Abstract
A previous study reported that a nondifferentiating myeloid leukemia cell line produced differentiation-inhibiting factors. One of the factors was purified as a homologue of the nm23 genes. Thenm23 genes were overexpressed in acute myelogenous leukemia (AML) cells, and a higher level of nm23 gene expression was correlated with a poor prognosis in AML. The present study determined the plasma levels of nm23-H1 protein by enzyme-linked immunosorbent assay and assessed the association between this level and the clinical outcome in 102 patients with AML. The plasma concentration of nm23-H1 was higher in patients with AML than in normal controls (P = .0001). Plasma nm23-H1 levels were correlated with the product of the intracellular nm23 messenger RNA (mRNA) level and the white blood cell count, but not with the mRNA level alone. Therefore, nm23-H1 plasma levels probably depend on the total mass of leukemic cells overexpressing the nm23-H1 gene. Overall survival was lower in patients with higher plasma nm23-H1 levels than in those with lower levels. Multivariate analysis using the Cox proportional hazard model showed that elevated plasma nm23-H1 levels significantly contributed to the prognosis of AML patients. Furthermore, the plasma nm23-H1 levels were investigated in 70 patients with other hematologic neoplasms, including 6 with acute lymphoblastic leukemia, 13 with chronic myelogenous leukemia, and 12 with myelodysplastic syndrome. Plasma nm23-H1 levels were significantly higher in all of these hematologic neoplasms than in normal controls. Increased plasma levels of nm23-H1 may have prognostic value in these hematologic malignancies as well as in AML.
We previously found that a differentiation-inhibiting factor (I factor) purified from a differentiation-resistant mouse myeloid leukemia cell line was identical to nm23 protein.1,2 The nm23 proteins inhibit the induction of differentiation of mouse and human leukemia cells,3 are involved in the regulation of tumor metastasis, and have nucleoside diphosphate (NDP) kinase enzyme activity.4,5 Two types of human nm23 gene have been identified: nm23-H1 andnm23-H2. The proteins encoded by nm23-H1 andnm23-H2 show 88% amino acid sequence homology and the genes are located in tandem on the same region of chromosome 17q21.6-10 The I-factor activity of nm23 proteins is independent of their NDP kinase activity and requires the presence of the 60 N-terminal amino acids.11 Based on the biologic activity of nm23 proteins as I factors, we previously investigated the relative levels of nm23-H1 and nm23-H2 transcripts in acute myelogenous leukemia (AML) and chronic myelogenous leukemia (CML) cells. Thenm23-H1 and nm23-H2 genes were overexpressed in AML and CML in blastic crisis (CML-BC) but not in CML in the chronic phase (CML-CP), and nm23-H1 gene expression predicted the prognosis of AML.12-14
Recently, we established an enzyme-linked immunosorbent assay (ELISA) technique to determine the plasma level of nm23-H1 protein. This assay is far simpler than that used to determine nm23 messenger RNA (mRNA) by reverse transcriptase-polymerase chain reaction (RT-PCR). Using this method, we recently found that an elevated plasma nm23-H1 protein concentration predicts a poor outcome in advanced non-Hodgkin lymphoma.15 In the present study, we determined the plasma nm23-H1 protein levels in patients with various hematologic malignancies and studied the association between the plasma nm23-H1 protein level and the prognosis of AML.
Patients, materials, and methods
Patients
Plasma samples were obtained from 172 patients who had been newly diagnosed with hematologic neoplasms, consisting of 102 with de novo AML, 6 with de novo acute lymphoblastic leukemia (ALL), 13 with CML-CP, 8 with CML-BC, 12 with myelodysplastic syndrome (MDS; 3 had refractory anemia, 6 had refractory anemia with excess of blasts [RAEB], and 3 had RAEB in transformation), 6 with MDS in the overt leukemic phase, 21 with multiple myeloma (MM), and 4 with chronic lymphocytic leukemia (CLL). Samples were obtained with informed consent at the diagnosis of the disease and before chemotherapy was administered. De novo AML, ALL, and MDS were classified according to the criteria devised by the French-American-British (FAB) classification and immunophenotypic studies.16 17 Of the 102 patients with AML, 52 were women and 50 were men. Their median age was 56 years, with a range between 16 and 72 years. The following FAB subtypes of AML were observed; 3 were M0, 18 were M1, 25 were M2, 14 were M3, 17 were M4, 16 were M5, 6 were M6, and 3 were M7 (Table 1). Leukocyte (WBC) counts varied widely among the patients (range, 1-286 × 109/L; median, 28).
The AML patients were treated with cytosine arabinoside (or behenoyl cytosine arabinoside) and daunorubicin with or without prednisolone or 6-mercaptopurine, and AML-M3 patients were consecutively treated with all-trans retinoic acid for remission induction therapy.18 19 Patients were judged to be in complete remission (CR) when bone marrow aspirates showed trilineage regeneration with fewer than 5% blasts by morphologic and immunocytochemical analyses in the presence of a normal blood count that persisted for at least 1 month. Patients who died from chemotherapy toxicity (infection or bleeding) before the time of expected marrow recovery were not evaluated. All other patients were considered to be nonresponsive (NR). The median follow-up time was 32 months (range, 11-48 months). Plasma samples were also obtained from 21 healthy volunteers with a mean age of 34 years (range, 24-52 years) and analyzed for comparison. None of the controls had had a fever within the past week, were receiving any medications, were known to be pregnant, or had a history of any chronic or acute illnesses.
Venous blood samples
Peripheral venous blood samples were collected in sterile test tubes with heparin and placed on ice for at least 10 minutes to avoid platelet activation. The samples were then centrifuged at 2000gfor 15 minutes at 4°C, filtered through a 0.22-μm microfilter (Millipore, Molsheim, France), and stored at −80°C.
RT-PCR
Quantitative RT-PCR was performed using a GeneAmp RNA PCR kit (Takara, Tokyo, Japan), as reported previously.12-14 The oligonucleotides used in PCR amplification were as follows: sense strand, 5′-ATGGCCAACTGTGAGCGTACC-3′; antisense strand, 5′-CATGTATTTCACCAGGCCGGC-3′ for nm23-H1; sense strand, 5′-ATGGCCAACCTGGAGCGCACC-3′; antisense strand, 5′-TCCCCACGAATGGTGCCTGGC-3′ for nm23-H2; and sense strand, 5′-ACATCGCTCAGACACCATGG-3′; antisense strand, 5′-GTAGTTGAGGTCAATGAAGGG-3′ for glyceraldehyde-3-phosphate dehydrogenase (gapdh). PCR consisted of 35 cycles fornm23-H1 and 25 for nm23-H2 and gapdh, with denaturing at 95°C for 1 minute, annealing at 60°C for 1 minute, and extension at 72°C for 0.5 minutes. The reaction was performed in a GeneAmp PCR system 9600 (Perkin Elmer, Norwalk, CT). The PCR products were then subjected to 6% polyacrylamide gel electrophoresis, and the dried gel was exposed to imaging plates (Fuji Film Co., Ltd., Tokyo, Japan) at room temperature for 15 to 20 minutes. The results of autoradiography were quantified using a Fuji Bio-Image Analyzer BAS2000 (Fuji Film).
ELISA for human nm23-H1
We previously established an ELISA procedure to determine nm23-H1 protein levels in plasma.15 Ninety-six-well plates (Costar 3369, Corning Inc, Corning, NY) were coated with monoclonal anti-nm23-H1 antibody (Seikagaku Kogyo Co, Tokyo, Japan), washed 4 times with phosphate-buffered saline (PBS) and incubated with 25% Block Ace (Dainihon Seiyaku, Osaka, Japan). Plasma samples were diluted 2-fold with PBS, and 50-μL aliquots were then added to the wells. After incubation at room temperature for 1 hour, the wells were washed 4 times with PBS containing 0.05% Tween 20 (T-PBS). The samples were incubated at room temperature for 1 hour with polyclonal anti-nm23-H1 antibody (Santa Cruz Biotechnologies, Santa Cruz, CA), washed 4 times with T-PBS, and incubated with alkaline phosphatase-conjugated antirabbit IgG (Bio Rad Laboratories, Hercules, CA). After washing 4 times with T-PBS, alkaline phosphatase activity was detected using diethanolamine as a substrate and an alkaline phosphatase-detecting kit (Bio Rad). The absorbance was measured at 405 to 415 nm with a correction wavelength of 620 to 630 nm using a Micro plate reader. Recombinant nm23-H1-GST protein (kindly provided by Prof H. Shiku, Mie University, Tsu, Japan) was used as a standard.3,20 Free plasma hemoglobin contents were measured photometrically with benzidine.21
Statistical analysis
Survival analysis was performed according to the Kaplan-Meier method.22 The statistical significance of differences among curves was determined by the log-rank test and the generalized Wilcoxon test.23 Differences between groups were evaluated by the Mann-Whitney U test (nonparametric analysis),24 andP < .05 was taken to indicate significance. The Pearson correlation coefficient (r) was used to evaluate the correlation between paired values. A multivariate analysis of prognostic factors was performed using the Cox proportional hazards regression model.25
Results
Plasma nm23-H1 protein levels in patients with AML and healthy controls
The plasma level of nm23-H1 protein was significantly elevated in AML patients (n = 102; mean ± SD, 61.8 ± 34.6 ng/mL) compared with the healthy controls (n = 21, 6.1 ± 4.1 ng/mL;P = .001). We analyzed the relationship between the levels of plasma nm23-H1 protein and the FAB classification of AML patients, except for AML-M0 and AML-M7 because there were only 3 such cases. All of the FAB subtypes showed higher levels of plasma nm23-H1 than the healthy controls (Table 1). The plasma level in AML-M6 was especially high and was significantly higher than that in the other FAB subtypes of AML (P < .05). The plasma level of nm23-H1 in AML-M5 was higher, and that in AML-M3 was lower, than that in the other FAB subtypes, but these differences were not statistically significant (data not shown).
Correlation of nm23-H1 protein level with WBC and blast levels
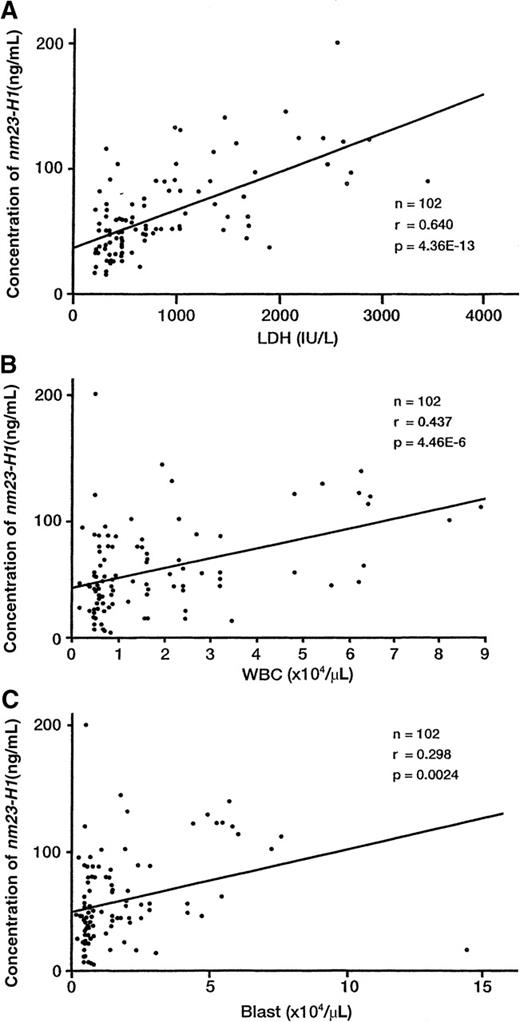
The plasma nm23-H1 level was weakly correlated with the WBC count (n = 102, r = 0.437, P < .00001, Figure1), the blast cell count in peripheral blood samples (n = 102, r = 0.298, P = .0024, Figure 1), and the plasma free-hemoglobin level (n = 102, r = 0.243,P = .0036). A significant positive correlation was also found between plasma nm23-H1 and lactic dehydrogenase (LDH) levels (n = 102, r = 0.640, P < .00001, Figure 1). In contrast, no relationship was observed between nm23-H1 and the platelet count (r = −0.08).
Correlation of the plasma nm23-H1 protein level with LDH, WBC count, and blast count.
(A) The plasma nm23-H1 level was correlated with the LDH level (n = 102, r = 0.640, P < .00001), (B) the WBC count (n = 102, r = 0.437, P < .00001), and (C) the blast count (n = 102, r = 0.298, P = .0024). Pearson's correlation coefficient (r) was used to evaluate the correlations between paired values.
Correlation of the plasma nm23-H1 protein level with LDH, WBC count, and blast count.
(A) The plasma nm23-H1 level was correlated with the LDH level (n = 102, r = 0.640, P < .00001), (B) the WBC count (n = 102, r = 0.437, P < .00001), and (C) the blast count (n = 102, r = 0.298, P = .0024). Pearson's correlation coefficient (r) was used to evaluate the correlations between paired values.
We previously reported that nm23 genes were overexpressed in AML cells and a higher level of nm23-H1 mRNA expression was correlated with a poor prognosis in AML. Therefore, we next examined the correlation of plasma nm23-H1 protein levels with nm23-H1 mRNA expression levels. We determined the plasma nm23-H1 protein levels of an additional 24 AML cases, for which we previously reported the nm23-H1 mRNA expression levels in AML cells by RT-PCR, and had stocked their plasma collected on the same day.12-14 There was no relationship between plasma nm23-H1 protein levels and intracellular nm23-H1 mRNA expression levels (r = 0.273, P = .197), as shown in Table 2. However, there was a significant correlation between the plasma nm23-H1 level and the product of the mRNA level and the WBC count (r = 0.712,P < .0005, Table 2). Therefore, the plasma nm23-H1 protein level probably depends on the total mass of leukemic cells overexpressing nm23-H1 gene.
Relationship between plasma nm23-H1 protein levels and clinicopathologic data in AML patients
Table 3 shows the clinicopathologic factors for the 102 AML patients. An increased WBC count was correlated with the nm23-H1 protein level (P = .0001). An increased LDH level was also correlated with the plasma nm23-H1 level (P = .0004); a good response to initial chemotherapy was inversely correlated with plasma nm23-H1 (P = .0001). All 102 patients were evaluable in terms of the response to treatment. Thirty-three patients failed to achieve remission after their initial chemotherapy. These patients with refractory AML had significantly higher plasma nm23-H1 protein levels (94.7 ± 36.1 ng/mL) than the 69 AML patients who achieved CR (40.1 ± 19.7 ng/mL). Because chromosomal aberrations are known to be the most useful prognostic indicators for AML, we separated patients into good [inv(16), t(8;21), and t(15;17)]: intermediate (normal karyotype and unknown), and poor (all others) cytogenetic groups. The plasma nm23-H1 protein level in the poor cytogenetic group was slightly higher than that in the other 2 groups, but this difference was not statistically significant (P = .1076, Table 3).
Level of nm23-H1 and survival
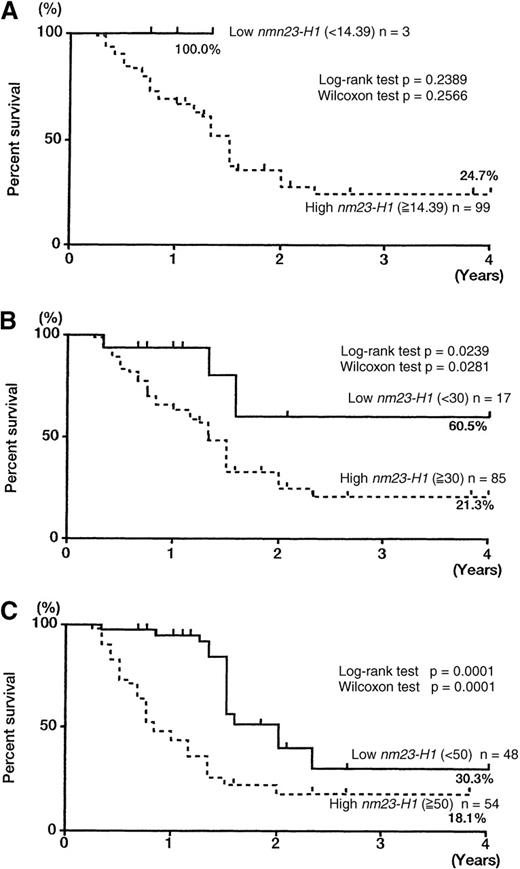
The 102 patients with AML were divided into 2 groups with different nm23-H1 levels to compare the overall survival of the 2 groups. First, we tried to set various cut-off points over the upper limit in control plasma (6.13 + 2SD = 14.39 ng/mL), as shown in Figure 2 and Figure3A: 14.39 ng/mL (< 14.39, n = 3 versus ≥ 14.39, n = 99), 30 ng/mL (< 30, n = 17 versus ≥ 30, n = 85), 50 ng/mL (< 50, n = 48 versus ≥ 50, n = 54), and 80 ng/mL (< 80, n = 73 versus ≥ 80, n = 29). We could not determine the statistical significance of the prognostic value using a cut-off level of 14.39 ng/mL because only 3 patients had levels lower than 14.39 ng/mL (Figure 2A). The cut-off levels of 30, 50, and 80 ng/mL showed significant prognostic value (Figures 2B-C and 3A). A level of 80 ng/mL was sufficient to severely affect survival and accurately identify intractable AML. All of the patients with levels higher than 80 ng/mL died within 2 years. Thus, we chose 80 ng/mL as a cut-off value for survival. The 2-year survival rates for the high nm23-H1 (≥ 80, n = 29) and low nm23-H1 (< 80, n = 73) groups were 0% and 33.3%, respectively (Figure 3A, P = .0001 for both the log-rank and generalized Wilcoxon tests).
Overall survival curves of patients with AML, according to different nm23-H1 levels.
The 102 patients with AML were divided into 2 groups with different nm23-H1 levels over the upper limit in control plasma (6.13 + 2SD = 14.39 ng/mL). (A) 14.39 ng/mL (< 14.39, n = 3 versus ≥ 14.39, n = 99). (B) 30 ng/mL (< 30, n = 17 versus ≥ 30, n = 85). (C) 50 ng/mL (< 50, n = 48 versus ≥ 50, n = 54).
Overall survival curves of patients with AML, according to different nm23-H1 levels.
The 102 patients with AML were divided into 2 groups with different nm23-H1 levels over the upper limit in control plasma (6.13 + 2SD = 14.39 ng/mL). (A) 14.39 ng/mL (< 14.39, n = 3 versus ≥ 14.39, n = 99). (B) 30 ng/mL (< 30, n = 17 versus ≥ 30, n = 85). (C) 50 ng/mL (< 50, n = 48 versus ≥ 50, n = 54).
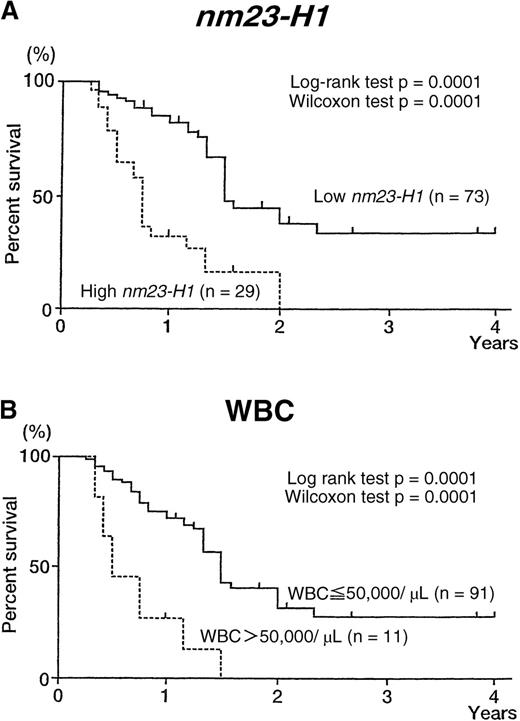
Overall survival curves of patients with AML, according to nm23-H1 level and WBC count.
(A) Patients with high nm23-H1 levels (≥ 80 ng/mL; n = 29) had a worse prognosis than those with low nm23-H1 levels (< 80 ng/mL; n = 73). (B) Patients with a high WBC count (> 50 000/μL; n = 11) had a worse prognosis than those with a low WBC count (≤ 50 000/μL; n = 91).
Overall survival curves of patients with AML, according to nm23-H1 level and WBC count.
(A) Patients with high nm23-H1 levels (≥ 80 ng/mL; n = 29) had a worse prognosis than those with low nm23-H1 levels (< 80 ng/mL; n = 73). (B) Patients with a high WBC count (> 50 000/μL; n = 11) had a worse prognosis than those with a low WBC count (≤ 50 000/μL; n = 91).
Prognostic factors for AML in univariate and multivariate analyses
The 102 evaluable AML patients were classified into groups based on age, gender, WBC count, LDH level, chromosomal aberrations, and plasma nm23-H1 protein level. The overall survival of each group is shown in Table 4. The univariate analysis showed that unfavorable prognostic factors for overall survival were WBC count over 50 000/μL (P = .0001), LDH level over 5 times normal (P = .0001), and nm23-H1 protein level over 80 ng/mL (P = .0001). Multivariate analysis (Table5) showed that plasma nm23-H1 level (≥ 80 ng/mL) was the strongest unfavorable factor (relative risk, 2.932;P = .0018), followed by WBC count (P = .0022) and LDH (P = .0333).
We believe that it is important to select a high-risk population at diagnosis to plan the best therapeutic strategy. Therefore, we tried to combine the 2 most important independent prognostic factors, plasma nm23-H1 level and WBC count (Table 5). As shown in Figure4A, the plasma nm23-H1 levels and WBC counts of all of the AML cases were plotted and divided into 4 groups (group A, only plasma nm23-H1 high; group B, both high; group C, both low; group D, only WBC high). A weak but statistically significant correlation was observed between the plasma nm23-H1 level and WBC count (Figure 1, n = 102, r = 0.437, P < .00001). The CR rate was 28.6% in group A (n = 21), 0% in group B (n = 8), 85.7% in group C (n = 70), and 100% in group D (n = 3). Analysis of survival probability between the groups showed that group B had significantly shorter survival than the other groups (Figure 4B). These data suggest that we can select a high-risk group such as group B by this combination.
Classification of 102 AML patients according to the 2 most important independent prognostic factors, plasma nm23-H1 level and WBC count.
(A) The plasma nm23-H1 levels and WBC counts of all of the AML cases were plotted and divided into 4 groups: group A, high nm23-H1 (≥ 80 ng/mL) and low WBC count (≤ 50 000/μ); group B, high nm23-H1 (≥ 80 ng/mL) and high WBC count (> 50 000/μL); group C, low nm23-H1 (< 80 ng/mL) and low WBC count (≤ 50 000/μL); and group D, low nm23-H1 (< 80 ng/mL) and high WBC count (> 50 000/μL). The CR ratio was 28.6% in group A (n = 21), 0% in group B(n = 8), 85.7% in group C (n = 70), and 100% in group D (n = 3). (B) Kaplan-Meier survival curves of groups A, B, C, and D.
Classification of 102 AML patients according to the 2 most important independent prognostic factors, plasma nm23-H1 level and WBC count.
(A) The plasma nm23-H1 levels and WBC counts of all of the AML cases were plotted and divided into 4 groups: group A, high nm23-H1 (≥ 80 ng/mL) and low WBC count (≤ 50 000/μ); group B, high nm23-H1 (≥ 80 ng/mL) and high WBC count (> 50 000/μL); group C, low nm23-H1 (< 80 ng/mL) and low WBC count (≤ 50 000/μL); and group D, low nm23-H1 (< 80 ng/mL) and high WBC count (> 50 000/μL). The CR ratio was 28.6% in group A (n = 21), 0% in group B(n = 8), 85.7% in group C (n = 70), and 100% in group D (n = 3). (B) Kaplan-Meier survival curves of groups A, B, C, and D.
Plasma nm23-H1 level in other hematologic malignancies
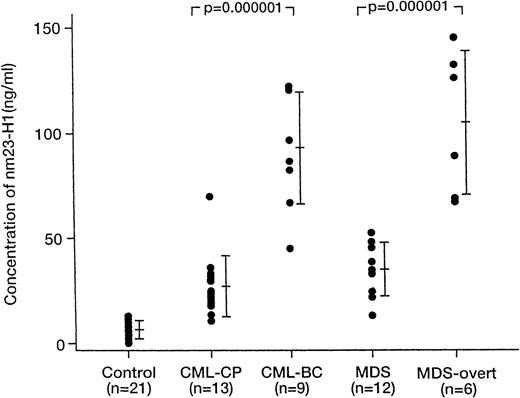
In an attempt to expand the prognostic use of this assay, we also determined the plasma levels of nm23-H1 protein in patients with various hematologic neoplasms other than AML (Table 1 and Figure5). The plasma nm23-H1 level was significantly elevated in ALL (n = 6; 65.7 ± 40.2 ng/mL;P = .0003), CML-CP (n = 13; 26.9 ±14.6 ng/mL;P = .0001), MDS (n = 12; 34.9 ± 12.9 ng/mL;P = .0001), MM (n = 12; 34.5 ± 19.6 ng/mL;P = .0001), and CLL (n = 4; 18.9 ± 4.1 ng/mL;P = .002) (Table 1). The level was increased to 99.0 ± 20.8 ng/mL (P = .000001) in CML-BC and to 104.7 ± 34.3 ng/mL in MDS with overt leukemia (P = .000001) (Figure 4).
Plasma nm23-H1 protein levels of patients with CML-CP, CML-BC, MDS, and MDS-overt leukemia.
The plasma nm23-H1 protein levels in CML-CP and MDS were increased in CML-BC and in MDS-overt leukemia, respectively. Mann-Whitney's U-test.
Plasma nm23-H1 protein levels of patients with CML-CP, CML-BC, MDS, and MDS-overt leukemia.
The plasma nm23-H1 protein levels in CML-CP and MDS were increased in CML-BC and in MDS-overt leukemia, respectively. Mann-Whitney's U-test.
Discussion
The nm23 gene was first identified as a gene that was expressed at lower levels by highly metastatic rodent tumors than by weakly metastatic tumors.5 Two closely related genes,nm23-H1 and nm23-H2, have been cloned.6,26The deduced amino acid sequences of the products of these genes were found to share a high level of homology with NDP kinase in a variety of species.7 The NDP kinases are ubiquitous enzymes that play a major role in the synthesis of nucleoside triphosphates other than adenosine triphosphate. Low nm23-H1 gene expression has been correlated with reduced overall survival or disease-free survival and with other histopathologic indicators of a high metastatic potential in cohorts of patients with breast, ovarian, cervical, gastric, and hepatocellular carcinoma, as well as melanoma. However, the opposite trend has been identified in patients with neuroblastoma and pancreatic carcinoma.4 We previously reported on the levels of nm23 mRNA expression in bone marrow and peripheral blood samples from 110 patients with newly diagnosed AML. The expression levels ofnm23-H1 and nm23-H2 genes in these AML samples were significantly higher than those in normal blood cells. The level ofnm23-H1 gene expression was significantly higher in the cells of patients with the M1, M2, M4, M5, and M6 subtypes of AML than in normal cells, whereas nm23-H2 gene expression was significantly higher in the cells of patients with the AML-M1, M4, M5, and M6 subtypes than in normal cells. In patients with the AML-M6 subtype, extremely high expression levels of both nm23 genes were observed, although only 2 cases were investigated.13,14 In the present study, all of the plasma nm23-H1 protein levels determined in patients with AML-M0 to AML-M7 were higher than those in normal controls (Table 1). Particularly, the level in AML-M6 was significantly higher than those in the M0, M1, M2, M3, M4, M5, and M7 subtypes (P < .05). On analysis using RT-PCR, we found no significant difference in nm23-H1 mRNA expression levels between AML-M3 patients and normal controls.13 However, the plasma nm23-H1 protein level was increased in AML-M3 patients (P = .0002). A significant correlation was found between the plasma nm23-H1 level and the product of its mRNA level and the WBC count or the blast count. Although we expected that the correlation coefficient between the plasma level of nm23-H1 protein with the product of mRNA of nm23-H1 and blast count would be higher than that with the product of mRNA and WBC count, there was no difference. Moreover, the plasma level of nm23-H1 protein was weakly correlated with the WBC count (r = 0.437,P < .00001) and with the blast count (r = 0.298,P = .0024) as shown in Figure 1. The plasma level of nm23-H1 protein seems to be more closely correlated with the WBC count than the blast count. We consider that WBC in AML might overexpress nm23-H1 mRNA, in addition to leukemic blast cells. There was also a positive correlation between the plasma nm23-H1 and LDH levels (Figure 1). Therefore, the plasma nm23-H1 protein levels probably depend on the total mass of leukemic cells overexpressing nm23-H1 gene.
We previously investigated the relative levels of nm23-H1 mRNA in AML cells by RT-PCR, and found that nm23-H1 mRNA levels predicted the prognosis of patients with AML.12-14 In these AML cases, we did not examine the relative levels of cellular nm23-H1 proteins. In cases in which we had enough bone marrow cells for protein analysis, we determined the nm23-H1 protein levels by Western blotting and immunohistochemistry. We could easily detect the proteins in AML cells, whereas we could not detect any evaluable protein levels in normal bone marrow cells under the same conditions (data not shown). The mRNA expression levels are roughly correlated with the cellular protein levels by these nonquantitative methods, but this has not been examined in all AML cases. We have some correlative data in leukemic cell lines. The relative levels of nm23-H1 mRNA in U937, HEL, and HL60 leukemic cells by RT-PCR were correlated with those of the cellular protein levels by ELISA (n = 3, r = 0.993, P = .037). These results suggest that cellular nm23-H1 protein levels may be correlated with mRNA expression levels in leukemia cells. In our present study, we compared the prognostic value of nm23-H1 gene transcripts by RT-PCR previously reported by us with that of the plasma nm23-H1 protein levels by a simple ELISA test. As shown in Table 2, there was no relationship between plasma nm23-H1 protein levels and intracellular nm23-H1 mRNA expression levels (perhaps intracellular nm23-H1 protein levels). It would be difficult to increase the concentration of nm23 protein in whole body plasma solely by the overexpression of intracellular nm23 in leukemic cells. This would require the overexpression of nm23-H1 gene by many cells, as shown by the significant correlation between the plasma nm23-H1 level and the product of the mRNA level and the WBC count. Therefore, we consider that the plasma nm23-H1 protein level is a prognostic factor that has a different meaning than intracellular nm23-H1 mRNA expression levels alone or the WBC count alone.
We have not yet determined the plasma nm23-H1 levels of patients with leukemoid reactions rather than malignancies. We had only 3 patients with leukemoid reactions. They had reactive lymphadenopathy with a WBC count of more than 30 000/μL. Their plasma nm23-H1 levels were 5, 5, and 7 ng/mL, respectively (data not shown), which are all within the normal range. We cannot draw any definite conclusions because there were only 3 cases. We consider that the increased plasma nm23-H1 level is an important marker that predicts a poor outcome in AML rather than a characteristic of leukemia.
An important challenge in the management of acute leukemia is to distinguish patients at high risk of relapse, who may benefit from high-dose chemotherapy followed by bone marrow transplantation, from patients in whom such treatments can be delayed. The WBC count, cytogenetic abnormalities, age, immunophenotypic characteristics, and the presence of Auer rods are accepted prognostic factors in AML.27-31 In the present study, the outcome of AML was poor in patients with high plasma nm23-H1 levels (Figures 2 and 3). Thus, elevation of the plasma nm23-H1 level can be used to predict a poor outcome. Because the overall survival rate of patients with a plasma nm23-H1 level of 80 ng/mL or more was lower than that of patients with an nm23-H1 level under 80 ng/mL, the former group had a higher risk of death. Multivariate analysis also showed that a high nm23-H1 level was a significant independent prognostic factor and the most potent prognostic factor among the variables studied, including WBC, LDH level, sex, and age. The combination of the 2 most important independent prognostic factors, plasma nm23-H1 level and WBC count, made it possible to select a high-risk population at diagnosis to plan the best therapeutic strategy (Figure 4). Recently, it has been reported that Wilms tumor gene (WT1) mRNA was expressed in most leukemic patients regardless of whether there was a specific marker gene such as AML1-MTG8 (ETO) or PML-RAR. The overall survival and disease-free survival periods are believed to be shorter when WT1 gene expression is high, and the WT1 gene expression level is considered to be a potential prognostic factor for acute leukemia.31 Because WT1 is hardly expressed in normal cells, it has been reported to be useful for detecting recurrence and for detecting very low tumor cell burdens. It might be important to compare the prognostic value of nm23 andWT1. The need for better prognostic factors is emphasized by the fact that certain consolidation regimens such as multiple cycles of high-dose cytarabine or bone marrow transplantation are highly toxic and are not beneficial for all patients. For these reasons, measurement of the plasma nm23-H1 level at diagnosis deserves further exploration with regard to evaluating the prognosis of AML.
Overexpression of the nm23-H1 gene has also been recognized in ALL, MDS, CML-BC and high-grade non-Hodgkin lymphoma. Therefore,nm23-H1 may be more widely useful as a prognostic factor for many neoplasms other than AML.13,14,32 In MDS, its expression increases before leukemic transformation. In CML-CP, neither nm23-H1 nor nm23-H2 gene expression was increased, but after acute transformation, the nm23 gene expression level increased and was comparable to that in AML.13 33 In this paper, we showed similar results using a determination of the plasma nm23-H1 level by ELISA (Figure 5). Consequently, it is possible that CML-BC or leukemic transformation of MDS may be detected early if the plasma nm23-H1 level is measured periodically in the course of CML or MDS.
On the other hand, the DR-nm23 gene has been isolated as a specific gene that is detected after acute transformation of CML.34DR-nm23 can inhibit the induction of differentiation, which is different from the action of I-factors. When 32Dc13 cells derived from the bone marrow of a normal mouse were transformed to express the DR-nm23 gene, the cells became resistant to differentiation induction by granulocyte colony-stimulating factor, and apoptosis was induced. Based on these findings, genes of the nm23 family, including DR-nm23, are considered to be useful for the diagnosis and treatment of CML-BC, as well as AML.
The present results suggest that determination of the plasma nm23-H1 protein level is a useful alternative to the determination of nm23-H1 mRNA by RT-PCR because it is simple and relatively quick, and a high level can be used to predict a poor outcome. In addition, because the nm23 level is closely related to the success rate for inducing remission by the initial chemotherapy, the development of a therapy that targets the nm23 gene may be desirable.
However, this analysis has limitations, including a short follow-up, retrospective establishment of cut-off points for biologic data, and a small number of patients examined. Therefore, we are now seeking to confirm these results in larger groups of patients.
Supported in part by a grant from the Ministry of Health and Welfare, and Grants-in-Aid for Scientific Research (C) and Cancer Research, from the Ministry of Education, Science, Sports and Culture, Japan.
Reprints:Junko Okabe-Kado, Saitama Cancer Center Research Institute, 818 Komuro, Ina, Kita-adachi, Saitama 362-0806, Japan; e-mail: jkado@cancer-c.pref.saitama.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.