Abstract
One of the major binding sites for factor VIII inhibitors is located within the A2 domain. In this study, phage display technology was used to isolate 2 human monoclonal antibodies, termed VK34 and VK41, directed toward the heavy chain of factor VIII. The VHdomain of a single-chain variable domain antibody fragment (scFv) VK34 is encoded by germline gene segment DP-10. Epitope-mapping studies revealed that scFv VK34 is directed against amino acid residues Arg484–Ile508 , a previously identified binding site for factor VIII inhibitors in the A2 domain. ScFv VK34 inhibited factor VIII activity with a titer of 280 BU/mg. The VH domain of VK41 was encoded by germline gene segment DP-47. A phage corresponding to VK41 competed with a monoclonal antibody for binding to amino acid residues Asp712–Ala736 in the acidic region adjacent to the A2 domain. Reactivity of VK41 with a factor VIII variant in which we replaced amino acid residues Asp712–Ala736for the corresponding region of heparin cofactor II was strongly reduced. In addition, substitution of Tyr718719723 for Phe abrogated binding of VK41 to factor VIII. ScFv VK41 did not inhibit factor VIII activity. This study not only defines the primary structure of human anti-factor VIII antibodies reactive with the A2 domain, it also describes an antibody with an epitope not previously identified in the antibody repertoire of hemophilia patients with an inhibitor.
Factor VIII is an essential cofactor in the intrinsic pathway of blood coagulation that enhances the activation of factor X by factor IXa in the presence of Ca++ ions and phospholipids. Based on internal sequence homology, the factor VIII molecule can be defined by the domain structure A1-a1-A2-a2-B-a3-A3-C1-C2 (for review, see Lenting et al).1 In plasma, factor VIII circulates as a heterodimer composed of a heavy chain (A1-a1-A2-a2-B domains) and a light chain (a3-A3-C1-C2 domains). The functional absence of factor VIII is associated with the X-linked bleeding disorder hemophilia A. In patients with hemophilia A, the bleeding tendency can be corrected by the administration of factor VIII concentrates. After multiple infusions, some patients with hemophilia A develop antibodies that neutralize the procoagulant activity of factor VIII.2
These antibodies, commonly termed factor VIII inhibitors, are directed against epitopes present in the A2, A3, and C2 domains of factor VIII.3 More detailed mapping of anti-C2 antibodies revealed a common binding site consisting of residues Val2248–Ser2312.4Using a series of active human/porcine factor VIII hybrids, a second determinant of the anti-C2 inhibitor epitope has been attributed to the region Glu2181-Val2243.5 Anti-C2 inhibitors prevent factor VIII from binding to phospholipids and von Willebrand factor.6,7 Two independent studies identified a binding site for factor VIII inhibitors in the A3 domain of factor VIII, which overlaps a previously identified binding site for factor IXa.8-10 Binding of these inhibitors interferes with assembly of the factor IXa–factor VIIIa complex.
Within the A2 domain, residues Arg484–Ile508have been shown to constitute a binding site for factor VIII inhibitors.11 Alanine scanning mutagenesis within this region indicated that amino acid residue Tyr487 is essential for binding most human inhibitors to the A2 domain.12 Anti-A2 inhibitors block the activation of factor X by the phospholipid bound factor VIIIa–factor IXa complex.13 Recently, it was shown that these antibodies abrogate the stimulatory effect of isolated A2 domain on factor IXa activity.14 These data indicate that anti-A2 inhibitors prevent the interaction of the A2 domain with factor IXa.
Previously, we have used phage display technology to isolate anti-C2 antibodies from the immunoglobulin repertoire of a patient with acquired hemophilia.15 Anti-C2 antibodies were characterized by an unusually long CDR3 of 20-23 amino acids and extensive somatic hypermutation. Surprisingly, the immunoglobulin heavy chain variable (VH) domains of all these antibodies were encoded by VH gene segments derived from the VH1 gene family. These findings suggest that a subset of VH gene segments is used to generate human anti-C2 antibodies. Here, we have used phage display technology to further define anti-A2 antibodies. The current study defines the molecular characteristics of a human antibody reactive with factor VIII sequence Arg484–Ile508, the major inhibitor binding site located within the A2 domain. Moreover, we provide evidence for the existence of an additional epitope for human anti-factor VIII antibodies located between residues Asp712–Ala736 in the a2 region.
Materials and methods
Materials
DNA restriction enzymes and Taq DNA polymerase were purchased from Life Technologies (Breda, The Netherlands) and New England Biolabs (Beverly, MA). Pwo DNA polymerase was obtained from Boehringer (Mannheim, Germany). Factor VIII heavy chain was purified from human plasma as described.16 Factor VIII-del(868-1562) (hereafter designated rFVIII), a B domain-deleted factor VIII, has been described previously.17 In rFVIII–HCII residues Asp712–Ala736 of factor VIII were replaced by residues Ile51–Leu80 of heparin cofactor II.18 Construction of a factor VIII variant in which amino acid residues Tyr718, Tyr719, and Tyr723 were replaced for Phe was performed by overlap extension mutagenesis using the previously described plasmid pCLB-dB695 as a template.17 rFVIII, rFVIII–HCII, and rFVIII–Tyr718,719,723→Phe were expressed in mouse C127 fibroblasts and purified as described.16 Monoclonal antibody (mAb) CLB–CAg 9 has been characterized previously.18 mAb ESH5 was purchased from American Diagnostica (Greenwich, CT).
FVIII assays
Construction of a hybrid FVIII/FV recombinant A2 domain
Plasmid pCLB-GP67B-A221 and factor V cDNA served as templates for the construction of a plasmid encoding the A2 domain and the a2 region (residues Ser373–Arg740) in which residues Arg484–Ile508 were replaced by the corresponding sequence of coagulation factor V. Primer combinations A2-1-484FV AS (5′-TCT TCA TAA GGG ACA TCA GTG ATT CCG-3′), 484FV S (TGA TGT CCC TTA TGA AGA TGA AGT, C-3′)-508FV AS (5′-TAT TTG AAT GTT TCC CCT GGT TGA AC-3′), and 508FV S (5′-CAG GGG AAA CAT TCA AAT ATA AAT GG-3′)-A2-2 were used to amplify 3 DNA fragments that were reassembled by overlap extension polymerase chain reaction using outer primers A2-1 and A2-2 in a second round of amplification.21The final product was cloned as NcoI–NotI fragment in pAc-GP67B to yield pCLB-GP67B-A2-FV484-508. Expression in insect cells and labeling of recombinant factor VIII fragments was performed as described previously.21
Phage display library construction and selection
In this study, peripheral blood mononuclear cells were used as a source of RNA for generation of the patient's IgG4-specific VH gene repertoire essentially as described previously.15 The obtained repertoire was combined with a VL gene repertoire of nonimmune origin in pHEN-1-VLrep and displayed as scFv on the surface of filamentous phage.23Phage were selected for binding to the factor VIII heavy chain using the following methods: microtiter wells were coated overnight at 4°C with 100 μL mAb CLB–CAg 9 (35 nmol/L in 50 mmol/L NaHCO3, pH 9.6). Subsequently, wells were blocked with Tris-buffered saline (150 mmol/L NaCl, 50 mmol/L Tris, pH 7.4) and 3% (wt/vol) human serum albumin (HSA) for 2 hours at 37°C. To reduce nonspecific binding, phage in Tris-buffered saline, 3% (wt/vol) HSA, and 0.5% (vol/vol) Tween-20 were preincubated for 2 hours at room temperature in blocked CLB-CAg 9–coated microtiter wells. Meanwhile, CLB-CAg 9–coated microtiter wells were incubated for 2 hours at 37°C with human factor VIII heavy chain (16 nmol/L in 1 mol/L NaCl, 50 mmol/L Tris, pH 7.4, 2% [wt/vol] HSA). Wells were blocked with HSA as outlined above and incubated for another 2 hours at room temperature with nonbound phage, which were transferred from the preincubations. After intensive washing, bound phage were eluted and rescued by reinfection of Escherichia coli TG1.24Alternatively, phage were selected on factor VIII heavy chain (40 nmol/L in 50 mmol/L NaHCO3, pH 9.6) immobilized to immunotubes (Maxisorp; Nunc, Breda, The Netherlands), thereby allowing the selection of phage directed toward epitopes blocked by antibody CLB–CAg 9. The library was subjected to 3 rounds of selection using the 2 procedures outlined above.
Screening and sequencing of selected clones
After 3 rounds, phages obtained from 20 single infected colonies of both selections were tested for binding to the factor VIII heavy chain immobilized to mAb ESH5. Bound phages were detected by anti-M13 antibody peroxidase conjugate (Pharmacia-LKB, Woerden, The Netherlands). VH and VL genes of factor VIII heavy chain binding clones were sequenced using the BigDye Terminator sequencing kit on a 377XL automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequences were compared with a database of germline V genes as compiled in the V-BASE sequence directory.25
Expression and purification of scFv
To facilitate the purification of scFv, a His-tag was introduced into the expressed protein by subcloning the V gene cassettes into the vector pUC119-Sfi/Not-His6.26 ScFv expression in E coli was induced with isopropyl β-D-thiogalactoside for 3 hours at 25°C. Purification of scFv by immobilized metal chelate affinity chromatography was performed as described previously.27Eluted fractions were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis under nonreducing conditions; protein concentrations were determined spectrophotometrically at A280.
Characterization of isolated clones
Immunoprecipitation of metabolically labeled factor VIII fragments by scFv was performed as described previously.15 Reactivity of phage derived from the isolated clones with plasma-derived factor VIII heavy chain, recombinant A2 domain, factor VIII heavy chain, rFVIII, rFVIII-HCII, and rFVIII-Tyr718,719,723→Phe was determined by enzyme-linked immunosorbent assay (ELISA). Factor VIII antigen was immobilized at a concentration of 1 nmol/L to ESH5-coated microtiter wells. Microtiter wells were incubated for 2 hours at room temperature with recombinant phage in 500 mmol/L NaCl, 50 mmol/L Tris, pH 7.4, 3% (wt/vol) HSA, and 0.5% (vol/vol) Tween-20. Bound phages were detected by anti-M13 antibody peroxidase conjugate.28 Experiments were performed in duplicate, and values were expressed as percentages of maximum binding.
Results
Characterization of anti-factor VIII antibodies in patient's plasma
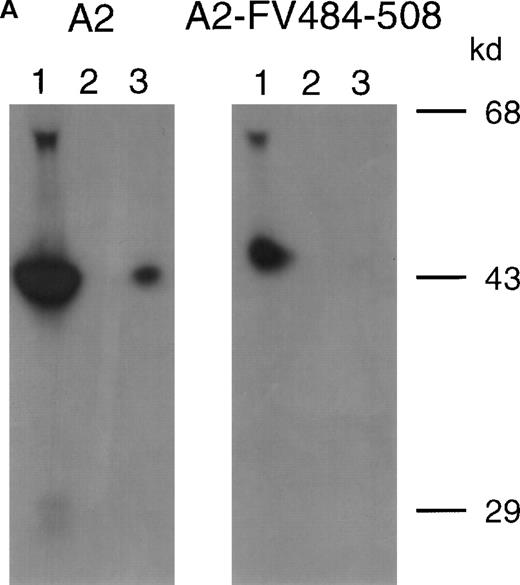
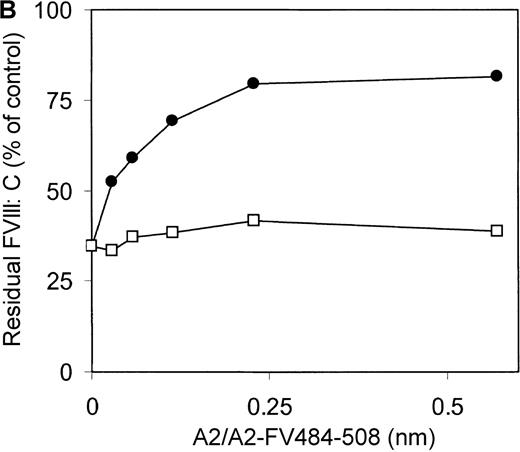
Previously, we reported on the domain specificity of anti-factor VIII antibodies in a patient (AMC-67) with mild hemophilia A caused by an Arg593→Cys substitution.22 The patient had a transient inhibitor with a maximum titer of 250 BU/mL. Plasma and peripheral blood mononuclear cells were isolated from blood samples collected when the inhibitor reached its peak value. Most factor VIII inhibitory antibodies in the patient's plasma were directed against the A2 domain. Here, we evaluated binding of these antibodies to a hybrid factor VIII/factor V recombinant A2 domain in which residues Arg484–Ile508 were substituted for the corresponding sequence of factor V. Immunoprecipitation analysis revealed that antibodies in the patient's plasma did not react with A2-FV484-508 (Figure 1A). Subsequently, an inhibitor neutralization assay using this fragment was performed. Limited neutralization was observed with the addition of A2-FV484-508, whereas the A2 domain almost completely neutralized factor VIII inhibitory activity (Figure 1B). These findings suggest that approximately 70% of the factor VIII inhibitory antibodies in the plasma of this patient are directed toward an epitope consisting of residues Arg484–Ile508.
Characterization of antibodies in patient plasma.
(A) Binding of antibodies to radiolabeled recombinant factor VIII fragments corresponding to the A2 domain (A2) and an A2 domain in which residues Arg484–Ile508 were replaced for the corresponding sequence of factor V (A2-FV484-508) was assessed by immunoprecipitation analysis. Lane 1, positive control (CLB–CAg 9); lane 2, negative control (normal plasma); lane 3, plasma of patient AMC-67. Molecular weight markers (in kd) are indicated at the right. (B) Neutralization of factor VIII inhibitory activity by recombinant factor VIII fragments. Inhibitor plasma was diluted to a final concentration of 2 BU/mL, corresponding to 25% of residual factor VIII activity. Samples were incubated for 2 hours at 37°C in the presence of increasing concentrations A2 domain (•) and A2-FV484-508 (□). After incubation for 1 additional hour at 37°C in the presence of normal plasma, residual factor VIII activity was determined relative to a sample that was incubated in the absence of an inhibitor.
Characterization of antibodies in patient plasma.
(A) Binding of antibodies to radiolabeled recombinant factor VIII fragments corresponding to the A2 domain (A2) and an A2 domain in which residues Arg484–Ile508 were replaced for the corresponding sequence of factor V (A2-FV484-508) was assessed by immunoprecipitation analysis. Lane 1, positive control (CLB–CAg 9); lane 2, negative control (normal plasma); lane 3, plasma of patient AMC-67. Molecular weight markers (in kd) are indicated at the right. (B) Neutralization of factor VIII inhibitory activity by recombinant factor VIII fragments. Inhibitor plasma was diluted to a final concentration of 2 BU/mL, corresponding to 25% of residual factor VIII activity. Samples were incubated for 2 hours at 37°C in the presence of increasing concentrations A2 domain (•) and A2-FV484-508 (□). After incubation for 1 additional hour at 37°C in the presence of normal plasma, residual factor VIII activity was determined relative to a sample that was incubated in the absence of an inhibitor.
Isolation and sequence analysis of antibodies directed toward the factor VIII heavy chain
V gene phage display was used to isolate human antibodies reactive with the factor VIII heavy chain from the immunoglobulin repertoire of the patient. Isotyping revealed that the factor VIII heavy chain-specific antibodies in the patient's plasma consisted predominantly of subclass IgG4 (data not shown). Therefore, a subclass-specific oligonucleotide primer was used for amplification of the patient's IgG4 VH gene repertoire. The IgG4-enriched VH gene repertoire was recombined with a nonimmune VL gene repertoire in pHEN-1–Vlrep, resulting in a library of 1.9 × 107 clones. To isolate anti-A2 antibodies, the library was selected for binding to the factor VIII heavy chain. After the third round of selection, phages derived from 40 single clones were analyzed for binding to the factor VIII heavy chain. Twenty-six of 40 clones reacted with factor VIII heavy chain (data not shown).
Sequence analysis of these 26 factor VIII heavy chain reactive clones revealed the presence of only 2 different VH domains. Two clones, VK34 and VK41, were selected for further study. The VH gene of clone VK34 was derived from germline gene segment DP-10, belonging to the VH1 gene family (Figure2). Comparison of the amino acid sequence of the VH segment of clone VK34 with that of the nonmutated germline gene segment DP-10 revealed 8 differences. The CDR3 of VK34 consists of only 5 amino acids. Interestingly, residues Ala93 and Arg94, located adjacent to the CDR3 and normally encoded by the VH germline gene segment DP-10, were replaced by Glu93 and Leu94. The amino acid sequence of the VH gene segment of clone VK41 differed at 13 positions from that of the most homologous germline gene segment DP-47 of the VH3 family (Figure 2). The CDR3 of VK41 comprises 12 amino acid residues. JH gene segments involved in immunoglobulin VDJ rearrangement in clones VK34 and VK41 were most homologous to JH3b and JH6b, respectively. Use of a particular D gene segment could not be ascertained. VL domains of VK34 and VK41 were both derived from gene segment DPL16, a member of the Vλ3 gene family (Figure 2).
Deduced protein sequences of isolated scFv.
Dashes indicate sequence identity to germline. Sequence numbering is in accordance with that of Kabat.29 FR, framework region; CDR, complementarity-determining region. Sequences are available from GenBank under accession numbers AF217789 (VH VK34), AF217790 (VH VK41),AF217791 (VL VK34), and AF217792 (VL VK41).
Deduced protein sequences of isolated scFv.
Dashes indicate sequence identity to germline. Sequence numbering is in accordance with that of Kabat.29 FR, framework region; CDR, complementarity-determining region. Sequences are available from GenBank under accession numbers AF217789 (VH VK34), AF217790 (VH VK41),AF217791 (VL VK34), and AF217792 (VL VK41).
Biochemical characterization of VK34 and VK41
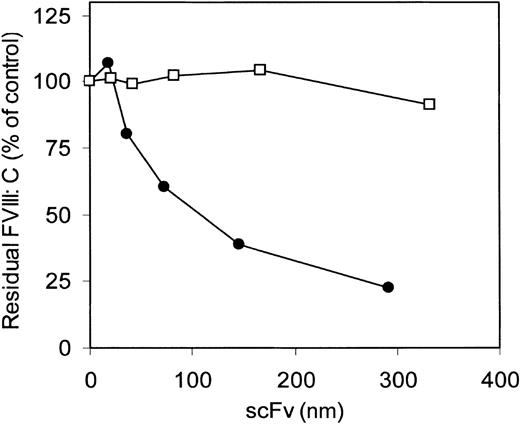
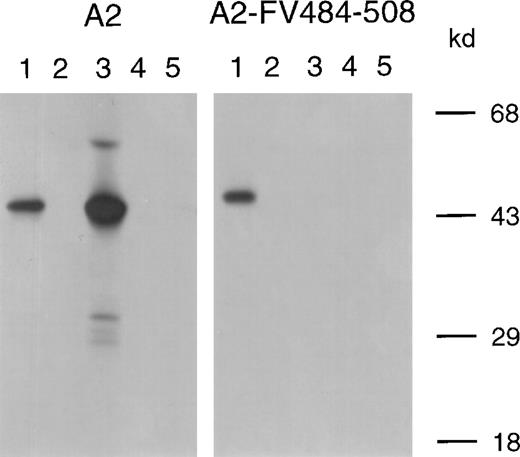
The inhibitory effect of antibody fragments of VK34 and VK41, expressed as scFv in E coli on factor VIII procoagulant activity, was evaluated in the Bethesda assay. ScFv VK34 had an inhibitor titer of 280 BU/mg. No inhibition of factor VIII activity was observed in the presence of scFv VK41 (Figure3). To define the epitopes of VK34 and VK41, scFv were tested for reactivity with different metabolically labeled A2 domain fragments by immunoprecipitation. ScFv VK34 reacted with the recombinant A2 domain (Figure 4; lane 3, left panel). A variant A2 domain, in which the region Arg484–Ile508 was replaced for the corresponding sequence of factor V, was not recognized by scFv VK34 (Figure 4; lane 3, right panel). Thus, binding of VK34 is dependent on the presence of Arg484–Ile508, a region previously identified as a major binding site for factor VIII inhibitors in the A2 domain. Surprisingly, neither recombinant A2 domain fragment was recognized by scFv VK41 (Figure 4; lane 4). Therefore, the epitope specificity of clone VK41 was examined using a different approach.
Inhibition of factor VIII activity in the 1-stage clotting assay.
ScFv VK34 (•) and VK41 (□) were tested for factor VIII inhibitory activity according to the Bethesda assay.20 Factor VIII activity is given in percentages relative to a control sample incubated in the absence of scFv.
Inhibition of factor VIII activity in the 1-stage clotting assay.
ScFv VK34 (•) and VK41 (□) were tested for factor VIII inhibitory activity according to the Bethesda assay.20 Factor VIII activity is given in percentages relative to a control sample incubated in the absence of scFv.
Immunoprecipitation of variant A2 domain fragments by isolated scFv.
Binding of scFv to recombinant A2 domain and A2-FV484-508 was assessed by immunoprecipitation. Lane 1, positive control (mAb CLB–CAg 9); lane 2, negative control (normal plasma); lane 3, scFv VK34; lane 4, scFv VK41; lane 5, scFv EL-14, directed toward the C2 domain of factor VIII. Molecular weight markers (in kd) are indicated at the right.
Immunoprecipitation of variant A2 domain fragments by isolated scFv.
Binding of scFv to recombinant A2 domain and A2-FV484-508 was assessed by immunoprecipitation. Lane 1, positive control (mAb CLB–CAg 9); lane 2, negative control (normal plasma); lane 3, scFv VK34; lane 4, scFv VK41; lane 5, scFv EL-14, directed toward the C2 domain of factor VIII. Molecular weight markers (in kd) are indicated at the right.
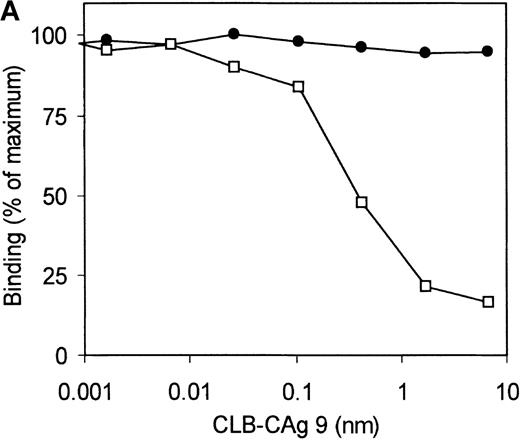
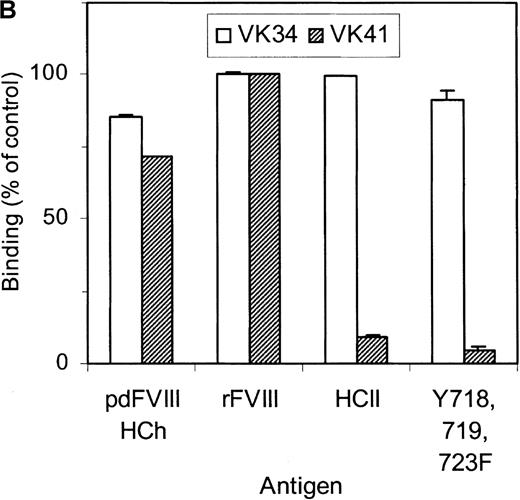
Selection of the library was performed using 2 different methods. Phages corresponding to clone VK41 were exclusively isolated from selection of the library using immunotubes coated with factor VIII heavy chain. Selection of the library on plasma-derived factor VIII heavy chain immobilized by mAb CLB–CAg 9 did not yield phages corresponding to clone VK41. The epitope of antibody CLB–CAg 9 has been localized to amino acid residues Asp712–Ala736.18 These results suggest that the epitope of VK41 may overlap with residues Asp712–Ala736, which constitute the epitope of CLB–CAg 9. Therefore, antibody CLB–CAg 9 was tested for its ability to compete with VK41 for binding to the factor VIII heavy chain. Because scFv VK41 reacted poorly with immobilized factor VIII heavy chain, phages corresponding to VK34 and VK41 were used for these studies. Phages at a concentration that corresponded to 75% of maximum binding were mixed with serial dilutions of CLB–CAg 9 and incubated with factor VIII heavy chain containing wells. Bound phages were detected as described in “Materials and methods.” Concentrations of 7 nmol/L CLB–CAg 9 were sufficient to reduce significantly the binding of VK41 to immobilized factor VIII heavy chain (Figure5A). In contrast, the binding of clone VK34 to factor VIII heavy chain was not affected by the addition of CLB–CAg 9. These data indicate that the epitope of VK41 is located within or close to a region bounded by residues Asp712–Ala736. Previously, we described a variant factor VIII in which amino acid residues Asp712–Ala736 were replaced by Ile51–Leu80 of heparin cofactor II. This variant, termed rFVIII–HCII, was not recognized by antibody CLB–CAg 9.18 Therefore, the reactivity of phages corresponding to VK34 and VK41 with rFVIII–HCII was evaluated. Phages corresponding to VK41 readily bound to rFVIII, whereas binding to rFVIII–HCII was strongly reduced (Figure 5B). Within region Asp712–Ala736, 3 tyrosine residues are present that are posttranslationally modified by tyrosine sulfation.30 We investigated the binding of VK41 to a factor VIII variant in which Tyr718, Tyr719, and Tyr723 were replaced by Phe (rFVIII–Tyr718,719,723→Phe). Only limited reactivity of VK41 with rFVIII-Tyr718,719,723→Phe was observed (Figure 5B). Our data suggested that Tyr718, Tyr719, and Tyr723 are part of a previously unidentified binding site for human anti-factor VIII antibodies in the acidic region adjoining the A2 domain.
Epitope mapping of clone VK41.
(A) Competition for binding to the heavy chain of factor VIII of phages corresponding to VK34 (•) and VK41 (□) with mAb CLB–CAg 9. Binding is expressed as a percentage of maximal binding. (B) Reactivity of phages corresponding to VK34 and VK41 with plasma-derived factor VIII heavy chain (pdFVIII HCh), rFVIII (rFVIII), rFVIII-HCII (HCII), and rFVIII-Tyr718,719,723→Phe (Y718, 719, 723F). Phage ELISAs were performed as described in “Materials and methods.” Binding of phage to rFVIII is expressed as a percentage relative to binding of VK41 to rFVIII.
Epitope mapping of clone VK41.
(A) Competition for binding to the heavy chain of factor VIII of phages corresponding to VK34 (•) and VK41 (□) with mAb CLB–CAg 9. Binding is expressed as a percentage of maximal binding. (B) Reactivity of phages corresponding to VK34 and VK41 with plasma-derived factor VIII heavy chain (pdFVIII HCh), rFVIII (rFVIII), rFVIII-HCII (HCII), and rFVIII-Tyr718,719,723→Phe (Y718, 719, 723F). Phage ELISAs were performed as described in “Materials and methods.” Binding of phage to rFVIII is expressed as a percentage relative to binding of VK41 to rFVIII.
Discussion
Epitope mapping studies revealed that a significant portion of factor VIII inhibitors binds to the A2 domain of factor VIII.3 Within the A2 domain, residues Arg484–Ile508 constitute a major determinant of the epitope of factor VIII inhibitors.11,12 In this study, we selected a phage display library of the IgG4-restricted VH gene repertoire derived from a patient with anti-A2 inhibitor for binding to the heavy chain of factor VIII. Two different antibodies (VK34 and VK41) reactive with the factor VIII heavy chain were isolated. Epitope mapping revealed that clone VK34 was directed toward the amino acid residues Arg484–Ile508 in the A2 domain. Antibodies directed toward this region account for most factor VIII inhibitory activity in the patient's plasma (Figure 1B). Furthermore, our study provides evidence for an additional binding site for anti-factor VIII antibodies in the a2 region, which comprises amino acid residues Asp712–Ala736. So far, anti-A2 antibodies are predominantly directed toward a major binding site that has been attributed to the region Arg484–Ile508.11,12,14 Anti-A2 inhibitors have been studied in functional assays, which only detect inhibitory anti-factor VIII antibodies.11 12 Because scFv VK41 does not inhibit factor VIII activity, antibodies in patient plasma corresponding to VK41 may have escaped detection using these assays. This may explain why amino acid region Asp712–Ala736 has not been identified previously as a binding site for anti-factor VIII antibodies. Alternatively, the plasma concentration of IgG corresponding to VK41 may be low in patients with an inhibitor. Competition experiments indicated that IgG in the patient's plasma was able to compete for binding to factor VIII heavy chain by scFv VK41 (data not shown). These findings suggest that IgG corresponding to VK41 is present in significant amounts in the plasma of patient AMC-67.
VK41 did not bind to a recombinant A2 fragment comprising residues Ser373–Arg740 in an immunoprecipitation assay, whereas it reacted with plasma-derived factor VIII heavy chain. This suggests that the properties of the recombinant fragment produced in insect cells are dissimilar to the corresponding region in plasma-derived factor VIII heavy chain. The acidic region carboxy-terminal of the A2 domain contains 3 potential tyrosine sulfation sites at positions Tyr718, Tyr719, and Tyr723. The inability of VK41 to react with variant rFVIII–Tyr718,719,723→Phe suggests that sulfated tyrosines in amino acid region Asp712–Ala736constitute at least part of the epitope of VK41. Interestingly, rFVIII-Tyr718,719,723→Phe is recognized normally by antibody CLB–CAg 9, indicating that the sulfated tyrosines are not required for the binding of this antibody (data not shown). Apparently, different amino acids within the a2 region contribute to the epitope of CLB–CAg 9 and VK41. The lack of reactivity of VK41 with the recombinant A2 fragment suggests that tyrosine sulfation at Tyr718, Tyr719, and Tyr723 occurs inefficiently in insect cells. Besides the acidic region carboxy-terminal of the A2 domain, acidic regions are present in the carboxy-terminal of the A1 domain and the amino-terminal of the A3 domain. Some inhibitor plasmas contain antibodies directed toward the a1 region of factor VIII.3,31 Recently, the acidic region a3, the amino-terminal of the A3 domain, has been identified as a binding site for factor VIII inhibitors.32 The presence of cleavage sites for thrombin and factor Xa at the borders of acidic regions adjacent to the A1, A2, and A3 domains indicates that these areas are exposed in factor VIII. This may explain the presence of binding sites for anti-factor VIII antibodies in the acidic regions a1, a2, anda3.
Factor VIII inhibitors directed toward the A2 domain are characterized by their restricted epitope specificity, suggesting that a limited number of VH genes participates in the assembly of antibodies that recognize Arg484–Ile508. It is of note that only a single clone reactive with region Arg484–Ile508 was isolated from the patient's repertoire. Clonal expansion of a single-memory B cell may be a particular feature of the patient analyzed. Isolation of anti-A2 antibodies from other patients should reveal whether a restricted number of VH germline genes encode for the VHdomains in anti-A2 antibodies. Recently, we have shown that anti-C2 antibodies are composed of multiple VH domains that are derived from germline genes of the VH1 family.15 Interestingly, the VH domain of clone VK34 is encoded by germline gene segment DP-10 of the VH1 gene family. Similar to VK34, the VH domain of the C2-specific scFv EL-14 was encoded by the DP-10 germline gene segment. In the human repertoire, the DP-10 germline gene segment is rearranged in less than 5% of the IgG-positive B cells.33 No cross-reactivity of scFv VK34 with the C2 domain (data not shown) or of scFv EL-14 with the A2 domain (Figure 4) was observed. The composition of the CDR3 may contribute to the differences in epitope specificity observed for VK34 and EL-14. The VH domain of VK34 is characterized by an extremely short CDR3 of only 5 amino acid residues, whereas the average length of a CDR3 is approximately 12 residues.34 In contrast, EL-14 contains an unusually large CDR3 of 21 amino acids.15 The VH domains of scFv VK34 and EL-14 displayed extensive somatic hypermutation, indicating that the VH genes are derived from antigen-stimulated B cells. For clones VK34 and EL-14, no homology in the patterns of somatic hypermutation were observed (data not shown). In addition, the VL domains of VK34 and EL-14, which may potentially contribute to antigen specificity, are derived from different VL germline gene families (DPL16 and DPK5). However, the VL domains are derived from a nonimmune source and are therefore unlikely to contribute to the epitope specificity of scFv. Based on the above considerations, we hypothesize that the binding of VK34 and EL-14 to distinct antigenic sites on factor VIII originates from differences in the somatic hypermutation and composition of CDR3 in the VH domains of these scFv.
The VH domain of clone VK41 is encoded by germline gene DP-47 of the VH3 gene family. Interestingly, DP-47 is the most frequently rearranged germline gene segment in the human repertoire, observed in approximately 12% of the IgG-positive peripheral B cells.33 Therefore, antibodies directed toward residues Asp712–Ala736, with molecular characteristics similar to those of VK41, may also be present in the repertoire of additional hemophilia A patients with inhibitors.
Acknowledgments
The authors thank G. van Stempvoort and P. H. N. Celie for providing the purified factor VIII variants and factor VIII heavy chain. They also thank R. C. Aalberse, W. G. van Aken, J. A. van Mourik, and K. Mertens for critical review of the manuscript.
Reprints:Jan Voorberg, Department of Plasma Proteins, CLB, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail:j_voorberg@clb.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.