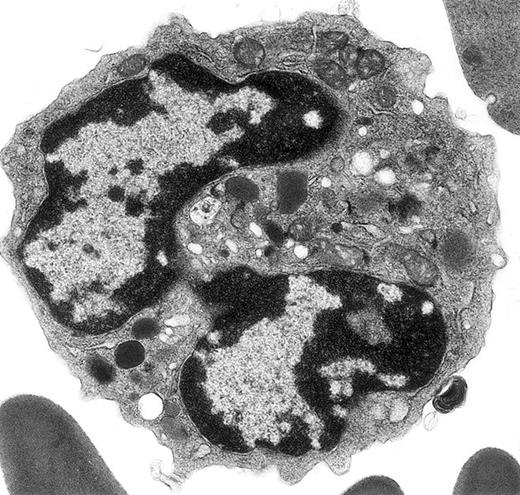
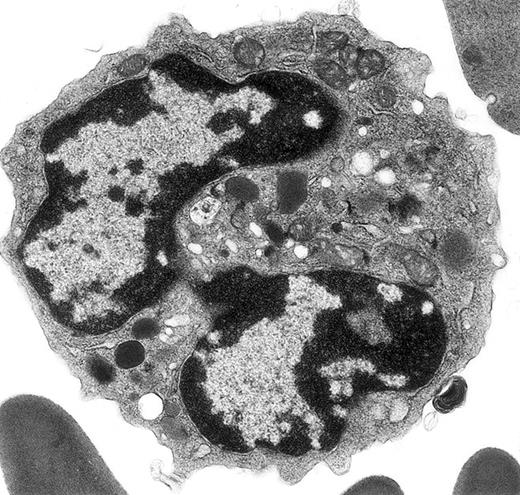
In a recent article,1 ultrastructural data were used to identify defects in terminal differentiation of neutrophils in the CCAAT enhancer binding protein family of transcriptional factors knock-out mice (C/EBPε −/−). Peripheral blood of C/EBPε −/− and control mice was obtained from the retro-ocular venous plexus and prepared for electron microscopy, whereas the remainder of the molecular, functional, and biochemical data in this report were obtained using bone marrow cells, peritoneal cells elicited with thioglycollate injections, or the cell line U937 stably transfected with a zinc inducible C/EBPε (p32) expression vector, or empty vector. The electron micrographs illustrated in figure 1A-B of Verbeek et al1 illustrate a classic mature mouse basophil, not an immature neutrophil as stated, in a C/EBPε −/− mouse. The granules shown are typical for histamine-containing mouse basophil granules,2-5 which are less numerous and larger than mature granules in mature mouse eosinophils and neutrophils2; basophils do not have secondary and tertiary granules, as do neutrophils, and the electron-lucent structures referred to as glutaraldehyde-extracted secondary granules are typical cytoplasmic vesicles and vacuoles, which are present in basophils from multiple species (see this letter's Figure).4-9 Figure 1C of Verbeek et al1 shows a classic neutrophil from a control mouse, as indicated in the legend. Whether or not the basophil illustrated in Verbeeek et al's figure 1A-B represents a real increase in this rare granulocyte class in C/EBPε −/− mice cannot be determined from the single cell presented here. Because much of the molecular, functional, and biochemical data for defective terminal neutrophil differentiation are based on studies of bone marrow and elicited peritoneal exudate cells in C/EBPε −/− mice, it would be important to do ultrastructural studies of these cell populations in these knock-out mice and their wild-type controls.
Electron micrograph shows a mature mouse basophil that was present in the FcεR+ cells sorted from normal mouse bone marrow.
(For more details, see Seder et al3and Dvorak et al.4) The mature nucleus has heavily condensed chromatin in 2 lobes. Small numbers of mature electron-dense granules are evident in the cytoplasm. Numerous mitochondria and electron-lucent secretory vesicles constitute the other cytoplasmic organelles in this view. Original magnification, × 23 000.
Electron micrograph shows a mature mouse basophil that was present in the FcεR+ cells sorted from normal mouse bone marrow.
(For more details, see Seder et al3and Dvorak et al.4) The mature nucleus has heavily condensed chromatin in 2 lobes. Small numbers of mature electron-dense granules are evident in the cytoplasm. Numerous mitochondria and electron-lucent secretory vesicles constitute the other cytoplasmic organelles in this view. Original magnification, × 23 000.
The mouse basophil lineage was defined by ultrastructural analysis, contrasted with developing mast cell, eosinophil, and neutrophil lineages, and reported in 1982 in this journal.2 These images clearly established the identification and evolution of these lineages in this species and provided the necessary background for the identification of mouse basophils as interleukin-4 (IL-4)–producing, high-affinity FcεR-positive, histamine-containing cells sorted from mouse bone marrow and spleen,3 previously referred to as non-B, non-T cells.10 The ultrastructural characteristics of these basophils are described and contrasted with those of FcεR-positive mast cells and with FcεR-negative neutrophils and eosinophils in detail.4 Further ultrastructural studies with this model revealed that mouse basophils are FcεR-positive and c-kit–negative, whereas mast cells are FcεR-positive and c-kit–positive, and that mouse basophils did not thrive in cultures supplemented with interleukin-3 (IL-3) or stem cell factor (SCF), on the one hand, or with a combination of these 2 mouse mast cell growth factors, on the other.5 In all of these references,2-5 ultrastructural images of mouse basophils identical to the basophil in Verbeek et al's figure 1A-B are included.