Abstract
Hypoxia is a strong stimulus for the transcription of a set of genes, including erythropoietin and vascular endothelial growth factor. Here we report on the cloning, functional significance, and expression of a complementary DNA (cDNA) that is involved in hypoxia-mediated expression of these 2 genes. The full-length cDNA encodes a predicted protein of 806 amino acids that contains a leucine zipper motif. This protein, termed HAF for hypoxia-associated factor, binds to a 17-base pair (bp) region of the erythropoietin promoter, which was shown earlier to participate in hypoxia-induced expression of the erythropoietin gene. In Hep3B cells, clones modified to express HAF antisense RNA showed an attenuated response to hypoxia-mediated induction of both erythropoietin and vascular endothelial growth factor transcription. HAF showed sequence-specific interaction with a DNA element in the 5′ untranslated region ofVEGF gene. The HAF 2.6-kilobase (kb) messenger RNA (mRNA) is expressed in most adult tissues. The highest expression occurs in fetal liver and the least in adult liver. HAF is the murine homolog of Sart-1, a 125-kd human protein expressed in the nuclei of normal and malignant cells.
Hypoxia is known to up-regulate expression of several genes, including erythropoietin (EPO), tyrosine hydroxylase, vascular endothelial growth factor (VEGF), platelet-derived growth factor B chain, phosphoglycerate kinase 1, and lactate dehydrogenase A (for review see Bunn and Poyton1). The physiologic importance of hypoxia-induced regulation of EPO andVEGF genes has been established in red blood cell formation and angiogenesis.1 2
EPO is primarily produced by fetal liver3 and adult kidneys4 and, to some extent, by brain cells5and hemopoietic progenitor cells.6 Hypoxia-induced expression of EPO is regulated by both the rate of gene transcription and posttranscriptional events.7Transcriptional regulation is achieved by the concerted action of several transacting factors interacting with the proximal promoter region and with the 3′ untranslated region of the EPOgene.8-12 In the 3′ untranslated region of theEPO gene, there is a 50-bp hypoxia-responsive–enhancer (HRE) element located approximately 120 bp 3′ of the polyadenylation site. This enhancer element is functionally tripartite. One site binds the hypoxia-inducible factor (HIF-1).13 The second is required for transactivation of the EPO gene mediated by HIF-1, but factors interacting with site 2 have not yet been described. The third is a binding site for the orphan receptors, hepatocyte nuclear factor 4 (HNF-4), and EAR3/COUP-TF-1; HNF-4 may act as a positive regulator and EAR3/COUP-TF-1 may play an antagonistic role in hypoxia-induced expression of the EPO gene.14 The role of the p300/CREB binding proteins was also described recently.15 HIF-1 has also been shown to be involved in the activation of VEGF gene transcription through its interaction with a 47-bp sequence located at nucleotides 985 to 939 5′ of the transcription start site.16
Several nuclear factors that recognize a sequence in the EPOpromoter have been shown to function synergistically with those interacting with the 3′ enhancer.12 We have earlier defined the role of the −61 to −45-bp sequence EP17 in hypoxia-inducible expression; the transcription start site of the murine EPO gene is numbered zero. Factors interacting with EP17 synergize with those binding to the 3′ enhancer for the maximal transcriptional response to hypoxia.17 In this paper we report the cloning and partial characterization of a factor that interacts with the EP17 sequence of the EPO promoter and the 5′ UTR of the VEGF gene. This factor acts in the regulation of hypoxia-induced expression of genes.
Materials and methods
Cell culture murine
NN10 cells18 were grown in Dulbecco's modified Eagle's medium (DMEM) low glucose (Sigma, St Louis, MO), supplemented with 5% fetal calf serum (FCS) (HyClone, Logan, UT), sodium bicarbonate (44 mmol/L), and gentamicin (40 mg/L). The human hepatoma cells Hep3B (ATCC, HB8064) were grown in minimum essential medium alpha (Sigma) supplemented with 10% FCS, sodium bicarbonate (26 mmol/L), and gentamicin (40 mg/L); incubated in an atmosphere of 5% CO2, 95% air, and passed every 3 to 4 days after trypsinization of the confluent cell layer. These cultures were grown at 37°C.
Preparation of a complementary DNA library
Total RNA was prepared from NN10 cells.19PolyA+ RNA was isolated by 2 rounds of affinity chromatography on oligo(dT)-cellulose20 (Life
Technologies, Grand Island, NY). Complementary DNAs (cDNAs) were synthesized using a random primer and Maloney murine leukemia reverse transcriptase as per the manufacturer's instruction (Stratagene, La Jolla, CA). After second strand synthesis with DNA polymerase 1 and RNase H, the double-stranded cDNA was size fractionated on Sephacryl S-400 columns (Pharmacia, Piscataway, NJ), and ligated to EcoR1 adapters. After digestion with EcoR1 (Boehringer Mannheim, Indianapolis, IN), the cDNA was ligated to the dephosphorylatedEcoR1 digested arms of the λgt11 vector and packaged using Giga pack II-gold packaging extract (Stratagene). The titer of the primary library was 1 to 2 × 106 pfu/μg of λgt11 DNA. The library was amplified to a titer of 2 × 109 pfu/μL.
Expression screening of the NN10 library
Screening of the library was performed as described by Singh et al.21 Briefly, after infection of Y1090 Escherichia coli cells (Stratagene), aliquots of the amplified cDNA library were plated in NZYM broth (Life Technologies), containing agarose (0.7% wt:vol), at a density of 50 000 pfu per 150-mm plate. The plates were incubated at 42°C for 3.5 hours until small plaques were just visible. The agarose was then overlaid with nitrocellulose membranes (Schleicher & Schuell, Keene, NH), which had been presoaked for 30 minutes in 10 mmol/L isopropyl thiogalactose (IPTG) and then air dried. Incubation was continued at 37°C for 3.5 hours after which the orientation of the filter on the plate was marked. The membranes were placed in blocking solution (5% nonfat dried milk, 50 mmol/L TrisHCl pH 7.5, 50 mmol/L NaCl, 1 mmol/L EDTA, and 1 mmol/L dithiothreitol (DTT)) at 4°C overnight. A duplicate membrane was prepared by incubation of the plate for 4 hours at 37°C. The probe used for screening the library was a 3 × tandem repeat of the EP17 sequence 5′CCCCCACCCCCACCCGC3′, generated as a complementary synthetic oligonucleotide (3EP17), with a BamH1 restriction site at the 5′ end and an EcoR1 site at the 3′ end. After gel purification and annealing, the double-stranded oligonucleotide was digested with EcoR1 and BamH1, and cloned into pBluescript SKII (Stratagene). The fragment of the resulting plasmid was end-labeled by first digesting the plasmid DNA with EcoR1 and filling in the ends using the Klenow fragment of DNA polymerase-1 and γ-32P dCTP (110 TBq/mmol [3000 Ci/mmol/L]) (Amersham, Arlington Heights, IL). After labeling, the fragment was released from the vector by digesting withBamH1 and gel purified on a 10% polyacrylamide gel. Hybridization of the membranes with the labeled probe was carried out for 3 hours at 25°C in 10 mmol/L TrisHCl pH 7.5, 50 mmol/L NaCl, 1 mmol/L EDTA, and 1 mmol/L DTT containing 2 × 106cpm/mL of labeled probe. Membranes were washed at 25°C for 30 minutes in the same buffer without the probe. Signal was detected by autoradiography on Kodak XAR-5 film (Eastman Kodak, Rochester, NY). The plaques showing signals in duplicate filters were cored, the phage DNA eluted in SM buffer (100 mmol/L NaCl, 8 mmol/L MgSO4, 50 mmol/L TrisHCl pH 7.5, 0.01% gelatin) and rescreened on duplicate membranes. After 3 additional rounds of screening, the DNA from purified phage was prepared.22 The cloned fragment was released after EcoR1 digestion and was subcloned at the EcoR1 site of pBluescript. Additional clones were obtained by screening the library with radiolabeled fragments representing 5′ and 3′ regions of the cloned DNA. The entire DNA was sequenced in both directions by using the dideoxy sequencing method and by automated polymerase chain reaction (PCR) sequencing at The University of Chicago sequencing facility using either T7, T3, or gene specific primers. We designate the product of this cloned gene as HAF, for hypoxiaassociated factor.
Bacterial expression of proteins
A crude extract of the recombinant phage-encoded protein was prepared after infection of Y 1089 E coli (Stratagene), isolation of the temperature-sensitive lysogenized colonies, and induction of protein expression with 10 mmol/L IPTG.23 The crude extract was analyzed for EP17 DNA binding activity using the mobility shift assay as described below.
For expression of recombinant protein as the glutathione-s-transferase (GST) fusion protein, 1.2-kilobase (kb) of HAF cDNA sequences (nucleotide [nt] 1-1200) were cloned in frame with the glutathione binding domain of Schistosoma japonicum GST in the pGEX-2T bacterial expression vector (Pharmacia). The orientation of HAF cDNA was confirmed by Pst1 digestion. The E coli strain BL-21 (DE3)pLysS was transformed with parental pGEX-2T or with the HAF pGEX-2T vector containing the 390 N-terminal amino acids of HAF fused in frame with GST. Bacteria were grown in 50 mL of Luria-Bertani (LB) medium (Life Technologies), supplemented with 100 μg of ampicillin per milliliter, for 12 hours at 30°C. The cultures were then diluted 10-fold in LB containing ampicillin and grown for 1 hour to an absorbance of 0.8 at 600 nm before induction with 0.1 mmol/L IPTG for 6 hours at 20°C. IPTG-induced bacteria were pelleted and washed with 10 mL of ice-cold phosphate-buffered saline with 20 mmol/L MgCl2 (PBSM) containing 100 μg each of aprotinin, leupeptin, and pepstatin, and 0.1 mmol/L PMSF. The pellet was resuspended in the above buffer and cellular proteins were extracted by sonication (a total of 6 bursts of 30 seconds each) at 4°C and incubation with 1% Triton X-100 (Eastman Kodak, Rochester, NY). Cellular debris was removed by centrifugation. Purification of the GST fusion protein by affinity chromatography on glutathione agarose beads was performed as follows; the bacterial lysate (300 mL) was incubated with 2.0 mL of a 50% (vol/vol) slurry of glutathione-Sepharose (Sigma) in PBSM at 4°C for 1 hour. After centrifugation, the Sepharose was thoroughly washed with PBSM plus 0.1% Triton X-100, and eluted with 100 mmol/L glutathione in 500 mmol/L TrisHCl pH 8.0, 10 mmol/L DTT, and 50 mmol/L MgCl2, for 20 minutes at 4°C.
Electrophoretic mobility shift assays
DNA binding activity of the crude lysate expressing the recombinant protein (5 μg), and of purified GST-HAF fusion protein (5-20 μg) were analyzed by mobility shift assays as described previously.8 25 Binding reactions with purified GST-HAF (10 μg) contained 2 μg of bovine serum albumin (BSA) (Sigma) as carrier and 0.1 to 0.5 μg of double-stranded poly (dI-dC). The probes used were double-stranded oligonucleotides corresponding to EP17 and to the +504 to +533-bp region of the VEGF gene (AGACACCGCCCCCAGCCCCAGCGCCCACCTC).
For the supershift assay, antibody against HAF was raised in rabbits using a synthetic peptide at positions 30 to 44, (H2N-PRHREHKKHKHRSSG-COOH). The peptide was synthesized and immunization was performed at Primm Laboratories (Cambridge, MA). The specificity of the antibody was tested by Western blotting where Hep3B lysate proteins (200 μg) and protein markers (high-molecular-weight rainbow marker, Amersham) were resolved by 10% SDS PAGE and electrotransferred to a Protran-nitrocellulose membrane (Schleicher & Schuell, Keene, NH). After blocking and washing, the membrane was incubated with HAF antibody (1:1000), followed by washing and incubation with secondary anti rabbit antibody (1:6000). The signal was generated by Supersignal Substrate Western Blotting Kit (Pierce, Rockford, IL) and detected by autoradiography. Nuclear extracts from the same cells were prepared and binding reactions were carried out with 5 μg of nuclear proteins in the presence of 1, 5, and 10 μL of immune or 10 μL of preimmune sera and 0.5 μg of double-stranded poly (dI-dC) as a nonspecific competitor. After 5 minutes of incubation, labeled EP17 was added and incubated an additional 15 minutes at 25°C before electrophoresis.
Production of clones stably expressing hypoxia-associated factor antisense RNA
The plasmid pCB6+HAF AS was generated by cloning the 700-bp 5′ region of HAF cDNA in the reverse orientation into the pCB6+ vector (a gift from Dr V. Sukhatme, Harvard Medical School, Boston, MA). The expression of the cloned cDNA was under the control of the cytomegalovirus (CMV) promoter. The vector also contained a neo-resistance gene. Plasmid DNA (pCB6+and pCB6+HAF AS, 20 μg each) were transfected into 106 Hep3B cells by the calcium-phosphate precipitation method.26 Twenty-four hours after transfection the medium was changed. Selection for stably transfected cells was made after 48 hours by replacing the medium with DMEM containing G418 (Life Technologies) at 0.5, 1, and 2.5 mg/mL. These concentrations are toxic to wild-type Hep3B cells. The selection in G418 continued for 3 weeks with a change of medium twice per week. Neo-resistant clones were isolated by partial trypsinization of the attached cells with 1 mL of 0.06% trypsin in Hanks balanced salt solution, supplemented with 11.6 mmol/L NaCl and 0.5 mmol/L EDTA, colonies were picked into 24-well plates in DMEM containing G418 at the same 3 concentrations. Twenty-four different clones were trypsinized, expanded into 25-cm2 flasks, grown to confluency for a period of 2 to 8 weeks, and maintained in G418 to prevent back mutations.
Expression of transfected HAF antisense RNA was confirmed by Northern blot analysis. Cells transfected with the pCB6+ vector alone and selected for neo resistance served as controls. Cells expressing antisense RNA and controls were exposed to 2% O2, 5% CO2, and 93% air for 48 hours. Total RNA was isolated, size fractionated on 1.2% formaldehyde agarose gels, and hybridized with either radiolabeled 642-bp monkey EPO cDNA or radiolabeled 400-bp human VEGF cDNA (3′ untranslated region, kindly provided by Dr C. Simon, The University of Chicago, IL).
Northern blot analysis
For study of HAF tissue distribution, liver, kidneys, brain, heart, spleen, and intestines were harvested from 20-g adult B6D2F1/J mice. The organs were quick frozen and powdered in liquid nitrogen. The tissue powder was homogenized in buffer (4 mol/L guanidinium isothiocyanate, 25 mmol/L sodium citrate pH 7.0, 0.1 mol/L β-merceptoethanol), and RNA was purified on a CsCl cushion.20
The study of HAF expression used kidneys and livers harvested from specified ages of fetuses and at different intervals after birth. Tissue samples for each data point (n = 6 to 10) were pooled and processed for RNA extraction as described above. Northern blot analysis of 20 μg of total RNA was as previously described. After prehybridization in a buffer containing 1 mol/L NaCl, 50% formamide, 10% dextran sulfate, and 1% sodium dodecyl sulfate at 42°C for 2 hours, the membrane was incubated overnight at 42°C in the same buffer with 1 × 106 cpm/mL of labeled HAF cDNA (nt 1-700) that was random primed using the Klenow fragment of DNA polymerase 1 and α-32P dCTP (specific activity 110 TBq/mmol [3000 Ci/mmol]) (Amersham). After final washing under stringent conditions (0.2XSSC, 0.1% SDS, 65°C for 30 minutes), the signal was detected by autoradiography.
Results
Cloning of HAF and its interaction with the EPO promoter
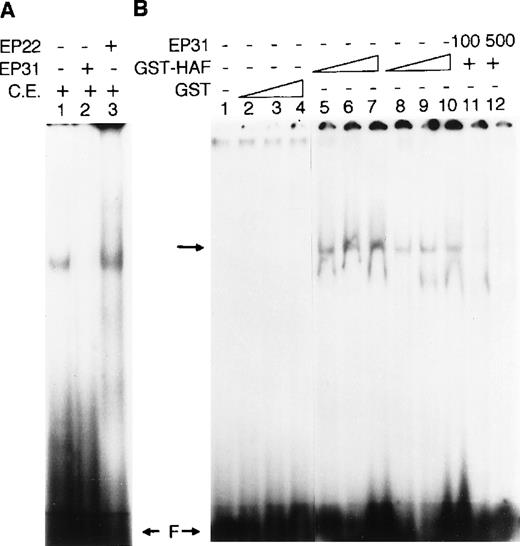
The messenger RNA (mRNA) of the murine erythroleukemic cell line NN10 was used as a source for constructing the cDNA expression library because those cells constitutively express EPO at high levels,18 and because NN10 nuclei contain factors that interact with EP17. Initially 1 × 106 plaques were screened with radiolabeled 3EP17. Of 5 clones obtained by 3 rounds of screening, 2 showed overlapping sequences that were tested for specific binding to EP17. These 2 clones were plated at 100 pfu per plate and lifted onto IPTG-saturated filters. Each filter was cut in half and each half screened with 32P-3EP17 in the presence of a 150-fold molar excess of either unlabeled wild-type EP17 or an unlabeled mutant EP17, where A at positions 6 and 12 was replaced by G. In our earlier study, this mutant did not compete with EP17 for the nuclear binding factor.17 For both clones, cold EP17 effectively competed for probe binding, whereas mutant EP17 did not (data not shown). The specificity of these clones for EP17 was confirmed further by performing a mobility shift assay using a crude extract prepared from a lysate of Y1089 E coli after infection with recombinant phage and lysis by expressing its temperature-sensitive phage repressor. The gel shift data (Figure1A) show specific complex formation with radiolabeled EP17 that was competed by EP31 (nucleotides −61 to −31) but not by EP22 (−91 to −69) of the EPOgene.
Gel-shift assay with HAF protein.
The probe used was the region −45 to −61 bp (EP17) of the epo promoter. (A) Gel-shift assay using a crude extract from HAF recombinant lysogen. The crude extract (CE) (5 μg) was incubated with radiolabel led EP17 alone (lane 1), or in the presence of a 500-fold molar excess of the specific competitor EP31 (lane 2) or nonspecific competitor EP22 (lane 3). (B) Gel-shift assay with HAF GST-fusion protein. Increasing amounts (5, 10, and 20 μg) of GST protein (lanes 2-4) or HAF GST-fusion protein (lanes 5-10) were incubated with 100 ng of poly dI-dC (lanes 5-7), or 500 ng of poly dI-dC (lanes 2-4 and 8-10). Lanes 11 and 12 represent binding reactions with 10 μg of GST-fusion protein in the presence of 100- and 500-fold molar excess of the specific competitor EP31. Lane 1 represents the binding reaction with no protein. Each of these reactions was carried out in the presence of 2 μg of BSA as carrier protein. The arrow indicates the shifted complex. F is free probe.
Gel-shift assay with HAF protein.
The probe used was the region −45 to −61 bp (EP17) of the epo promoter. (A) Gel-shift assay using a crude extract from HAF recombinant lysogen. The crude extract (CE) (5 μg) was incubated with radiolabel led EP17 alone (lane 1), or in the presence of a 500-fold molar excess of the specific competitor EP31 (lane 2) or nonspecific competitor EP22 (lane 3). (B) Gel-shift assay with HAF GST-fusion protein. Increasing amounts (5, 10, and 20 μg) of GST protein (lanes 2-4) or HAF GST-fusion protein (lanes 5-10) were incubated with 100 ng of poly dI-dC (lanes 5-7), or 500 ng of poly dI-dC (lanes 2-4 and 8-10). Lanes 11 and 12 represent binding reactions with 10 μg of GST-fusion protein in the presence of 100- and 500-fold molar excess of the specific competitor EP31. Lane 1 represents the binding reaction with no protein. Each of these reactions was carried out in the presence of 2 μg of BSA as carrier protein. The arrow indicates the shifted complex. F is free probe.
The DNA binding ability of the encoded HAF protein, outside the β galactosidase domain, was confirmed by gel shift assay with GST-HAF fusion protein containing GST and the 390 amino terminal sequence of HAF and radiolabeled EP17. The data show interaction of the GST-HAF fusion protein with labeled EP17, in a sequence specific manner (Figure 1B). GST alone (Figure 1B) or a GST fusion protein encoded by HAF in reverse orientation did not show any interaction with EP17 (data not shown). These data clearly show that HAF binds to EP17 in a sequence-specific manner.
The complete nucleotide sequence of murine HAF cDNA derived from several overlapping clones contains an open reading frame encoding 806 amino acids. The first ATG codon encoding the putative start methionine shows favorable context for translational initiation based on the Kozak consensus criteria.27,28 The sequence of HAF cDNA was submitted to GenBank under the accession number AF129931. A search of the GenBank database revealed that the nucleotide sequence encoding HAF cDNA is homologous (94% identity/96% similarity) to the cDNA sequence of Sart-1 (accession number AB006198) isolated from a human squamous cell carcinoma.29 In comparison to Sart-1, the HAF protein has an insertion of 6 amino acids from position 454 to position 459. In addition, HAF cDNA shows 31% identity and 47% similarity with the Caenorhabditis eleganschromosome III locus CELF19F10 (accession number U97005).
Analysis of the full length HAF protein using Prosite software (Swiss Institute of Bioinformatics, Geneva, Switzerland) identified several possible sites for posttranslational modification, including N-glycosylation, protein kinase A, casein II kinase, protein kinase C, tyrosine kinase, and amidation.
Further computer analysis of the encoded protein revealed the presence of a leucine zipper motif (amino acids 365-386), comprising 4 leucines each separated by 6 residues. Immediately adjacent to this domain, the HAF protein has several clusters of basic amino acids, which probably can serve as DNA binding domains. Additional analysis of the possible secondary structure of the putative HAF protein revealed that it may be about 58% alpha helix. No stretches of hydrophobic amino acids, consistent with the presence of a transmembrane domain, are present.
Supershift of nuclear protein-EP17 complex with antihypoxia-associated factor
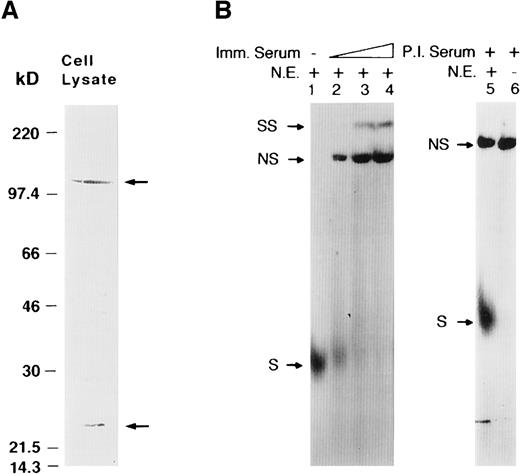
The antibody raised to the N-terminal region of HAF recognizes endogenous HAF with 2 possible degradation products of approximate molecular weight of 105 and 25 kd, which together may represent approximately the 130 kd endogenous HAF. No cross-reactivity with other cellular proteins was observed (Figure 2A). A supershift assay using this antibody confirms the role of endogenous HAF in binding to EP17. Nuclear proteins from Hep3B cells bound to labeled EP17, as indicated by S in Figure 2B. When HAF antiserum was included in the binding reaction, we found 2 additional complexes, in addition to the S complex, 1 nonspecific (NS) and 1 supershift (SS). With increased concentration of HAF antiserum, the amount of the SS complex increased, the S decreased, and the NS complex remained unaffected. When nuclear proteins were incubated with preimmune serum in place of HAF immune serum, a nonspecific complex with mobility similar to that of NS was formed. Preimmune serum had no effect on the formation of the specific complex and did not produce SS. In the binding reaction, when labeled EP17 was incubated alone with the immune or preimmune serum in the absence of nuclear factors, it produced a complex with identical mobility to NS but no SS or S complexes were formed (Figure 2B). These data demonstrate that an antibody raised against HAF recognized the nuclear proteins of Hep3B cells that bind to the EP17 probe. The NS band is generated by nonspecific interaction of the probe with serum proteins.
Super-shift assay with HAF antibody and Hep3B nuclear extract.
(A) Western blot showing the specificity of HAF antibody utilized in the supershift assay. Arrows indicate the two proteins discussed in the text. (B) Increasing amounts (1, 5, and 10 μL) of anti-HAF serum were incubated with Hep3B nuclear proteins (5 μg) in the presence of the EP17 probe (lanes 2-4). Lane 1 is a control reaction with nuclear proteins alone. Control reactions with preimmune serum in the presence (lane 5) or absence (lane 6) of nuclear extract. Imm. Serum and P.I. Serum refers to immune and preimmune serum respectively, N.E. refers to nuclear extract. S, NS, and SS represent specific, nonspecific, and super-shifted complexes, respectively.
Super-shift assay with HAF antibody and Hep3B nuclear extract.
(A) Western blot showing the specificity of HAF antibody utilized in the supershift assay. Arrows indicate the two proteins discussed in the text. (B) Increasing amounts (1, 5, and 10 μL) of anti-HAF serum were incubated with Hep3B nuclear proteins (5 μg) in the presence of the EP17 probe (lanes 2-4). Lane 1 is a control reaction with nuclear proteins alone. Control reactions with preimmune serum in the presence (lane 5) or absence (lane 6) of nuclear extract. Imm. Serum and P.I. Serum refers to immune and preimmune serum respectively, N.E. refers to nuclear extract. S, NS, and SS represent specific, nonspecific, and super-shifted complexes, respectively.
Role of hypoxia-associated factor in hypoxia-induced gene expression
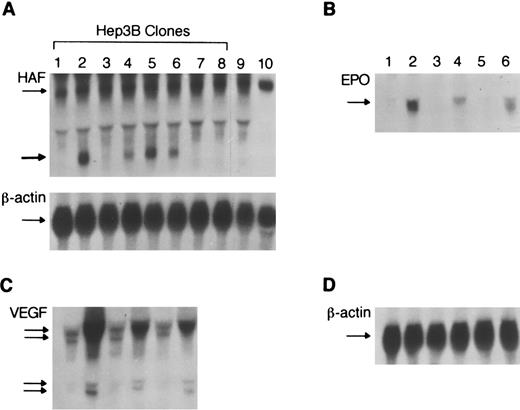
We used an antisense strategy to determine the functional role of HAF in hypoxia-induced expression of the EPO gene. HEP3B cells were transfected with the plasmid expressing the first 700-bp of HAF antisense RNA driven by the CMV promoter. Cells transfected with the vector alone served as controls. After G418 selection, expression of HAF antisense RNA was confirmed by Northern blot analysis. Of 10 clones analyzed, 4 (clones 2, 4, 5, and 6) showed a positive signal for partial HAF antisense RNA (0.7 kilobase [kb]) along with the full-length endogeneous HAF mRNA (Figure 3A). To examine the effect of HAF antisense RNA on hypoxia-induced expression of the EPO gene, clones 4 and 5, along with a control (clone 8) were analyzed further. Each of these clones was grown in duplicate, overnight, at 5% CO2, and 95% air. One set of plates was transferred to 2% oxygen, 5% CO2, and 93% N2 for 48 hours. The other set of plates was incubated in 5% CO2 and 95% air for 48 hours. Total RNA was analyzed for EPO message. Quantitation of EPO mRNA normalized to β-actin mRNA in response to hypoxia showed 73 ± 10-fold induction of EPOmRNA in the control clone (no 8). There were 3 separate determinations each with a pool of 5 plates. In contrast, induction of EPOmRNA in clones 4 and 5, containing HAF antisense was only 27 ± 8-fold and 19 ± 11-fold. Again, each was the result of 3 separate experiments with a pool of 5 plates. A representative radiogram is shown in Figure 3B. Wild-type Hep3B cells showed a 81 ± 14-fold induction in EPO mRNA in response to hypoxia.
Functional role of HAF in hypoxia-induced expression of the epo gene.
(A) Northern analysis of Hep3B clones expressing HAF-antisense RNA. Individual clones (1-8) stably transfected with either HAF cDNA in reverse orientation (lanes 1-7) or the vector alone (lane 8); lanes 9 and 10 represent controls with non-transfected Hep3B cells and NN10 cells. Total RNA was isolated, size fractionated and hybridized with the HAF cDNA probe. The thin arrow indicates the 2.6-kb HAF transcript. The thick arrow indicates the presence of the 0.7-kb HAF antisense-RNA in clones 2, 4, 5, and 6 but not in wild-type (clone 9) or clones transfected with vector alone (8). The bottom panel shows β-actin mRNA. (B) Effect of HAF-antisense-RNA on hypoxic induction of epo mRNA. Two clones (4 and 5 of panel A) expressing HAF antisense RNA (lanes 3-6), and a control clone (8 of panel A) (lanes 1 and 2) transfected with vector alone were exposed to hypoxia (2% O2) for 48 hours (lanes 2, 4, and 6), or under normal oxygenation (lanes 1, 3, and 5). The probe used was a monkey epo cDNA. The arrow indicates 1.4-kb epo mRNA. (C) Effect of HAF antisense RNA on hypoxia induced expression of VEGF mRNA. The membrane in panel B was stripped of the epo probe and hybridized to human VEGF cDNA. Arrows indicate different transcripts of VEGF. (D) The same membrane was stripped and rehybridized with mouse β-actin cDNA to control for loading differences.
Functional role of HAF in hypoxia-induced expression of the epo gene.
(A) Northern analysis of Hep3B clones expressing HAF-antisense RNA. Individual clones (1-8) stably transfected with either HAF cDNA in reverse orientation (lanes 1-7) or the vector alone (lane 8); lanes 9 and 10 represent controls with non-transfected Hep3B cells and NN10 cells. Total RNA was isolated, size fractionated and hybridized with the HAF cDNA probe. The thin arrow indicates the 2.6-kb HAF transcript. The thick arrow indicates the presence of the 0.7-kb HAF antisense-RNA in clones 2, 4, 5, and 6 but not in wild-type (clone 9) or clones transfected with vector alone (8). The bottom panel shows β-actin mRNA. (B) Effect of HAF-antisense-RNA on hypoxic induction of epo mRNA. Two clones (4 and 5 of panel A) expressing HAF antisense RNA (lanes 3-6), and a control clone (8 of panel A) (lanes 1 and 2) transfected with vector alone were exposed to hypoxia (2% O2) for 48 hours (lanes 2, 4, and 6), or under normal oxygenation (lanes 1, 3, and 5). The probe used was a monkey epo cDNA. The arrow indicates 1.4-kb epo mRNA. (C) Effect of HAF antisense RNA on hypoxia induced expression of VEGF mRNA. The membrane in panel B was stripped of the epo probe and hybridized to human VEGF cDNA. Arrows indicate different transcripts of VEGF. (D) The same membrane was stripped and rehybridized with mouse β-actin cDNA to control for loading differences.
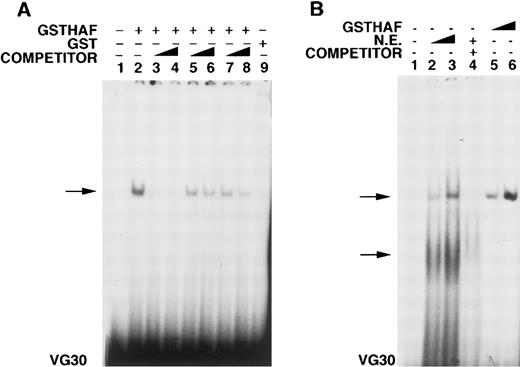
When the same membranes were stripped of the EPO probe and hybridized with a VEGF cDNA probe, we found a reduced induction of VEGF mRNA in the antisense clones, compared with the control clone after hypoxic exposure. Quantitation of the major transcript ofVEGF mRNA (normalized for β-actin mRNA) showed a 51 ± 10-fold increase in VEGF mRNA in the control clone (no 8) in response to hypoxia, whereas antisense clones 4 and 5 showed only a 30 ± 12-fold and a 21 ± 14-fold induction, respectively, each representing the mean of 3 separate experiments with a pool of 5 plates. A representative radiogram is shown in Figure3C. Additional evidence for the role of HAF in VEGF gene expression comes from a gel shift experiment in which we show that the GST-HAF fusion protein interacts with a probe (VG30) consisting of bases +504 to +533 of the VEGF 5′UTR. The VEGFsequence also competes with EP17 for HAF binding (Figure4A). To examine whether the VG30 sequence recognizes native proteins in Hep3B nuclear extract and how the mobility of the native proteins compares with that of recombinant protein, an additional gel shift was conducted using VG30 as a probe. Data presented in Figure 4B show that VG30 interacts with Hep3B nuclear factors in a sequence specific manner with 2 shifted complexes. The mobility of the higher shifted complex corresponds to the mobility of the complex obtained with HAF recombinant protein. Thus, collectively our data indicate that (a) HAF interacts with the regulatory regions of the EPO and VEGF genes, (b) a 30-bp sequence of theVEGF gene in the 5′UTR interacts with nuclear factors of Hep3B cells, and (c) expression of antisense HAF mRNA attenuates the hypoxia-induced expression of EPO and VEGF mRNA.
Gel shift assay with VEGF UTR binding to HAF protein and nuclear proteins of Hep3B cells.
(A) Lane 1, probe alone (30 mer); lane 2, GST-HAF + probe; lanes 3, 4, self competition at 100 and 500 molar excess; lanes 5, 6, lack of competition with EP22 at 100 and 500 molar excess; lanes 7, 8, competition with EP17 at 100 and 500 molar excess; lane 9, GST only. (B) Lane 1, probe alone (30 mer); lane 2, 3, nuclear proteins (5 and 10 μg) + probe; lanes 4, self-competition at 100 fold molar excess using 10 μg nuclear proteins; lanes 5, 6, GST-HAF (2 and 5 μg) + probe. The bottom of the gel contains the VEGF 30 mer probe (VG30). The arrows indicate the specific complex.
Gel shift assay with VEGF UTR binding to HAF protein and nuclear proteins of Hep3B cells.
(A) Lane 1, probe alone (30 mer); lane 2, GST-HAF + probe; lanes 3, 4, self competition at 100 and 500 molar excess; lanes 5, 6, lack of competition with EP22 at 100 and 500 molar excess; lanes 7, 8, competition with EP17 at 100 and 500 molar excess; lane 9, GST only. (B) Lane 1, probe alone (30 mer); lane 2, 3, nuclear proteins (5 and 10 μg) + probe; lanes 4, self-competition at 100 fold molar excess using 10 μg nuclear proteins; lanes 5, 6, GST-HAF (2 and 5 μg) + probe. The bottom of the gel contains the VEGF 30 mer probe (VG30). The arrows indicate the specific complex.
Tissue distribution and development-stage–specific expression of hypoxia-associated factor messenger RNA
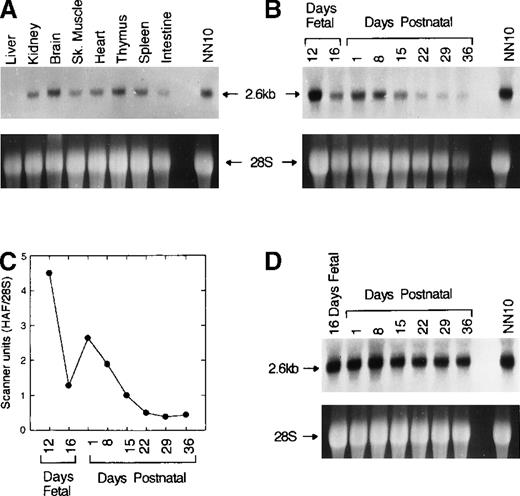
The expression of HAF was studied by Northern blot analysis of total RNA isolated from adult mouse liver, kidney, brain, heart, skeletal muscle, and spleen. RNA from NN10 cells was used as a positive control. A major transcript of about 2.6 kb was detected in those adult tissues studied, with minimal expression in liver (Figure5A).
Northern analysis of HAF expression in different tissues and at different development stages in liver and kidney.
The probe used was 700 bp (nt 1-700) of HAF cDNA. Total RNA from NN10 cells was used as a positive control. (A) A 2.6-kb single transcript was identified in all the tissues indicated above each lane with the exception of the liver. The bottom panel shows ethidium bromide stained 28 S rRNA. (B) Expression of HAF in liver at indicated development ages. The bottom panel shows ethidium bromide stained 28 S RNA. (C) Quantitative analysis of panel B using densitometer scanning. Scanner units were normalized for 28 S RNA. (D) Expression of HAF in kidneys at different development stages. The arrow indicates 2.6-kb HAF mRNA in upper panel and 28 S rRNA in bottom panel.
Northern analysis of HAF expression in different tissues and at different development stages in liver and kidney.
The probe used was 700 bp (nt 1-700) of HAF cDNA. Total RNA from NN10 cells was used as a positive control. (A) A 2.6-kb single transcript was identified in all the tissues indicated above each lane with the exception of the liver. The bottom panel shows ethidium bromide stained 28 S rRNA. (B) Expression of HAF in liver at indicated development ages. The bottom panel shows ethidium bromide stained 28 S RNA. (C) Quantitative analysis of panel B using densitometer scanning. Scanner units were normalized for 28 S RNA. (D) Expression of HAF in kidneys at different development stages. The arrow indicates 2.6-kb HAF mRNA in upper panel and 28 S rRNA in bottom panel.
Because EPO production occurs in fetal liver3 and switches to adult kidney,4 we analyzed HAF mRNA expression in liver and kidney from 12- and 16-day mouse fetuses and from mice at 1, 8, 15, 22, 29, and 36 days after birth. Twelve-day fetal liver had considerably higher HAF expression than did 16-day fetal liver. Hepatic expression decreased gradually and it was barely detectable at 36 days after birth (Figure 5B and C). Expression in kidneys was relatively constant over all the times studied (Figure 5D).
Discussion
In this report we describe the cloning of a cDNA encoding a protein (HAF) that shows sequence-specific interaction with a 17-bp sequence (EP17) in the proximal promoter region of the EPO gene and modulates expression of EPO and VEGF mRNA in response to hypoxia.
Comparison of the HAF cDNA sequence with those in the GenBank showed that HAF is a murine homolog of human Sart-1 cDNA. Sart-1 cDNA was cloned as an antigenic peptide recognized by cytotoxic T lymphocytes and was reported while this study was in progress.29 Sart-1 mRNA is suggested to encode 2 proteins, one 43 kd and the other 125 kd. The 43-kd protein is cytosolic, whereas 125-kd protein (HAF homolog) is nuclear. It was also suggested that the 43-kd but not the 125-kd protein may be the major protein recognized by cytotoxic T lymphocytes.
HAF mRNA was detected in all murine tissues studied with the lowest expression in adult liver. The highest level was found in fetal liver at day 12; it declined with time and almost none was detected at 36 days past birth.
In rodents, the liver is the major site of EPO synthesis during fetal life,3 whereas the kidney is the major source late in gestation and after birth.4 Mechanisms behind this switch or those involved in suppression of EPO production by liver are poorly understood. Earlier, experiments with transgenic mice carryingEPO-LacZ constructs identified a silencer sequence in the 1.2-kb 3′-flanking region of the EPO gene. The silencer element showed interaction with different sets of nuclear factors in fetal liver, compared with adult liver.30 The expression of HAF mRNA in fetal but not adult liver and the switch in EPOexpression from fetal liver to adult kidney suggest that HAF may play a role in hepatic expression, as well as a permissive role in renal expression.
Hypoxia-regulated expression of the EPO gene is shown to involve HIF-1, HNF4, and the COUP family of transcription factors through their interaction with the HRE in the 3′ untranslated region. Earlier, other investigators have reported a cooperative interaction of factors binding to HRE with those binding to the proximal promoter of the EPO gene.12 We have reported that HRE requires the presence of EP17 to produce its maximal effect.17 In the same study, we also showed that EP17 does not play a role in the basal expression of the EPO gene by Hep3B cells. HIF-1 is a well-characterized transcription factor and is a heterodimeric complex of HIF-1α and HIF-1β that belongs to the bHLH family of transcription factors containing a PAS domain.31,32 HIF-1α is a 826-residue protein, whereas HIF-1β is identical to the ARNT protein with 2 isoforms of 774 and 789 amino acids generated by alternative splicing, HIF-1α and HIF-β protein levels and their DNA-binding activities are induced by hypoxia.31-33
Besides EPO, HIF-1 has been shown to regulate hypoxia-mediated expression of VEGF.16 Other similarities between the EPO and VEGF gene regulation include their induction by CoCl2 and suppression of this induction by carbon monoxide.34,35 The evidence presented here suggests that HAF also plays a role in VEGF gene expression. With computer analysis, we found in the VEGF gene a sequence (CCCCCAGCCCCA) with close similarity to EP17. By using the numbering used in Genbank accession number U41383, this sequence is located at position +512 to +523 bp in the 5′ untranslated region of theVEGF gene.36 This 12-base sequence has one difference from the 5′ 12 bases of the EP17 sequence in that the seventh base from the 5′ end is a G rather than a C. The 5 bases at the 3′ end do not match those in EP17. Despite these differences, HAF interacts with the VEGF 5′UTR similar to the EP17 in EPO promoter. The importance of poly C repeats in EP17 for nuclear factor binding was established earlier by our point mutation analysis, where replacement of Cs by A and G was sufficient to decrease competition for factor binding.16
In this paper, we establish that HAF, a mouse homolog of the Sart-1 125-kd protein, through its interaction with the EP17 regulatory element of the EPO promoter region, plays a role in hypoxia-induced regulation of EPO gene expression. This is supported by the following observations: (1) HAF protein interacts with EP17 in a sequence-specific manner, (2) anti-HAF antibody produces a supershift in the EP17 DNA/protein complexes obtained using Hep3B nuclear extracts, and (3) HAF antisense RNA causes a reduction in mRNA for EPO and VEGF in response to hypoxia in Hep3B cells. The fact that HAF antisense does not completely suppress hypoxia-induced EPO expression is not surprising in view of our finding17 that basal EPO expression seems to not depend on EP17 function. Thus, HAF in conjunction with other transacting factors may modulate the hypoxia-induced expression ofEPO and VEGF genes.
Supported in part by grant HL30121 from the National Heart, Lung, and Blood Institute; by a gift from Kirin-Amgen, Inc, to the University of Chicago and, for sequencing, by the Cancer Research Center support grant P30 CA14599 from the National Cancer Institute.
HAF cDNA GenBank accession number AF129931.
Reprints:Eugene Goldwasser, Department of Biochemistry and Molecular Biology, University of Chicago, 920 E 58th St, Chicago, IL 60637; e-mail: egoldwas@midway.uchicago.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.