Abstract
The translocation of chromosome 11, long arm, region 2, band 1, to chromosome 18, long arm, region 2, band 1 (t(11;18)(q21;q21)) represents a recurrent chromosomal abnormality in extranodal marginal zone B-cell lymphoma (MZBCL) of mucosa-associated lymphoid tissue (MALT) type and leads to a fusion of the apoptosis inhibitor-2 (API2) gene on chromosome 11 and the MALT lymphoma-associated translocation (MLT) gene on chromosome 18. A 2-color fluorescence in situ hybridization (FISH) assay, which can be used for the detection of t(11;18) in interphase nuclei and metaphase chromosomes on fresh and archival tumor tissue, was developed. The P1 artificial chromosome (PAC) clone located immediately telomeric to the MLT gene and the PAC clone spanning the API2 gene were differentially labeled and used to visualize the derivative chromosome 11 resulting from t(11;18), as evident by the overlapping or juxtaposed red and green fluorescent signals. The assay was applied to interphase nuclei of 20 cases with nonmalignant conditions and 122 B-cell non-Hodgkin's lymphomas (NHLs). The latter group comprised 20 cases of nodal follicle center cell lymphoma and diffuse large B-cell NHL, 10 cases of gastric diffuse large B-cell lymphoma, 10 cases of hairy cell leukemia, and 82 cases of MZBCL (41 extranodal from various locations, 19 nodal, and 22 splenic MZBCL) including 35 cases with an abnormal karyotype, 2 of which revealed t(11;18). By interphase FISH, t(11;18) was detected in 8 gastrointestinal low-grade MALT-type lymphomas including the 2 cytogenetically t(11;18)+ cases. In the 8 t(11;18)+ cases, the FISH results were confirmed by reverse transcriptase–polymerase chain reaction (RT-PCR) usingAPI2 and MLT specific primers. Our results indicate that t(11;18)(q21;q21) specifically characterizes a subgroup of low-grade MZBCL of the MALT-type and that the FISH assay described here is a highly specific and rapid test for the detection of this translocation.
Introduction
Mucosa-associated lymphoid tissue (MALT)–type extranodal marginal zone B-cell lymphoma (MZBCL) represents a distinct subtype of B-cell non-Hodgkin's lymphoma (NHL), accounting for about 5% of all NHLs and the vast majority of lymphomas arising at extranodal sites.1,2 The clinical course of most extranodal MALT lymphomas is remarkably indolent. In contrast to other low-grade B-cell lymphomas, they tend to remain localized for a long period of time. In this condition, patients with MALT lymphomas display a favorable prognosis even with local treatment such as resection, radiotherapy, or eradication of Helicobacter pylori by antibiotic treatment (in the case of early-stage gastric MALT lymphomas). The cytogenetic analysis of MALT lymphomas is hampered by the low yield and poor quality of metaphase spreads. In spite of this, the translocation of chromosome 11, long arm, region 2, band 1, to chromosome 18, long arm, region 2, band 1 (t(11;18)(q21;q21)) has been shown to represent a recurrent chromosomal abnormality in these neoplasms.3-5 However, the prevalence of this translocation and the spectrum of disease characterized by it are not well defined.
We have recently shown that the relevant fusion resulting from the t(11;18) involves the derivative chromosome 11 and leads to the expression of a chimeric transcript fusing the 5′-end of apoptosis inhibitor-2 (API2), also known as cIAP2, HIAP1, and MIHC, on chromosome 11 to 3′-sequences of a noval gene, MALT lymphoma-associated translocation (MLT), located on chromosome 18.6 As a tool to rapidly detect t(11;18) in clinical samples, we have now developed a 2-color FISH assay using DNA probes specific for the API2 gene and a region immediately telomeric to the MLT gene. This assay detects theAPI2-MLT fusion both in metaphase spreads and interphase nuclei and can be applied to fresh and archival tumor tissue.
Material and methods
Preparation of the specimens
For the present study we used cells that were fixed with methanol–acetic acid and had been previously cultured and freshly prepared cytospin preparations of cells isolated by manual disaggregation from lymphoma-infiltrated snap-frozen tissue sections of 15- to 20-μm thickness. The cytospin preparations were fixed for 10 minutes in methanol–acetic acid; incubated for 5 minutes in a solution with 1% formaldehyde, 1 times phosphate-buffered saline (PBS), and 50 mmol/L magnesium dichloride (MgCl2) solution; shortly washed in 1 times PBS; dehydrated in a graded ethanol series; and then subjected to FISH analysis.
Fluorescence in situ hybridization
DNA labeling with biotin-dUTP (uridine 5′-triphosphate) (P1 artificial chromosome [PAC] 59N7) and digoxigenin-dUTP (PAC 166G16) (both from Roswell Park Memorial Institute Library, Buffalo, NY) by nick translation and FISH were performed according to standard methods as previously described.7 For the cytospin preparations, we dissolved 80 ng PAC 59N7, 50 ng PAC 166G16, and 11 μg Cot-1 DNA in 5 μL hybridization mixture and covered it with a round 13-mm coverslip. For the fixed-cell preparations, 140 ng PAC 59N7, 140 ng PAC 166G16, and 11 μg Cot-1 DNA were used.
In each case, we analyzed 300 well-preserved, separately located interphase cells with clearly visible distinct signals. The cells were considered t(11;18)+ when 1 green and 1 red signal were fused (overlapping signals) or colocalized (touching signals). Analyses were performed independently by 3 experienced investigators (J.D., M.S., and K.H.), and the respective results were averaged.
FISH probes
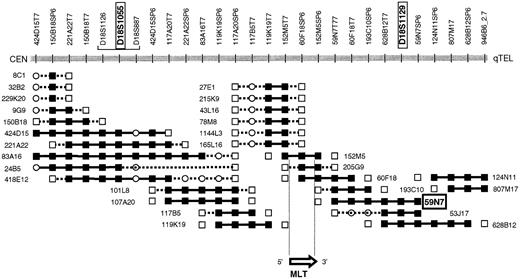
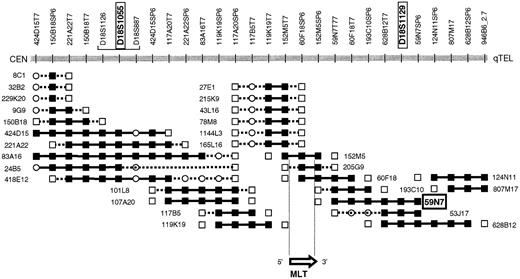
PAC 59N7 was identified by chromosome walking. The chromosome 18 breakpoint of t(11;18) could be located between D18S1055 and D18S1129.6 A walking strategy starting from both STSs yielded the PAC contig covering the chromosome 18 breakpoint region (Figure 1). The presence of the STS in the PACs was confirmed by polymerase chrome reaction (PCR), and each PAC was analyzed by FISH on normal metaphase spreads. PAC clone 166G16 (Roswell Memorial Park Institute Library), spanning theAPI2 gene, was selected by PCR-based screening.
PAC contig spanning the chromosome 18 breakpoint of t(11;18)(q21;q21).
PAC 57N9, which has been applied in the present interphase FISH assay, hybridizes telomerically to the breakpoint and is translocated to the derivative chromosome 11 resulting from t(11;18). PACs 205G9 and 152M5 span the breakpoint and the MLT gene. STS present, ▪; STS absent, ■; STS not tested, ○.
PAC contig spanning the chromosome 18 breakpoint of t(11;18)(q21;q21).
PAC 57N9, which has been applied in the present interphase FISH assay, hybridizes telomerically to the breakpoint and is translocated to the derivative chromosome 11 resulting from t(11;18). PACs 205G9 and 152M5 span the breakpoint and the MLT gene. STS present, ▪; STS absent, ■; STS not tested, ○.
Normal controls
For normal controls, we used interphase FISH with PACs 59N7 and 166G16 to analyze lymph node tissue from 20 patients with reactive lymphadenitis and splenic tissue from 2 patients with idiopathic thrombocytopenic purpura; 10 specimens were from fixed cells, and 10 cytospin preparations were obtained from frozen tissue. The cut-off levels were defined by the mean plus or minus 3 SD of the respective results in the experiments with the normal controls.8
Patient samples
FISH was performed in 122 cases of B-cell NHL including 10 cases of nodal follicle center cell lymphoma (grade 1 and 2), 10 cases of nodal diffuse large B-cell lymphoma, 10 cases of hairy cell leukemia (densely infiltrated spleen specimens), 10 cases of gastric diffuse large B-cell lymphoma, and 82 cases of MZBCL (41 extranodal cases from various locations, 19 nodal, and 22 splenic MZBCL) (Table1). For FISH analysis, we collected samples from the following cases, based on the availability of frozen tissue or fixed cells: 36 extranodal MZBCL cases including 6 t(11;18)+ cases, as shown by FISH, 17 nodal MZBCLs, 12 splenic MZBCLs, 10 gastric diffuse large B-cell lymphomas, and 10 hairy cell leukemias (University of Leuven, Leuven, Belgium); 4 extranodal MZBCL cases including 1 case involving the gut (which revealed t(11;18) by FISH), 2 nodal MZBCLs, and 10 splenic MZBCLs (the Centre Hospitalier Universitaire Henri Mondor, Créteil, France); 10 nodal follicle center cell lymphoma cases and 10 nodal diffuse large cell lymphomas (the University of Hamburg, Hamburg, Germany); and 1 extranodal MZBCL case of the stomach that carried t(11;18) (the University of Salamanca, Salamanca, Spain).
For FISH, fixed cells that were additionally cytogenetically analyzed were used in all cases, and cells obtained from the frozen sections were studied in the remaining cases (Table 1). Most of the cytogenetically analyzed MZBCLs have been included in previous studies.6,9,10 The diagnoses of the MZBCLs were established according to the Revised European-American Classification of Lymphoid Neoplasms.1 We documented 20 cases of nonmarginal zone B-cell NHL by abnormal karyotypes that did not reveal t(11;18). Of the 82 MZBCLs, 35 cases (6 extranodal, 18 nodal, and 11 splenic MZBCLs) revealed clonal karyotypic abnormalities including t(11;18), which occurred in 2 cases (Table 1).
Results
Selection of FISH probes
We previously showed that the relevant genetic lesion in NHL with t(11;18)(q21;q21) is the 5′-API2/3′-MLT fusion encoded on the derivative chromosome 11.6 The breakpoint within API2 occurred consistently in the intron separating exon 7 and exon 8 of API26 (M.B. and P.M., unpublished data, February 2000). In one case, t(11;18) was accompanied by a cryptic deletion involving the API2sequences distal to the breakpoint.6 The break in theMLT gene occurred in different introns6,11 (M.B. and P.M., unpublished data, February 2000). The molecular cytogenetic analysis of a MALT lymphoma with t(11;18) recently reported revealed a concurrent deletion of the 5′-end of the MLTgene.12
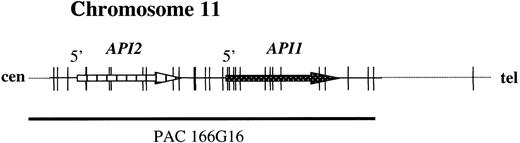
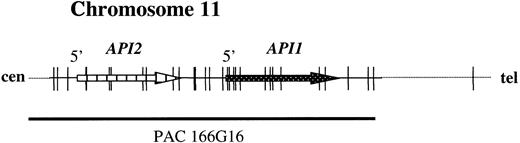
To develop a FISH assay that reliably detects theAPI2-MLT fusion, 2 new probes were selected. PAC 59N7 contains genomic sequences derived immediately downstream of theMLT gene (Figure 1). This probe is translocated to the derivative chromosome 11 resulting from t(11;18). PAC clone 166G16 spans approximately 100 kilobases (kb) and contains both the API2 gene and the highly homologous API1 gene, which resides 7 kb telomeric toAPI213 (Figure 2).
The structure of the
API2 gene and the adjacent API1gene on chromosome 11q22. The figure illustrates modifications from Young et al.13 PAC 166G16 spans both genes and is split by the translocation (Figure 3B). In the case of a deletion of chromosome 11 sequences distal to the breakpoint, the third hybridization signal on the derivative chromosome 18 may not be visible (Figure 3C).
The structure of the
API2 gene and the adjacent API1gene on chromosome 11q22. The figure illustrates modifications from Young et al.13 PAC 166G16 spans both genes and is split by the translocation (Figure 3B). In the case of a deletion of chromosome 11 sequences distal to the breakpoint, the third hybridization signal on the derivative chromosome 18 may not be visible (Figure 3C).
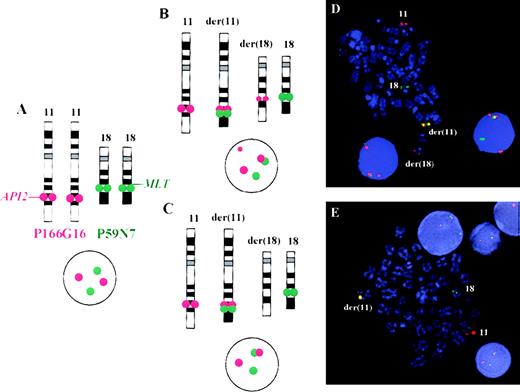
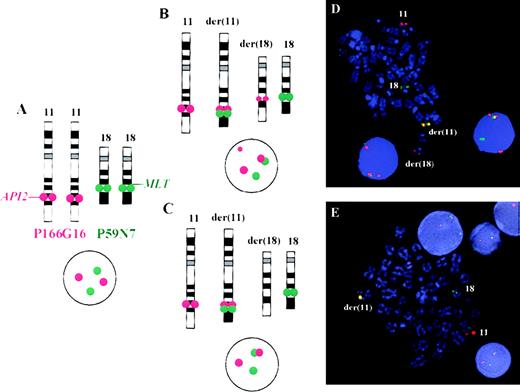
The disruption of the API2 gene by t(11;18) thus leads either to (1) a split hybridization signal of PAC 166G16, resulting in 3 hybridization signals located on the derivative chromosomes 11 and 18 and the normal chromosome 11 (Figure 3B) or (2) alternatively, in the case of a concomitant deletion of 11q sequences, the presence of 2 hybridization signals, 1 retained on the derivative chromosome 11 and the other located on the normal chromosome 11 (Figure 3C). In both instances, a fusion (overlapping) signal or colocalization (touching) signal of PACs 166G16 and 59N7 is clearly visible on the derivative chromosome 11 resulting from t(11;18). In our assay this was regarded as an indicator for the presence of the t(11;18)/API2-MLT fusion (Figure3).
Fluorescence in situ hybridization assay for the detection of t(11;18)(q21;q21) on metaphase chromosomes and interphase nuclei.
(A) On the left side, the normal localization of PACs 166G16 and 59N7 can be seen. As shown in panels B and C, t(11;18) leads to a fusion or colocalization signal of both probes on the derivative chromosome 11. (D) PAC 166G16 is split by the translocation leading to either 3 hybridization signals of this probe located on the derivative chromosomes 11 and 18 and the normal chromosome 11 (case 8) or, (E) in the case of a deletion of chromosome 11 sequences, to 2 signals of this probe located on the derivative chromosome 11 and the normal chromosome 11 (case 6).
Fluorescence in situ hybridization assay for the detection of t(11;18)(q21;q21) on metaphase chromosomes and interphase nuclei.
(A) On the left side, the normal localization of PACs 166G16 and 59N7 can be seen. As shown in panels B and C, t(11;18) leads to a fusion or colocalization signal of both probes on the derivative chromosome 11. (D) PAC 166G16 is split by the translocation leading to either 3 hybridization signals of this probe located on the derivative chromosomes 11 and 18 and the normal chromosome 11 (case 8) or, (E) in the case of a deletion of chromosome 11 sequences, to 2 signals of this probe located on the derivative chromosome 11 and the normal chromosome 11 (case 6).
Normal controls and cut-off values for diagnosing t(11;18)
The “cut-off values” for diagnosing t(11;18), which were established by analyzing the normal controls, were 4.1% for the fixed cell preparations and 4.4% for the cells obtained from the frozen tissue sections.
Detection of t(11;18) by interphase FISH in B-cell NHL
In the 122 B-cell NHL cases analyzed by FISH, t(11;18) was exclusively detected in 8 cases of low-grade extranodal MALT-type MZBCL (Table 1). The percentage of fusion and colocalization signals in the latter cases ranged from 19%-73% and for cases 1-8, the percentages were, respectively: 36%, 19%, 20%, 73%, 45%, 39%, 63%, and 52%. In 5 cases, 3 hybridization signals of PAC 166G16 were visible (cases 1, 3, 4, 7, and 8), whereas in 3 cases, 2 signals of this probe were observed in t(11;18)+ cells (cases 2, 5, and 6) (Figure 3). These results suggest that in the latter cases, a deletion of material of chromosome 11, as shown for case 6 in our previous paper,6 might have occurred. In all 8 t(11;18)+ cases, the FISH results were confirmed by reverse transcriptase–PCR (RT-PCR) using API2 and MLT specific primers.14 The cytogenetic and FISH data correlated well. The 2 cases (cases 6 and 8) cytogenetically characterized by t(11;18) were also t(11;18)+ by FISH, whereas none of the cytogenetically t(11;18)− cases was found to be positive by FISH. The clinical data of the 8 t(11;18)+ patients are summarized in Table 2.
Discussion
We describe a novel 2-color FISH assay using API2 and MLT specific probes for the detection of t(11;18)(q21;q21) and demonstrate, by applying this assay to a large number of B-cell NHL cases including nodal and splenic MZBCLs, that t(11;18) specifically characterizes a subgroup of low-grade extranodal MALT-type MZBCL cases. This assay detects the 5′-API2/3′-MLT fusion encoded on the derivative chromosome 11 produced by t(11;18). It can be used to rapidly and specifically detect this rearrangement on metaphase chromosomes or interphase nuclei obtained from fresh as well as archival tumor tissue including formalin-fixed paraffin-embedded tissue (data not shown). By analyzing a series of cases with nonmalignant conditions and B-cell lymphomas that did not show t(11;18) by karyotypic analysis, we demonstrated the high specificity of the FISH assay described here.
The t(11;18) FISH assay can detect 5 or more t(11;18)+ cells in 100 cells, which corresponds to the so called cut-off value obtained by analyzing a series of normal control samples. The cut-off value assessed for the t(11;18)-FISH assay is comparable with other interphase FISH assays detecting fusion signals.8 Extranodal low-grade MALT lymphomas differ in their clinical behavior and the way they are treated fundamentally from other low-grade NHLs. Therefore it is of particular importance to define genetic markers to characterize these lymphomas. Moreover, the detection of specific genetic abnormalities may be helpful in distinguishing inflammatory or regressive changes from lymphomatous tissue. The present study suggests that t(11;18) specifically characterizes a subgroup of low-grade extranodal MALT lymphomas and could thereby serve as a genetic determinant characterizing this disease entity.
Our findings are supported by the sparse cytogenetic data reported in the literature. Researchers have described t(11;18) in 21 of 58 low-grade MALT lymphomas with abnormal karyotypes,3but t(11;18) has not been described in other malignancies including high-grade gastric MALT lymphomas.5 Along the same lines, exclusively in a subgroup of extranodal low-grade MALT lymphomas, Rosenwald and coworkers15 detected t(11;18) by interphase FISH using 2 yeast artificial chromosome (YAC) clones flanking the chromosome 11 breakpoint. This t(11;18) occurred in all cases as a sole cytogenetic abnormality.3 Correspondingly, none of our t(11;18)+ cases revealed trisomy 3 or trisomy 18 by interphase FISH (data not shown).
Of particular interest is the absence of t(11;18) in nodal and splenic MZBCL and hairy cell leukemia, which all share, at least in part, morphologic and immunophenotypic features with extranodal MZBCL.1,9,16-18 Some nodal MZBCLs likely represent nodal manifestations of a concomitant unrecognized extranodal MALT lymphoma.17,18 Given this fact, one could hypothesize that either t(11;18)+ neoplasms do not tend to spread to distant lymph nodes or, alternatively, that t(11;18) evolves in cells that are not present or are not prone to acquire this translocation in nodal or splenic localizations. Similarly, in this present study, the absence of t(11;18) in large B-cell lymphomas of the stomach5 15suggests that t(11;18)+ MALT lymphomas do not usually transform to higher histological grades or that high-grade and t(11;18)+ low-grade gastric lymphomas are pathogenetically different.
The application of the interphase FISH assay described here will enable the prospective and retrospective investigation of large numbers of MALT lymphomas treated homogeneously within clinical studies and thereby help to further define the clinical and pathological characteristics of t(11;18)+ lymphomas.
Supported by grant 70-2175DiI (J.D.) from the Deutsche Krebshilfe/Dr Mildred Scheel Stiftung für Krebsforschung, Germany; the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming, Belgium; the Flemish government in the frame of the action Kom op tegen Kanker/Vlaamse Kanker Liga, Belgium; and grants G.0153.96 and G.0377.97 (P.M. and A.H.) from the Fonds voor Wetenschappelijk Onderzoek—Vlaanderen (F.W.O.). (P.M. is a research director and M.B. is a postdoctoral fellow of the F.W.O.)
Submitted March 3, 2000; accepted May 3, 2000.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter Marynen, Human Genome Laboratory, Center for Human Genetics and Flanders Interuniversity Institute for Biotechnology, Herestraat 49, B-3000 Leuven, Belgium; e-mail:Peter.Marynen@Med.KULeuven.ac.be.