31 hematopoietic cell lines, representing most major lymphoma subgroups of B- and T-cell origin, was assessed by hybridizing labeled complementary DNA to Atlas human expression arrays containing 588 genes. Genes known to be specific for B, T, or myelomonocytic lineages were appropriately identified in the arrays, validating the general utility of this approach. One gene,clusterin, not previously known to be expressed in lymphoid neoplasms, was specifically found in all 4 anaplastic large-cell lymphoma (ALCL) cell lines, but not in any of the 27 remaining tumor lines. Using a monoclonal antibody against clusterin, its differential expression was confirmed by Western blotting and immunohistochemistry. A total of 198 primary lymphomas (representing most major lymphoma subtypes), including 36 cases of systemic ALCL, were surveyed for clusterin expression by immunohistochemistry and Western blotting. All of the 36 ALCL cases marked for clusterin, with most cases showing moderate to strong staining in the majority of neoplastic cells. Clusterin expression was not related to expression of anaplastic lymphoma kinase-1. With 2 exceptions, none of the remaining 162 non-ALCL cases marked with the clusterin antibody, including Hodgkin disease and primary cutaneous ALCL. In reactive lymphoid tissues, only follicular dendritic cells and fibroblastic reticular cells exhibited staining. Clusterin is a highly conserved glycoprotein implicated in intercellular and cell matrix interactions, regulation of the complement system, lipid transport, stress responses, and apoptosis. Although its function in ALCL is unknown, the unique expression of clusterin within this category of lymphoma provides an additional marker for the diagnosis of ALCL. This study illustrates the enormous potential of gene array technologies for diagnostic marker discovery.
The identification of tumor-specific targets for diagnostic or therapeutic use has been a principal goal for both clinicians and biomedical researchers. Traditionally, investigators have accumulated this information through comparative expression studies of individual gene products that have been identified during the course of biochemical, physiologic, or molecular biologic studies of various neoplasms. In this way, a series of diagnostic and prognostic markers have been developed that can distinguish among many types of cancers and provide useful clinical information.
Recent advances in DNA sequencing technology and the development of gene expression arrays have provided investigators with a powerful tool to study the expression of thousands of genes in parallel.1-3 This methodology promises to revolutionize the search for tumor-specific markers. Gene expression arrays are created by depositing unique complementary DNA (cDNA) fragments on a solid matrix that can be either a glass slide or a nylon filter. The slide or filter is then hybridized with labeled cDNA from a tissue of interest. The readout is performed by high throughput fluorescence scanners for fluorescent probe hybridizations or by traditional phosphoimagers when radioactively labeled probes are used. Large format macroarrays containing hundreds of genes as well as high-density microarrays with more than 20 000 different known genes and expressed sequence tags (ESTs) can be generated using high-speed robotic printers.4-6 For the analysis of the hybridization signals on high-density microarrays, it is absolutely essential to have computer assistance, and sophisticated algorithms for this purpose have been developed.7-9
Array technology has broad applications, and its power is being directed toward the study of global gene expression changes in both physiologic and pathologic states. Such diverse physiologic conditions as aging in mice,10 gene expression during sleep cycles,11 serum stimulation of fibroblasts,12irradiation-induced stress,13 yeast sporulation,14 and the diauxic shift from aerobic to anaerobic metabolism in yeast3 have already been put under the microscope of this technology. Various pathologic conditions are also being scrutinized, from inflammatory diseases15 to neoplasia.8,16-21 Array technology is particularly well suited for studying gene expression changes between normal tissues and their corresponding cancers, and there have already been several studies of gene expression in colon cancer,8,16 renal cell carcinoma,17 rhabdomyosarcoma,18leukemias,19 cervical cancer,20 and ovarian carcinoma.21 Most of these latter studies have focused on global differences between normal and cancer in an attempt to better understand the pathogenesis of the particular neoplasm. Validation of array results is generally performed by assessing RNA expression of the sample by traditional Northern blot or reverse transcriptase-polymerase chain reaction because the identification of tumor-specific markers has not been a primary goal of most of these studies.
We have begun to apply this technology for identification of tumor-specific markers in lymphomas. As a first approach we have compared the gene expression profiles of a series of lymphoma cell lines derived from most of the common lymphoma subtypes, using commercial nylon filter macroarrays from Clontech Laboratories. These arrays are composed of 588 known genes grouped into several functional categories. Our initial goal was to validate the use of these arrays by an analysis of genes contained on the filters already known to be differentially expressed among the selected hematopoietic cell lines. After successful validation, we then searched for the presence of previously unknown differentially expressed genes that could potentially represent new diagnostic markers. Differential gene expression was then validated at the protein level on selected cell lines, and finally on a series of primary lymphoma tissues, using a combination of immunoblotting and immunohistochemistry.
We now report a general approach for the identification of tumor-specific markers using gene macroarray technology and describe a new marker, clusterin (Apo J), that is specifically expressed in a subtype of large-cell lymphoma, anaplastic large-cell lymphoma (ALCL).
Materials and methods
Cell lines and growth conditions
Thirty-one hematopoietic cell lines, comprised of 19 B-cell, 10 T-cell, and 2 myeloid cell lines, were selected for comparative gene expression studies. The B- and T-cell lines were chosen to represent a broad range of primary lymphoid neoplasms, and within each neoplastic subtype we attempted to obtain a minimum of 2 examples (Table1).
All cell lines were cultured in RPMI 1640, supplemented with 10% heat-inactivated fetal calf serum, l-glutamine, and antibiotics. To limit the effect of cell culture conditions on gene expression as much as possible, all cell lines were harvested during their exponential growth phase (0.3-0.7 × 106/mL). Centrifugations were carried out at 4°C and total RNA was extracted immediately thereafter.
cDNA arrays
Atlas cDNA expression arrays from Clontech Laboratories (Palo Alto, CA) were chosen for these studies. These arrays consist of 588 human cDNA fragments, organized into broad functional groups. A complete list of the genes included on the membranes is available on the Clontech Web site (http://www.clontech.com). All cDNAs used for printing on the array have been sequence verified by the company.
RNA extraction, labeling, and hybridization of Atlas human cDNA expression arrays
Total RNA was extracted from the cell lines using a guanidium isothiocyanate-based method (Ultraspec II RNA isolation system, Biotex Lab, Houston, TX) according to the manufacturer's instructions. Subsequently, polyA + messenger RNA (mRNA) was isolated from 500 μg of total RNA using 2 rounds of Oligotex beads purification (Qiagen, Valencia, CA), according to the manufacturer's instructions. The polyA + mRNA was then subjected to DNase I treatment to minimize genomic DNA contamination. The quality of the mRNA was assessed by gel electrophoresis, as well as by OD 260/280 ratios.
cDNA labeling, hybridization, and washing of the cDNA Atlas array membranes were carried out according to the instructions accompanying the macroarrays. Briefly, 1 to 2 μg polyA+ RNA was used as template for cDNA synthesis, which was done in the presence of α-P-32–labeled deoxyadenosine triphosphate (dATP) (Amersham Pharmacia Biotech, Piscataway, NJ). The labeled probes were purified by spin column centrifugation (Chroma Spin-200, Clontech Laboratories), and hybridization was carried out at 68°C in a rotation hybridization oven (Robbins Scientific, Sunnyvale, CA) using 1 × 106 cpm/mL (Cherenkov) of radioactive probe. The membranes were then washed at 68°C (4 times with 2 × standard sodium citrate [SSC], 1% sodium dodecyl sulfate [SDS], followed by 2 times 1 × SSC, 0.1% SDS) and exposed for 1 to 3 days and analyzed by a phosphoimaging system (Molecular Dynamics, Sunnyvale, CA). Stripping of the membranes was carried out according to the manufacturer's instructions and they were re-used up to 3 times.
Array exposures on the phosphoimager were normalized by equalizing the intensity of the signals from a set of housekeeping genes provided on the arrays. Following this normalization step, the array images were printed and the hybridization signals of all 588 genes were scored visually on a 0 to 3 scale, with 0 being no signal, and 1 to 3 representing increasing signal intensities. However, to simplify interpretation in our initial analysis reported here, all positive signal intensities (1-3) were grouped together. Therefore, each gene on the array was scored as either positive or negative.
Preferential lineage expression was arbitrarily defined as any gene overrepresented in the cell lines of 1 lineage (B, T, or myeloid) by a ratio of 3:1 or greater, at any signal intensity.
Lymphoma subtype-restricted expression was defined as any gene expressed at any signal intensity in all or all but 1 member of a specific subtype of lymphoma, but not in more than 1 example of the other lymphoma cell line subtypes.
Primary lymphomas
A group of 198 well-characterized lymphomas including 90 T-cell lymphomas, 78 B-cell lymphomas, and 30 cases of Hodgkin lymphoma (Table2) were retrieved from the files of the Hematopathology Section, Laboratory of Pathology, National Cancer Institute, Bethesda, MD. These lymphomas were chosen to represent all major lymphoma subtypes and included 36 cases of primary ALCL and 9 cases of cutaneous ALCL. Among the 78 B-cell non-Hodgkin lymphomas were 12 cases that showed weak or focal CD30 expression. In addition, 30 representative examples of nonneoplastic lymphoid tissues including tonsil, lymph node, and spleen were studied to determine the occurrence and distribution of clusterin in normal lymphoid tissues. All diagnoses were confirmed by immunohistochemistry performed on paraffin-embedded tissue sections, using selected members of a panel of monoclonal antibodies comprised of CD20, CD3, CD4, CD8, CD5, CD10, CD15, CD30, CD43, CD23, ALK-1, TIA-1, perforin, CD56, cyclin D1, and Bcl-2, performed during the diagnostic workup. Anaplastic lymphoma kinase-1 (ALK-1) (Dako Corp, Carpinteria, CA) immunohistochemistry was performed on all cases of ALCL.
Analysis of clusterin expression
To verify clusterin expression, all cases and cell lines were evaluated using a monoclonal anticlusterin antibody (Upstate Biotechnologies, Lake Placid, NY). This antibody specifically recognizes the human α subunit of the clusterin heterodimer. Immunohistochemistry with anticlusterin was performed as follows: 5-μm paraffin sections of primary cases and paraffin-embedded cell line clots were mounted on Fisherbrand/plus Superfrost Precleaned slides (Fisher Scientific, Pittsburgh, PA) and dried overnight at 58°C. After deparaffinization, the slides were placed in 10 mmol/L citrate buffer pH 6.0 containing 0.1% Tween 20 and antigen retrieval was performed in a microwavable pressure cooker for 8 minutes of boiling time. The sections were rinsed in 0.05 mol/L Tris-HCl, pH 7.6 with 3% goat serum and incubated with the primary anticlusterin antibody (50 ng/mL) overnight at room temperature. The remainder of the procedure (secondary antibody application and avidin-biotin detection) was carried out with an automated immunostainer (Ventana Medical System, Tucson, AZ).
Western blot analysis
Total cell lysates were prepared by lysing cell line pellets or 6-μm thick frozen sections in Laemmli sample buffer.22The protein concentrations were determined using a BCA-200 protein assay kit (Pierce Chemical, Rockford, IL). Thirty micrograms of each extract was separated on 12% SDS-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. After blocking with a 10-mm TRIS-saline solution (pH 7.4) containing 3% bovine serum albumin, the membranes were incubated overnight with the anticlusterin antibody at 1μg/mL. The blots were developed using an131I-labeled goat antimouse immunoglobulin secondary antibody, followed by overnight exposure on Kodak film or a phosphoimager screen.
Results
Differential gene expression in hematopoietic cell lines
Macroarray hybridization data from all 31 hematopoietic cell lines (Table 1) were collected and scored as described in “Materials and methods.” Of the 588 genes represented on the Atlas arrays, 380 (65%) genes showed detectable levels of expression, whereas 208 genes were not expressed (35%). A total of 267 genes were expressed across B-, T-, and myeloid cell lineages, whereas 113 genes were preferentially expressed within a single lineage. Not unexpectedly, a high percentage of genes expressed across all cell lineages were genes involved in basic biologic processes such as cell proliferation and signal transduction, stress response, and DNA repair.
Of the 113 genes that showed preferential expression within a single restricted lineage, 74 were preferentially expressed in T-cell lines, 36 preferentially expressed in B-cell lines, and 3 in myeloid cell lines. Several of these included genes known to be lineage restricted, and these showed the expected restriction pattern. For example, CD19, CD40, and CD27 were expressed exclusively in B-cell lines; CD4 and CD100 (semaphorin) were confined to T-cell lines; and CD33 and granulocyte colony-stimulating factor (G-CSF) receptor expression were limited to the 2 myeloid leukemia cell lines (Table3).
A relatively small number of genes were specifically or preferentially expressed in all of the representatives of a particular lymphoma subset (Table 3). For example, cyclin D1 was expressed only in the 2 mantle cell lymphoma lines and in 1 t(11;14)-bearing myeloma cell line, and CD30 was expressed only in the 4 ALCL cell lines. N-Cadherin was expressed in all 4 primary effusion lymphoma (PEL) cell lines and in 3 other cell lines, each derived from a different lymphoma subtype. Another example was clusterin, a gene included on the array because of its putative role in apoptosis. Clusterin was specifically expressed in all 4 ALCL cell lines and not in any of the other cell lines. Representative portions of the Atlas arrays containing the clusterin hybridization signal are shown in Figure 1.
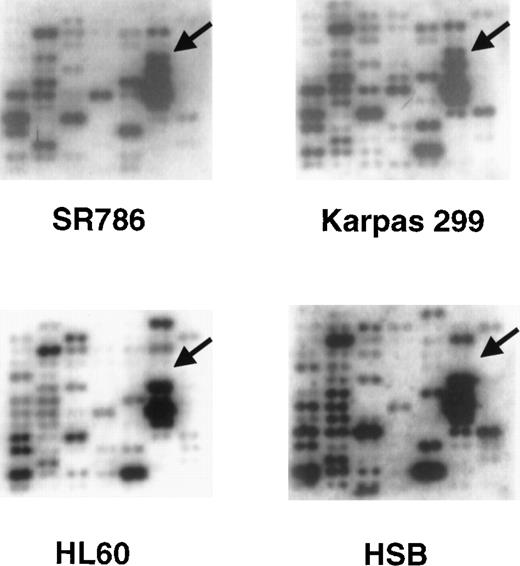
Representative Clontech human Atlas macroarray analyses.
The cDNA hybridization patterns from 2 ALCL cell lines (SR786 and Karpas 299) and 2 non-ALCL cell lines (HL-60 and HSB) from 1 field of the Atlas array are shown. The arrows identify the position of theclusterin gene cDNA and show strong hybridization signals in the 2 ALCL cell lines and no signals in the non-ALCL cell lines.
Representative Clontech human Atlas macroarray analyses.
The cDNA hybridization patterns from 2 ALCL cell lines (SR786 and Karpas 299) and 2 non-ALCL cell lines (HL-60 and HSB) from 1 field of the Atlas array are shown. The arrows identify the position of theclusterin gene cDNA and show strong hybridization signals in the 2 ALCL cell lines and no signals in the non-ALCL cell lines.
Confirmation of clusterin as a differentially expressed gene in ALCL cell lines
To confirm the differential expression of clusterin at the protein level, we obtained a commercially available monoclonal antibody to human clusterin and studied a subgroup of cell lines by Western blot analysis. Clusterin is a heavily glycosylated protein that is synthesized as a proprotein and cleaved into α and β subunits that remain covalently linked by disulfide bonds.23 The clusterin antibody that we used reacts specifically with the 35- to 40-kd α subunit.
Immunoblot analysis revealed clusterin expression only in the ALCL cell lines (Figure 2). All ALCL cell lines, and a human serum control for clusterin, showed a strong band of approximately 40 to 42 kd consistent with the α subunit of the clusterin heterodimer. In addition, the primary ALCL cases, but not the human serum control, showed a weaker high molecular weight band of approximately 70 kd possibly representing a proprotein form of clusterin containing the α-subunit precursor. Two ALCL cell lines (Karpas 299 and SR786) also displayed a 50- to 55-kd band of unknown significance, and a smaller 30-kd band was present in the serum control and in 1 ALCL cell line (Karpas 299). These weak reacting bands may represent partially degraded or, in the case of the 50- to 55-kd band, an alternatively glycosylated form of the α chain. The identification of clusterin protein by immunoblot was also confirmed by immunohistochemistry on paraffin-embedded cell line pellets, using the same antibody (data not shown).
Expression of clusterin in ALCL cell lines.
Western analysis of representative hematopoietic cell lines was performed using a clusterin antibody specific to the α subunit. A major reaction product of approximately 40 kd is present in the 4 ALCL cell lines (Karpas 299, KI-JK, SUDHL-1, and SR-786) and the serum control, but not in 2 representative non-ALCL cell lines (KMS-12 [myeloma] and Molt-4 [T-lymphoblastic leukemia]).
Expression of clusterin in ALCL cell lines.
Western analysis of representative hematopoietic cell lines was performed using a clusterin antibody specific to the α subunit. A major reaction product of approximately 40 kd is present in the 4 ALCL cell lines (Karpas 299, KI-JK, SUDHL-1, and SR-786) and the serum control, but not in 2 representative non-ALCL cell lines (KMS-12 [myeloma] and Molt-4 [T-lymphoblastic leukemia]).
Clusterin specifically marks cases of classical ALCL
Because clusterin had not previously been reported to be differentially expressed in lymphoid tissues, we were interested in pursuing the possibility that it could be used as a diagnostic marker for ALCL. For this purpose, we assembled a panel of 198 cases of primary lymphomas, including 90 T-cell lymphomas, 78 B-cell lymphomas, 30 Hodgkin lymphomas (Table 2), and 30 representative examples of nonneoplastic lymphoid tissues and stained them with the clusterin antibody. Among the 90 T-cell lymphomas were 36 primary nodal ALCL cases and 9 cutaneous ALCL cases.
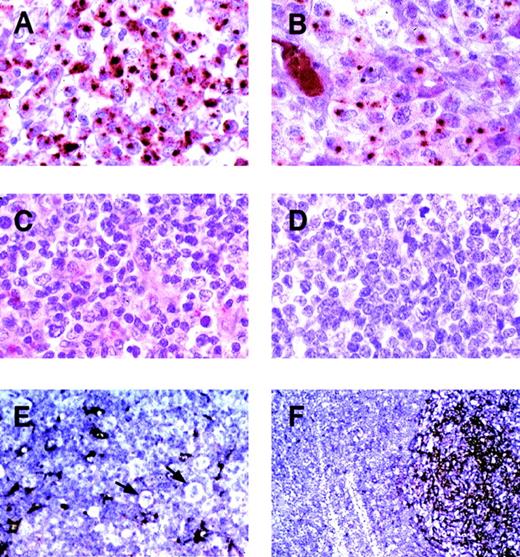
In the reactive lymphoid tissues (tonsils, lymph node, and spleen), clusterin was identified in follicular dendritic cells (FDC) and fibroblastic reticular cells (Figure 3). The FDCs showed strong cytoplasmic staining that extended into the cell processes. Fibroblastic reticular cells showed a weaker but discernable marking of their cytoplasmic processes. None of the lymphoid elements displayed staining.
Expression of clusterin in representative primary lymphoid neoplasms.
Immunohistochemistry was performed using the anticlusterin antibody. (A) and (B) Two anaplastic large cell lymphomas, positive for clusterin. Note the strong characteristic Golgi staining (original magnification × 400). (C) Peripheral T-cell lymphoma, negative for clusterin (original magnification × 400). (D) Nasal NK/T cell lymphoma, negative for clusterin (original magnification × 400). (E) Nodular sclerosis Hodgkin disease (original magnification × 200). Reed-Sternberg cells and variants are negative for clusterin (arrows). Residual dendritic cells or fibroblastic reticulum cells are positive. (F) Reactive lymphoid hyperplasia (original magnification × 100). A reactive follicle shows intense staining of dendritic cells with weaker staining of interfollicular reticular cells. Normal B and T lymphocytes show no staining.
Expression of clusterin in representative primary lymphoid neoplasms.
Immunohistochemistry was performed using the anticlusterin antibody. (A) and (B) Two anaplastic large cell lymphomas, positive for clusterin. Note the strong characteristic Golgi staining (original magnification × 400). (C) Peripheral T-cell lymphoma, negative for clusterin (original magnification × 400). (D) Nasal NK/T cell lymphoma, negative for clusterin (original magnification × 400). (E) Nodular sclerosis Hodgkin disease (original magnification × 200). Reed-Sternberg cells and variants are negative for clusterin (arrows). Residual dendritic cells or fibroblastic reticulum cells are positive. (F) Reactive lymphoid hyperplasia (original magnification × 100). A reactive follicle shows intense staining of dendritic cells with weaker staining of interfollicular reticular cells. Normal B and T lymphocytes show no staining.
All of the 36 systemic ALCL cases stained for clusterin. The staining was primarily cytoplasmic and characterized by moderate to strong dot-like staining in the Golgi areas in the majority of tumor cells (Figure 3). Occasional cases showed membranous staining as well. Twenty-three of the 36 ALCL cases were positive for ALK-1; 13 were negative.
Only 2 non-ALCL cases stained with the clusterin antibody. One was a T-cell–rich B-cell lymphoma with CD30+ neoplastic B cells. Unlike the staining in ALCL, this case showed weak cytoplasmic staining without the characteristic Golgi staining of the ALCL cases. The pale and diffuse quality of the cytoplasmic staining suggested that it could be an artifact; however, we conservatively scored it as positive. The second case was a diffuse large B-cell lymphoma that showed moderately strong cytoplasmic staining in a subplasma membrane pattern, again without any Golgi staining. In all cases of Hodgkin lymphoma, the Reed-Sternberg cells were negative. It is also of interest that all 9 cases of cutaneous ALCL were negative for clusterin. This finding is consistent with the current concept that cutaneous ALCL represents a different clinicopathologic entity from systemic ALCL. When present in the primary lymphoma cases, residual follicular dendritic cells or fibroblastic reticular cells (or both) were highlighted by their reactivity with the clusterin antibody. Representative cases of ALCL, other non-Hodgkin lymphomas, Hodgkin disease, and reactive follicular hyperplasia are shown in Figure 3.
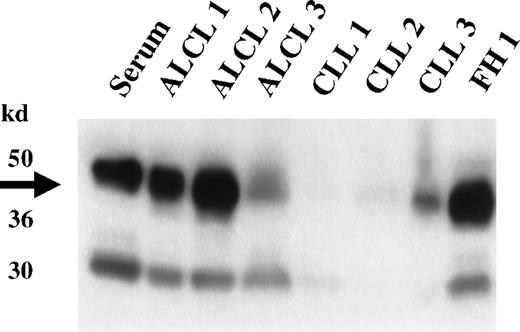
Western blot analysis of selected ALCL cases and non-ALCL cases confirmed the expression of clusterin in primary ALCL (Figure4). The molecular weight of the α-clusterin reactive bands was identical to those found in the ALCL cell lines; both the higher molecular weight band presumably corresponding to the uncleaved precursor form of clusterin and the 40- to 42-kd α chain were present. Some of the non-ALCL cases showed weak signals for clusterin. The clusterin present in these cases was presumably derived from trapped blood and residual dendritic or fibroblastic reticular cells.
Expression of clusterin in primary lymphoid neoplasms.
Western analysis of representative lymphoid neoplasms was performed using a clusterin antibody specific to the α subunit. High-level expression of clusterin is detected in the 3 primary ALCL cases (ALCL 1-3); none or lesser amounts are seen in 3 representative cases of CLL (CLL 1-3). The limited expression of clusterin seen in the non-ALCL cases is presumably derived from trapped serum or from residual dendritic cells present in the biopsy specimens. High-level expression of clusterin is also seen in a reactive lymph node (FH1), showing marked follicular hyperplasia with accompanying follicular dendritic cell proliferation.
Expression of clusterin in primary lymphoid neoplasms.
Western analysis of representative lymphoid neoplasms was performed using a clusterin antibody specific to the α subunit. High-level expression of clusterin is detected in the 3 primary ALCL cases (ALCL 1-3); none or lesser amounts are seen in 3 representative cases of CLL (CLL 1-3). The limited expression of clusterin seen in the non-ALCL cases is presumably derived from trapped serum or from residual dendritic cells present in the biopsy specimens. High-level expression of clusterin is also seen in a reactive lymph node (FH1), showing marked follicular hyperplasia with accompanying follicular dendritic cell proliferation.
Discussion
The dramatic advances in human genome sequencing and the development of gene array technologies have given investigators a new powerful tool to assess the expression of large numbers of genes in a single experiment. The initial uses of array technology have been to decipher global gene expression differences resulting from physiologic changes or changes related to disease state. For clinical oncology, this tool promises to accelerate the identification of diagnostic and prognostic markers. In this study, we show that analysis of gene expression using tumor cell lines can be used to identify novel tumor-specific markers.
In the current study, we chose a series of hematopoietic cell lines, primarily representing specific subtypes of lymphoid neoplasms, to demonstrate the utility of array technology in identifying tumor-specific markers. The arrays from Clontech Laboratories are relatively low-density nylon filter macroarrays containing 588 genes representing a broad range of functional classes. These macroarrays can be analyzed using common laboratory equipment, in contrast to the microarrays, which require specialized detection systems not widely available.24 Despite the modest number of genes contained on these macroarrays (588), we were able to verify previously known, differentially expressed genes as well as identify a previously unknown differentially expressed gene, clusterin, that we have shown to be specifically expressed in ALCL.
We chose to study tumor cell lines derived from representative subtypes of lymphomas rather than primary lymphoma specimens for the following reasons. Primary tumors are, in reality, complex tissues that are comprised not only of tumor cells but also of varying percentages of infiltrating lymphocytes, blood vessels, and other stromal components. The use of cell lines avoids the problem of having mixed populations of cells contributing to the extracted mRNA and complicating the analysis of the hybridization signals. Secondly, it is difficult, if not impossible, to control the condition of the primary tissues with regard to both in vivo factors and the results of degradation occurring between the time of the biopsy and the extraction of the RNA. Cell culture conditions are easily standardized, eliminating many of the differences that might exist between primary tumors as a result of specimen handling and local conditions affecting the tumor in vivo. Thirdly, material from primary tumors is often limited, and frequently the RNA extracted is of poor quality, whereas cell line RNA is essentially unlimited and of good quality. Although cell lines may not be perfect replicates of primary tumors in some respects, most cell lines retain a majority of known phenotypic and differentiation-related markers possessed by the parent primary tumor. We reasoned that there would likely be many unstudied proteins shared between the derived cell lines and primary tumors that could serve as tumor-specific markers.
To assess whether we could identify known differentially expressed markers in the cell lines, we first looked at a series of markers contained on the blot and known to be differentially expressed. This exercise revealed that, in general, the results from the arrays were capable of identifying previously reported differentially expressed genes. As examples, CD19, a B-cell lineage-specific marker, was specifically expressed only in B-cell lines. CD4, a T-cell–restricted marker within lymphoid neoplasms, was expressed only in T-cell lines. Cyclin D1, a specific marker for mantle cell lymphomas and t(11;14)-carrying myelomas, was expressed only in the 2 mantle-cell–lymphoma cell lines and in 1 myeloma cell line having a t(11;14) translocation.
Having satisfied ourselves that the Clontech macroarrays were reliable in assessing differential expression of known genes, we next wished to assess their ability to identify previously unknown, differentially expressed genes, because these would be potential new diagnostic markers. For this initial demonstration, we used a fairly strict definition of differential gene expression. We required that the candidate gene be expressed in all, or all but one, of the representative members of the lymphoma subtype and that it be expressed in no more than one example of another lymphoma subtype. Among the genes that showed differential expression limited to a particular group of cell lines, we identified a single gene, clusterin, which was strongly expressed in all 4 ALCL cell lines and in none of the remaining cell lines. We obtained an antibody to human clusterin and confirmed expression at the protein level by Western blotting and immunohistochemistry. We next studied by immunohistochemistry and in some cases by Western blotting, a series of primary lymphomas including 36 ALCL cases. This study confirmed the differential expression of clusterin in primary cases of ALCL and illustrated its potential use as a diagnostic marker.
Anaplastic large-cell lymphoma is a form of T-cell lymphoma that is defined by CD30 positivity and anaplastic morphology.25Recently it has become apparent that there are at least 2 forms of ALCL based on the presence or absence of a characteristic cytogenetic abnormality, the t(2;5), that leads to the overexpression of an unusual protein kinase designated ALK-1.26-31 ALK-1–positive cases occur more frequently in children and young adults and have a relatively good prognosis. ALK-1–negative ALCL occurs in older individuals and has a poorer prognosis. It is interesting that clusterin marks both forms of primary nodal ALCL and is not correlated with the presence or absence of ALK protein. On the other hand, clusterin did not mark 9 cases of cutaneous ALCL, a lymphoma resembling nodal ALCL, but now thought to be distinct because of its specific presentation, indolent clinical course, and lack of ALK-1 staining in virtually all cases.27
It is also of interest that clusterin expression does not appear to be correlated with CD30 expression, because none of the strong CD30+ Reed-Sternberg cells in cases of Hodgkin lymphoma marked for clusterin. Furthermore, although 1 of the 2 possible clusterin-expressing tumors among the B-cell non-Hodgkin lymphomas weakly expressed CD30, none of 11 other diffuse large B-cell lymphomas (Table 3), showing weak or focal CD30 positivity, expressed clusterin.
The functional significance of clusterin expression in ALCL is unclear, although in other tissues it has been implicated in a variety of functions.23,32 Clusterin was originally isolated from ram rete testis fluid33 and so named because of its ability to induce clustering of Sertoli cells.34 It has since been shown to be widely expressed in the epithelial cells of many other organs including liver, stomach, and brain, and is secreted in numerous body fluids such as semen, urine, breast milk, cerebrospinal fluid, and plasma.23 Clusterin has been implicated in lipid transport, reproduction, complement regulation, tissue remodeling, cell–cell interaction, programmed cell death (apoptosis), and cell survival.23,32 Most intriguing is its proposed role in apoptosis. Initially it was believed to be proapoptotic because of its accumulation in tissues undergoing apoptosis.35 However, recent data suggest that clusterin is accumulated in the surviving cells adjacent to areas undergoing apoptosis, leading investigators to reassess the role of clusterin in apoptosis.36 In accord with these more recent observations, clusterin has been shown to have potent chaparone-like activities and protects a wide variety of proteins from heat or mercaptoethanol-induced denaturation.37 38 These data are consistent with a generalized protective antiapoptotic function rather than a proapoptotic one.
Few studies have investigated clusterin expression in lymphoid cells because early studies of its tissue distribution failed to identify clusterin in peripheral blood T cells.39 Because of its hypothesized role in apoptosis, clusterin had been proposed to play a role in the generation of T-cell tolerance in the thymus. However, its expression was limited to the epithelial cells of the medulla, whereas thymocytes lacked clusterin expression.40,41 These findings suggested that clusterin was not an important regulator of programmed cell death in thymocytes and confirmed the lack of clusterin expression in a second source of T-lineage lymphocytes. Finally, in their attempts to develop inducible models of clusterin expression, French et al36 were not able to show clusterin expression or induction in a T-cell lymphoblastic leukemia cell line. Although most ALCL cases have a T-cell phenotype, we also were not able to identify a normal T-cell population expressing clusterin, and at the present time we have no clues regarding its differential expression in ALCL. We have previously shown that ALCL displays some unusual phenotypic features in that the majority of cases express both CD4 and cytotoxic granular proteins.42 Thus, it is possible that the normal cellular counterpart is a rare cell or is present only at particular stages of differentiation or development. Alternatively, clusterin may be a true tumor antigen in ALCL and may not be expressed in the normal cellular counterpart.
In summary, we have described an approach for the identification and confirmation of novel tumor-specific markers, using a combination of gene array technology on selected representative cell lines and immunochemical screening of primary case material. Using relatively small format gene expression macroarrays with 588 genes and stringent criteria for differential gene expression, we were able to identify differentially expressed gene products, including clusterin, a new specific marker for ALCL. Undoubtedly, the use of larger format microarrays with many thousands of genes will increase the yield of new markers as will more sophisticated computer-assisted analysis of quantitative differences in expression levels. In the future, improvements in microdissection and mRNA/cDNA amplification technology will allow the direct use of amplified cDNA derived from primary tumors, without the confounding effect of contaminating mRNA from nonneoplastic cells. The use of primary tumor material will be particularly important for the identification of prognostic markers.
Acknowledgments
We wish to thank Dr G. Jokhadze (Clontech Laboratories) for helpful discussions regarding the Clontech Atlas arrays, Ms T. Davies-Hill and Dr L. Quintanella-Martinez for assistance with the Western blotting, and Drs A. Karpas and M. Shimakage for kindly providing the Karpas 299 and Ki-JK cell lines, respectively.
A.W. and C.T. contributed equally to this work.
Reprints:Mark Raffeld, Laboratory of Pathology, National Cancer Institute, Bldg 10, Rm 2N110, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: mraff@box-m.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 2. Expression of clusterin in ALCL cell lines. / Western analysis of representative hematopoietic cell lines was performed using a clusterin antibody specific to the α subunit. A major reaction product of approximately 40 kd is present in the 4 ALCL cell lines (Karpas 299, KI-JK, SUDHL-1, and SR-786) and the serum control, but not in 2 representative non-ALCL cell lines (KMS-12 [myeloma] and Molt-4 [T-lymphoblastic leukemia]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/2/10.1182_blood.v96.2.398/5/m_bloo01446002w.jpeg?Expires=1767847957&Signature=yvuZYm14sqfqK7Wrw4KlxjzWZFGLp-Mt8jXQnQvkz9ShIAaWzOWcHkgQylX5sWVUDzWvUmfUayh3CLLJYenLMMOBLvl6oE7s3ZLNY5rMH2vVj6d6BDYbV-i-J1H1al~Q9mtQlzPzYPY8xDrGIC4VTYT1yAfj2cAgBwBo32Mxp-kDuStOF~YTmmYV1FDhlYjlVwirO65Wq94EK1ZlbzOQPTIFxn-pDQZmw6KvEf37Ku6Gafle381U0AhJIRVtI4-U87st3Or01-4Loaku0AyoW8Y5L5i1UdJCAiAPZwRJTl~Z3Y1snKBuoqJpOb4RPgvr3b1en-tvRKUIIUS4acCsaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)