Abstract
The AML1 gene, situated in 21q22, is often rearranged in acute leukemias through t(8;21) translocation, t(12;21) translocation, or less often t(3;21) translocation. Recently, point mutations in the Runt domain of the AML1 gene have also been reported in leukemia patients. Observations for mutations of the Runt domain of the AML1 gene in bone marrow cells were made in 300 patients, including 131 with acute myeloid leukemia (AML), 94 with myelodysplastic syndrome (MDS), 28 with blast crisis chronic myeloid leukemia (CML), 3 with atypical CML, 41 with acute lymphoblastic leukemia (ALL), and 3 with essential thrombocythemia (ET). Forty-one of the patients had chromosome 21 abnormalities, including t(8;21) in 6 of the patients with AML, t(12;21) in 8 patients with ALL, acquired trisomy 21 in 17 patients, tetrasomy 21 in 7 patients, and constitutional trisomy 21 (Down syndrome) in 3 patients. A point mutation was found in 14 cases (4.7%), including 9 (22%) of the 41 patients with AML of the Mo type (MoAML) (none of them had detectable chromosome 21 rearrangement) and 5 (38%) of the 13 myeloid malignancies with acquired trisomy 21 (1 M1AML, 2 M2AML, 1 ET, and 1 atypical CML). In at least 8 of 9 mutated cases of MoAML, both AML alleles were mutated: 3 patients had different stop codon mutations of the 2 AML1 alleles, and 5 patients had the same missense or stop codon mutation in both AML1 alleles, which resulted in at least 3 of the patients having duplication of the mutated allele and deletion of the normal residual allele, as shown by FISH analysis and by comparing microsatellite analyses of several chromosome 21 markers on diagnosis and remission samples. In the remaining mutated cases, with acquired trisomy 21, a missense mutation of AML1, which involved 2 of the 3 copies of the AML1 gene, was found. Four of the 7 mutated cases could be reanalyzed in complete remission, and no AML1 mutation was found, showing that mutations were acquired in the leukemic clone. In conclusion, these findings confirm the possibility of mutations of the Runt domain of the AML1 gene in leukemias, mainly in MoAML and in myeloid malignancies with acquired trisomy 21. AML1 mutations, in MoAML, involved both alleles and probably lead to nonfunctional AML1 protein. As AML1 protein regulates the expression of the myeloperoxidase gene, the relationship between AML1 mutations and Mo phenotype in AML will have to be further explored.
Introduction
The AML1/CBFA2 gene belongs to the Runt domain gene family that encodes the major subunit, α, of heterodimeric transcription factor PEBP2/CBF.1 The Runt domain, named after the Drosophila segmentation gene,runt, is an evolutionarily conserved protein motif that consists of 128 amino acids (AAs) and is responsible for both DNA binding and heterodimerization with the non–DNA-binding regulatory subunit, β.2 AML1 is normally expressed in all hematopoietic lineages and acts to regulate the expression of various genes specific to hematopoiesis, including the granulocyte colony-stimulating factor receptor, interleukin 3, T-cell receptor β, and myeloperoxidase (MPO) genes.3Furthermore, mice lacking either AML1 or PEBP2β/CBFB have no fetal liver hematopoiesis,4-6 showing that PEBP2/CBF is essential for definitive hematopoiesis of all lineages.
The AML1 gene is one of the genes most frequently deregulated in leukemias,7,8 generally through translocations that produce chimeric messenger RNA and proteins: AML1-ETO in t(8;21) translocation; AML1-ETV6 in t(12;21) translocation; and less often AML1-EVI1, AML1-MDS1, and AML1-EAP in t(3;21) translocation. The chimeric AML1-ETO and AML1-ETV1 proteins contain the entire Runt domain, but the transactivation domain region of the AML1 gene is largely deleted or replaced by foreign proteins. Both chimeric proteins appear to interfere with transactivation by the normal AML1 in a transdominant manner.9-11
Recently, Osato et al12 reported another mechanism of AML1 gene inactivation in acute myeloid leukemia (AML), by point mutation of the Runt domain. In this study, we confirmed the presence of point mutations in the Runt domain of the AML1 gene in a small number of leukemic patients, but in a relatively high proportion of AML of the Mo type (MoAML) and to a lesser extent of myeloid malignancies with acquired trisomy 21.
Patients, materials, and methods
Analysis of point mutations of the Runt domain of the AML1 gene was made in 25 healthy subjects (bone marrow donors) and in 300 leukemia patients, including 123 cases of AML (41 cases of MoAML, 19 cases of M1 AML, 29 cases of M2 AML including 6 with t(8;21), 1 case of M3 AML, 14 cases of M4 AML including 5 cases of M4eo, 17 cases of M5 AML, 1 case of M6 AML, and 1 case of M7 AML); 20 cases of pre-B–acute lymphoblastic leukemia (ALL) without chromosome 21 abnormalities; 94 cases of myelodysplastic syndrome (MDS; 13 refractory anemia, 4 refractory anemia with ringed sideroblasts, 23 refractory anemia with excess of blast, 27 refractory anemia with excess of blast in transformation, and 27 chronic myelomonocytic leukemia); 28 chronic myeloid leukemia (CML) in blast crisis, including 22 myeloid and 6 lymphoid blast crises. Because the AML1 gene is located on chromosome 21, we studied 35 additional cases of hematologic malignancies with chromosome 21 abnormalities, including 17 cases of acquired trisomy 21 (6 M2 AML, 3 essential thrombocythemia [ET], 3 atypical CML, and 5 ALL), 6 ALL with tetrasomy 21, 1 M1 AML with tetrasomy 21, 8 ALL with t(12;21), and 3 patients (2 ALL and 1 AML) with constitutional trisomy 21 (Down syndrome). Diagnosis of acute leukemia and MDS was based on morphologic and immunophenotypic analysis according to French-American-British (FAB) criteria.13 A high number of MoAML (41 cases) were included in the study because Osato et al12 and our preliminary findings had shown a high incidence of mutations in MoAML. Diagnosis of MoAML was based on FAB proposals14: cytochemical reaction of MPO with benzidine base positive in less than 3% blasts, negative lymphoid markers except either CD7 or CD4, and at least one of the following myeloid markers: CD13, CD33, and MPO antigen. All patients had given informed consent.
Cytogenetic analysis
Conventional cytogenetic analysis was performed on bone marrow cells, after short-term (24 hours) culture without stimulation. Chromosomes were identified by RHG and GTG banding and classified according to the International System for Human Cytogenetic Nomenclature.15 t(12;21) Translocation was detected by using two different techniques: reverse transcriptase–polymerase chain reaction (RT-PCR) and fluorescent in situ hybridization (FISH).16 17 The FISH technique was performed on metaphase spreads with AML1 and ETV6 dual-color probes (Vysis, Woodcreek Drive, IL).
Analysis of AML1 mutations
Detection of AML1 gene mutations in the Runt domain was made on DNA extracted from bone marrow cells, by single-stranded conformation polymorphism (SSCP) analysis18,19 of exons 3 to 5 of the AML1 gene (Table 1). PCR was performed in a total reaction volume of 20 μL that contained 50 ng DNA, 2.5 mmol/L MgCl2, 0.3 μmol/L of each primer (primer names and sequences in Table 1), 10 mmol/L Tris HCl, 50 mmol/L KCl, 200 μmol/L of each dNTP (Pharmacia, Stockholm, Sweden), 1 U of Taq DNA polymerase (Promega, Charbonnières, France), and 1μCi32P-labeled deoxycytidine trisphosphate. Samples were heated for 5 minutes at 94°C, then they underwent 34 cycles with 1 minute at 94°C, 1 minute at 58°C, 1 minute at 72°C, followed by final elongation of 7 minutes at 72°C. After amplification, PCR products were loaded on a 2% agarose gel stained with ethidium bromide. The size of the PCR products was 312 base pairs (bp), 249 bp, and 246 bp for exon 3 to 5, respectively. For SSCP analysis, 4 μL of PCR product was diluted in 16 μL of a solution that contained 0.1% SDS, 20 mmol/L EDTA; 3 μL of this mixture was mixed with 3 μL of a solution of 95% formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol, then heated at 95°C during 2 minutes and cooled in ice. Finally, 3 μL was loaded on MDE gel (Tebu, Le Perray en Yvelines, France) containing 5% glycerol. Electrophoresis was performed at room temperature for 12 hours.
DNA sequencing of the AML1 gene
DNA sequencing was performed directly on new PCR products obtained as for SSCP analysis but without 32P αdCTP and was subjected to cycle sequencing (Perkin Elmer, Forster City, CA). In patients 1, 2, 9, and 14, the PCR products were subcloned into plasmid PCR II. The sequence of each identified mutation was confirmed by using sense and antisense primers and was reconfirmed on another independent PCR product.
Analysis of loss of heterozygosity (LOH) for AML1 gene
FISH analysis.
To determine if patients with AML1 mutation had loss of one AML1 allele, we performed FISH analysis with the dual-color AML1-ETV6 probe (Vysis) on interphase and/or metaphase cells. The size of the AML1 probe that contained the whole genomic sequence of the AML1 gene was 500 kilobase (kb); the ETV6 probe was used as internal control hybridization. Dual-color probes were directly labeled by spectrum green and spectrum orange. Probes were hybridized according to the conditions of the manufacturer. After hybridization, DNA was counterstained with DAPI (0.2 μg/mL). We also performed a second hybridization with the cCMP21 probe (a kind gift by Ferguson-Smith, Cambridge, UK).20 The size of this probe is 55 kb, and it is located at the D21S19 locus (21q22.3 region). For the 2 probes, fluorescence signals were visualized on a Zeiss axioskope epifluorescence microscope (Zeiss, Oberkerchen, Germany) equipped with a triple bandpass filter. Fluorescence images were optimized by using the quips smart capture FISH Imaging software (Vysis). For each slide, 500 nuclei were analyzed.
PCR analysis of chromosome 21 DNA polymorphic markers.
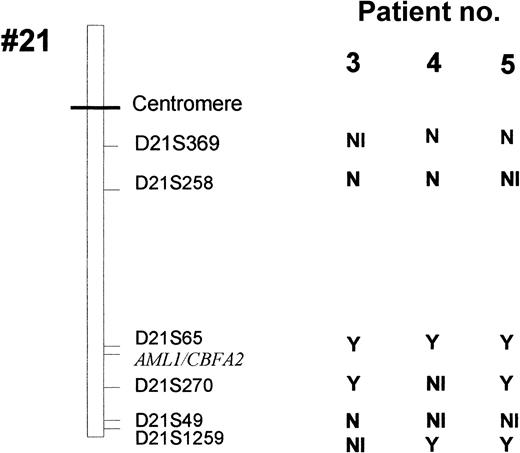
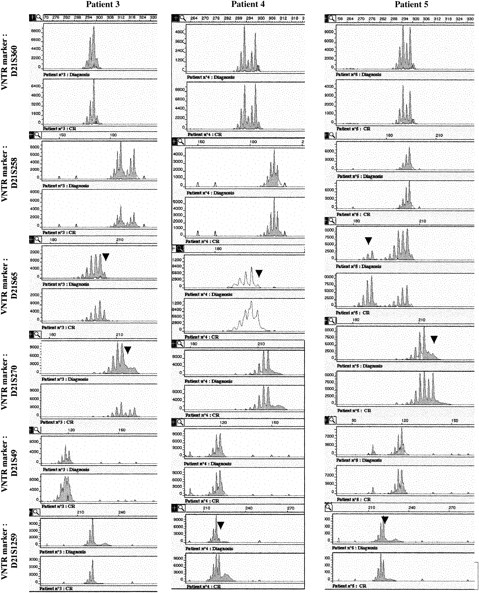
Six highly polymorphic markers (heterozygotes in more than 70% of subjects) localized near the AML1 gene (D21S65 and D21S270) or more distant from the AML1 locus (D21S369, D21S258, D21S49, and D21S1259) were studied (Figure 1). PCR analysis was performed on a total reaction volume of 50 μL containing 50 ng DNA, 2.5 mmol/L MgCl2, 200 μmol/L of each dNTP, 50 mmol/L Tris HCl, 10 mmol/L KCl, 1 U of Taq DNA polymerase (Promega), and 0.4 μmol/L of each primer (see Table 1).
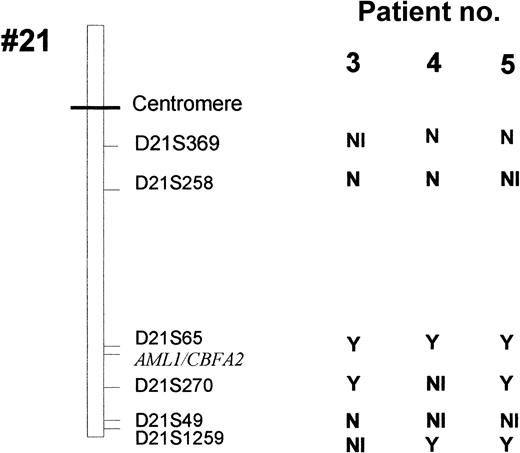
LOH for chromosome 21 polymorphic markers in patients 3, 4, and 5.
Y indicates a loss of heterozygosity; N, no loss of heterozygosity; NI, not informative.
LOH for chromosome 21 polymorphic markers in patients 3, 4, and 5.
Y indicates a loss of heterozygosity; N, no loss of heterozygosity; NI, not informative.
The forward primers were 5′ labeled with FITC. Samples were heated 3 minutes at 94°C, then underwent 30 cycles with heating at 94°C for 1 minute, at 58°C for 1 minute, and at 72°C for 1 minute, followed by final elongation at 72°C for 3 minutes. PCR products were mixed with a formamide gel loading solution (Perkin Elmer, Forster City, CA), denatured at 94°C, ice cooled, and loaded on denaturing 8% polyacrylamide gel (Perkin Elmer). Gel electrophoresis was performed on an ABI 377 (Perkin Elmer), and allele losses were determined by using GENESCAN software (Perkin Elmer).
Results
In the 25 healthy subjects, no point mutation or polymorphism of the Runt domain of the AML1 gene was detected.
In the 300 patients, no abnormal SSCP profile was detected in patients with MDS, blast crisis CML, and ALL. Fourteen patients showed an abnormal SSCP profile (Figure 2), including 9 Mo AML (patients 1-9), 1 M1 AML (patient 10), 2 M2 AML (patients 11 and 14), 1 patient with ET (patient 12), and 1 patient with atypical CML (patient 13; Table 2). The last 5 patients had acquired trisomy 21 (patients 11-14) or tetrasomy 21 (patient 10), whereas no chromosome 21 rearrangement was detected cytogenetically in the 9 MoAML cases with abnormal SSCP findings.
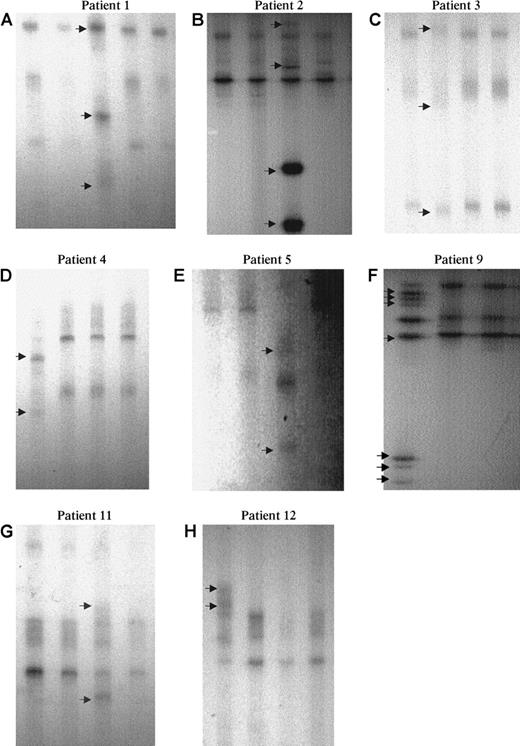
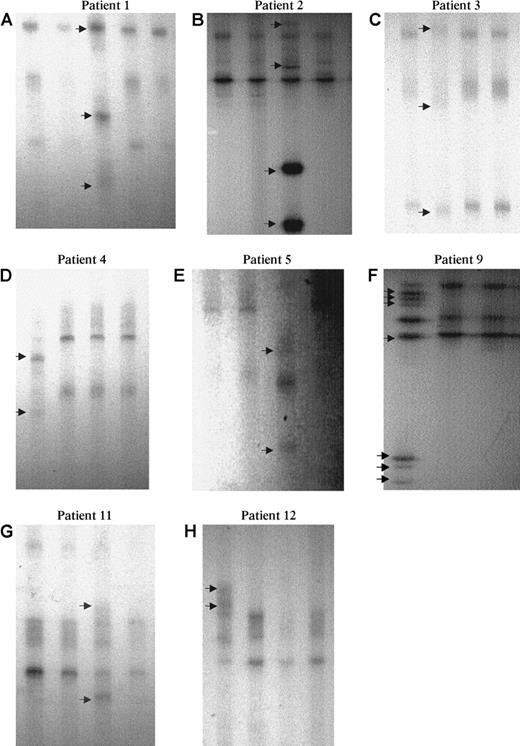
SSCP analysis of AML1 point mutations.
Eight examples (6 MoAML and 2 cases with inquired trisomy 21) showed an abnormal profile (arrow): A lane 3, B lane 3, C lane 2, D lane 1, E lane 3, F lane 1, G lane 3, and H lane 1. Normal residual bands were absent or very faint in patients 1 to 5 and 9.
SSCP analysis of AML1 point mutations.
Eight examples (6 MoAML and 2 cases with inquired trisomy 21) showed an abnormal profile (arrow): A lane 3, B lane 3, C lane 2, D lane 1, E lane 3, F lane 1, G lane 3, and H lane 1. Normal residual bands were absent or very faint in patients 1 to 5 and 9.
In patient 1, 2 different mutations in exon 4, consisting of a 14-bp deletion, including AAs 132 to 135, the last base of the AA 131, and the first base of AA 136 on one allele and 22-bp insertion on the other allele were detected. The 22-bp insertion corresponded to a tandem duplication of AAs 115 to 121 and the last base of the AA 114. Both mutations induced a stop codon. Patient 2 had duplication of 6 bp on one allele at AA 140, corresponding to an insertion of Ser-Arg, and a large insertion of 14 bp on the second allele, corresponding to a noncoding sequence of chromosome 21 located at the splicing site that started with the first base of intron 4. Patients 3 to 5 had a missense mutation: Gly 138 to Asp (G138D), Arg 135 to Gly (R135G), and Asp171 to Gly (D171G), respectively; patients 6 and 7 had a stop codon mutation (Arg 139 to stop; R139 ter). Patient 8 had a mutation at the splicing site between intron 3 and exon 4. This mutation was described by Song et al21 and induces a stop codon. Finally, patient 9 had a missense mutation Arg 174 to Gln on one allele and a large insertion (23 bp) at AA 135, inducing a stop codon on the second AML1 allele. This insertion corresponded to a part of intron 4 of AML1.
Thus, patients 1, 2, and 9 had 2 different mutations of the 2 AML1 alleles. In patients 3, 4, 5, 6, and 8, an alteration of the second AML1 allele was suspected on the SSCP profile (Figure 2). Patients 3, 4, 5, and 8, with more than 90% marrow blast cells, had no normal residual band, and, in patient 6, the intensity of the abnormal band was much greater than that of the normal band (this patient had 75% marrow blast cells).The alteration of the second allele was confirmed on the sequence profile. In patients 3, 4, 5, and 8, the peak corresponding to the normal allele was absent or very faint. The same results were observed with the forward and reverse primers on direct sequence (data not shown). In patient 6, the peak corresponding to the normal allele was present but was smaller than the mutated allele, also confirming the biallelic alteration of AML1. In patient 7, no conclusion could be drawn because the sequence peaks corresponding to the normal and mutated allele were of similar size in this patient with near tetraploid karyotype and 3 copies of chromosome 21.
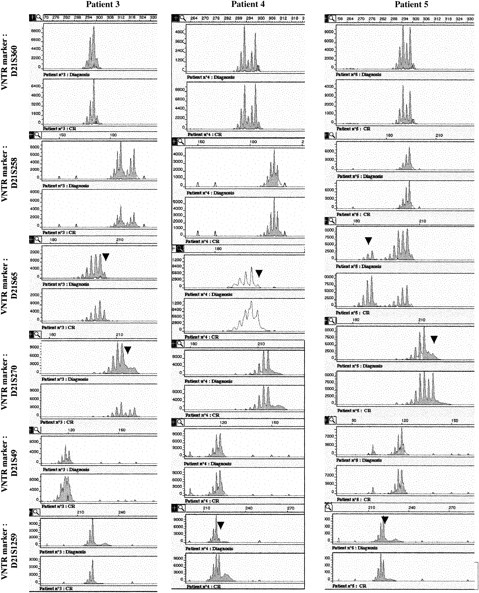
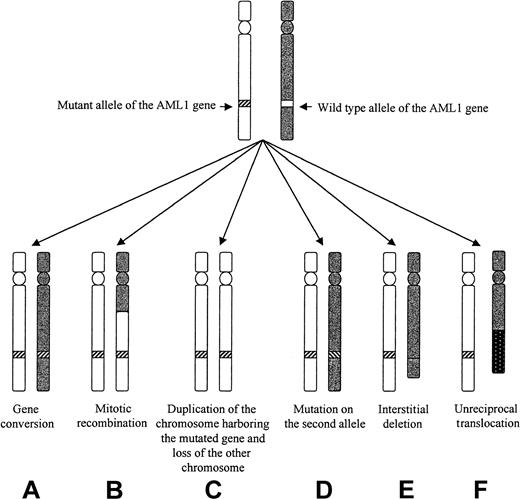
Leukemic cells from patients 3 to 6 (but not patients 7 and 8) could be analyzed by FISH with the dual-color AML1-ETV6 probe and the cCMP21 probe situated at 21q22.3. All 4 cases had 2 signals with the cCMP21 probe and 2 copies of the AML1 gene, ruling out a single deletion of the nonmutated AML1 allele (Figure 3). This finding suggested either the same mutation on the 2 AML1 alleles or the total or partial duplication of the mutated allele with deletion of the normal residual AML1 allele. Three of those patients (3-5) could be reanalysed in complete remission with 6 chromosome 21 DNA polymorphic markers localized near the AML1 gene (D21S65 and D21S270) or more distant from the AML1 gene (D21S369, D21S49, D21S258, and D21S1259; Figure 1). Patients 3 and 5 were heterozygous for the 2 markers close to the AML1 locus (D21S65 and D21S270), and patient 4 was heterozygous for only one of them (D21S65). As seen in Figure4, all 3 patients showed LOH for those markers at diagnosis, by comparison to samples obtained in complete remission. These findings showed that leukemic samples of those patients had duplication of the mutated AML1 allele and deletion of the normal residual allele rather than the same mutation on both AML1 alleles. Furthermore, patients 3, 4, and 5 were heterozygous for one or several of the following markers: D21S369, D21S258 localized near the centromere, and D21S49 and D21S1259 localized at the extremity of 21q. No LOH was found for the centromeric markers. LOH was found for the telomeric markers in patients 4 and 5, but not in patient 3. These findings suggested that duplication of the mutated AML1 resulted from duplication of a short chromosome segment (gene conversion) in patient 3 and from a larger chromosome duplication, including most of 21q with its telomeric region (mitotic recombination), in patients 4 and 5 (Figures 1 and 5).
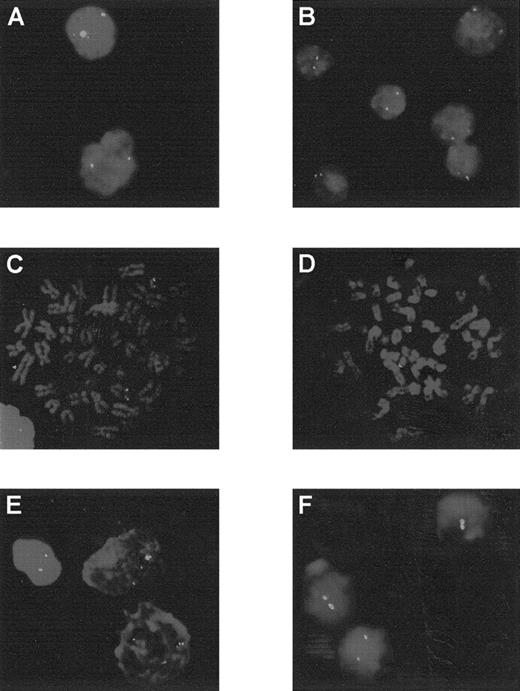
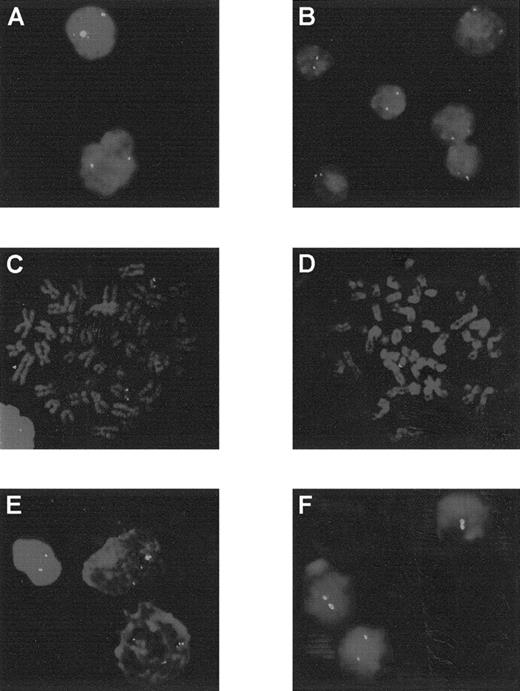
Analysis by FISH using the cCMP21 probe.
Patients 3 to 6 and 12 are at diagnosis (A to E) and patient 12 in AML evolution (F). Patients 3 to 6 showed 2 green spots, corresponding to 2 copies of chromosome 21 in each cell: interphase cells for patients 3 and 4 (A and B) or metaphase cells for patients 5 and 6 (C and D). At diagnosis, patient 12 presented 3 spots in each cells (E) and 2 spots in AML evolution (F).
Analysis by FISH using the cCMP21 probe.
Patients 3 to 6 and 12 are at diagnosis (A to E) and patient 12 in AML evolution (F). Patients 3 to 6 showed 2 green spots, corresponding to 2 copies of chromosome 21 in each cell: interphase cells for patients 3 and 4 (A and B) or metaphase cells for patients 5 and 6 (C and D). At diagnosis, patient 12 presented 3 spots in each cells (E) and 2 spots in AML evolution (F).
Analysis of polymorphic markers.
Six polymorphic markers (D21S396, D21S258, D21S65, D21S270, D21S49, and D21S1259) are located in 21q11.1, 21q22.11, 21q22.1-q22.2, 21q22.2, 21q22.3, and 21q22.3, respectively. The heterozygosity values are 0.67, 0.76, 0.80, 0.86, 0.72, and > 0.7, respectively. Microsatellite analyses were analyzed in patients 3 to 5 at the time of diagnosis and at complete remission (CR). At diagnosis, LOH was observed in patient 3 for markers D21S65 and D21S270, markers D21S258 and D21S49 were heterozygote, and markers D21S1259 and D21S369 were not informative. In patient 4, only markers D21S369 and D21S258 were heterozygote (markers D21S49 and D21S270 were not informative), and LOH was seen for markers D21S65 and D21S1259. In patient 5, LOH was observed for all the studied markers except D21S369, and D21S258 and D21S49 were informative. Analysis of length of fluorescent fragments was performed on ABI-PRISM 377 P.E. Arrows show the LOH at diagnosis.
Analysis of polymorphic markers.
Six polymorphic markers (D21S396, D21S258, D21S65, D21S270, D21S49, and D21S1259) are located in 21q11.1, 21q22.11, 21q22.1-q22.2, 21q22.2, 21q22.3, and 21q22.3, respectively. The heterozygosity values are 0.67, 0.76, 0.80, 0.86, 0.72, and > 0.7, respectively. Microsatellite analyses were analyzed in patients 3 to 5 at the time of diagnosis and at complete remission (CR). At diagnosis, LOH was observed in patient 3 for markers D21S65 and D21S270, markers D21S258 and D21S49 were heterozygote, and markers D21S1259 and D21S369 were not informative. In patient 4, only markers D21S369 and D21S258 were heterozygote (markers D21S49 and D21S270 were not informative), and LOH was seen for markers D21S65 and D21S1259. In patient 5, LOH was observed for all the studied markers except D21S369, and D21S258 and D21S49 were informative. Analysis of length of fluorescent fragments was performed on ABI-PRISM 377 P.E. Arrows show the LOH at diagnosis.
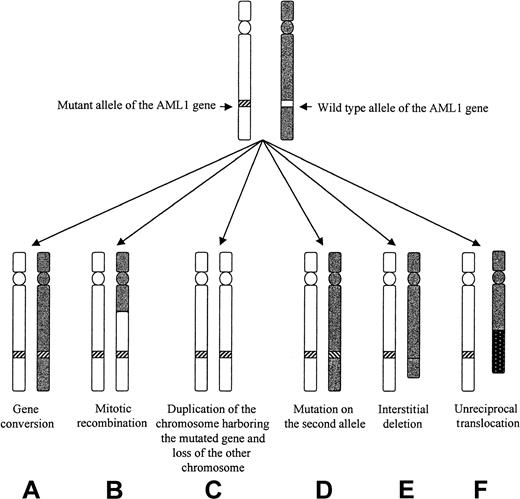
Different mechanisms that could lead to LOH of the AML1 gene in patients 3 to 5.
Mechanism C, D, E, and F could be ruled out in the 3 patients. Mechanism A seemed to apply to patient 3, and mechanism B applied to patients 3 and 5.
Different mechanisms that could lead to LOH of the AML1 gene in patients 3 to 5.
Mechanism C, D, E, and F could be ruled out in the 3 patients. Mechanism A seemed to apply to patient 3, and mechanism B applied to patients 3 and 5.
The 5 remaining patients with mutations (10-14) had M1 AML (patient 10), M2 AML (patients 11 and 14), ET (patient 12), and atypical CML (patient 13), respectively. Three of them (patients 10, 11, and 12) had a missense mutation: Asp 171 to Gly (D171G), Arg 177 to Gln (R177Q), and Arg 174 to Gln (R174Q). Patient 13 had a stop codon mutation Arg 139 to stop (R139 ter). Finally, patient 14 had, on the same allele, a deletion of 4 bp followed by an insertion of 76 bp, corresponding to a part of intron 4. This insertion induced a stop codon. Interestingly, those 5 patients had acquired trisomy 21 (patients 11-14) or tetrasomy 21 (patient 10). Analysis of the profiles of direct sequencing confirmed that patients 11 to 14 had 2 mutated alleles and 1 normal allele (data not shown). In patient 10, the normal band was very faint, but it could not be concluded that this patient (with 33% blasts in the sample studied) had loss of all normal alleles.
Patient 12, with an initial diagnosis an ET, progressed to MPO-negative AML with a blast cell immunophenotype corresponding to MoAML: CD13+, CD33+. During the AML phase, the karyotype was complex and showed only 2 chromosomes 21. The SSCP profile and direct sequence confirmed the presence of the mutation R174Q observed during the ET phase but with loss of the normal allele. FISH analysis confirmed trisomy 21 during the ET phase and the presence of only 2 chromosomes 21 after progression to AML (Figure 3).
Discussion
This is the second study that shows nontranslocation generated acquired mutations of the AML1 gene in patients with leukemias. In the first study, Osato et al12 described 8 mutations in 160 leukemia patients (6 with AML, 1 with blast crisis CML, and 1 with ALL). Although the whole AML1 coding region was studied, all 8 reported mutations that involved the Runt domain of the gene. Two of the mutations were silent (in 1 MoAML and in the ALL case), 4 were heterozygous missense mutations (in 1 M3 AML, 1 M5a AML, 1 M4 AML, and 1 CML blastic phase), and 2 were biallelic nonsense frameshift mutations (2 MoAML). Heterozygous missense mutations lead to partial defects in AML1 function (with impaired DNA binding and transactivation, but intact heterodimerization properties), whereas biallelic nonsense mutants encoded truncated AML1 proteins that had lost almost all functions. Mutations were particularly frequent in MoAML (3 of the 8 cases of AML studied).
By restricting our study to the Runt domain of the AML1 gene, we found a rather similar incidence of mutations (14 in 300 patients). We confirmed the high incidence of AML1 mutation in MoAML (9 of 41 cases, as compared with 1 of 85 in the other FAB subtypes of AML). At least 8 of the 9 mutations that occurred in our MoAML were biallelic, as 2 of the 3 mutated MoAML cases reported by Osato et al.12 By contrast, as in the report of Osato et al,12 we found that the mutations occurring outside MoAML were heterozygous. Osato et al.12 found that nonsilent mutations detected in MoAML induced premature termination mutations. Six of the mutations we observed in MoAML lead to premature termination codon mutations, whereas the remaining 3 were biallelic missense mutations, with duplication of the mutated AML1 allele and deletion of the normal AML1 allele.
An alteration of both AML1 alleles was indeed demonstrated in 8 of our 9 mutated cases of MoAML. This was easily observed in 3 cases in which a different mutation was seen on the 2 alleles. In 5 other mutated cases, an alteration of the other AML1 allele was suspected by SSCP and direct sequencing. Three of them were studied both at diagnosis and in complete remission by 6 DNA polymorphic markers localized on chromosome 21. The 3 patients were heterozygous in remission and homozygous at diagnosis for flanking markers situated close to the AML1 gene. This strongly suggested, when combined to the presence by FISH analysis of 2 AML1 alleles and of 2 signals for the cCMP21 probe, that deletion of the normal AML1 allele with duplication of the mutated AML1 allele had occurred in those 3 cases. Analysis of LOH at diagnosis for markers more distant from the AML1 gene (either centromeric or telomeric) showed different results in the 3 cases. No LOH was seen in patient 3 for proximal and distal markers, showing that duplication of the mutated AML1 gene resulted from duplication of a small chromosome area (gene conversion, Figure 4). Gene conversion has been described in many tumor types, especially for the Rb and met gene in retinoblastoma and renal carcinoma, respectively.22,23 To our knowledge, our patient 3 is the first description of a similar event in acquired hematologic malignancies. Gene conversion in 7q region has been suspected by another group in AML and MDS, using markers flanking a minimal commonly deleted region in 7q.24 However, no specific gene was analyzed in that study. In patients 4 and 5, duplication of the mutated AML1 gene resulted from a larger recombination of 21q. These patients had simultaneous LOH of AML1 locus and the telomeric marker but not of the centromeric marker on the chromosome 21, suggesting mitotic recombination of 21q. Mitotic recombination is an event frequently observed in tumors.25
The mutation of both AML1 alleles in at least 8 of our 9 mutated cases of MoAML occurred, although no chromosome 21 abnormality was detected by conventional cytogenetic testing. Our findings and those of Osato et al,12 therefore, suggest that MoAML is often associated with the absence of normal AML1 protein in leukemic cells, through premature terminating codon mutations or point mutations of this protein.
Because of the localization of the AML1 gene on chromosome 21, and the relatively high frequency of acquired trisomy 21 in hematologic malignancies, we studied a relatively large number of patients with this chromosomal abnormality. Three patients with Down syndrome, in which a highly increased risk of acute leukemia exists, were also studied. Five of the 27 patients with acquired trisomy or tetrasomy 21 carried an AML1 mutation. More precisely, 5 of the 13 patients with myeloid malignancies and acquired trisomy or tetrasomy 21 had an AML1 mutation. Interestingly, in those 4 patients with trisomy 21 and a mutation, 2 of the 3 AML1 alleles had the same mutation, and in the patient with tetrasomy 21 the same mutation was found in at least 3 of the 4 chromosomes 21. This also suggested, in those 5 patients, duplication of the mutated AML1 allele. None of the 8 mutated cases reported by Osato et al12 had trisomy or tetrasomy 21, but karyotype was not available in 3 of the mutated cases. However, Osato et al12 found 20q deletion in 2 of the mutated cases, including 2 of the 3 mutated MoAML. Our study included 4 patients with 20q deletion and none of them had detectable AML1 gene mutation.
Finally, AML1 mutation was no more found in the 4 patients with biallelic mutation that could be restudied in complete remission. This confirmed that AML1 mutations were acquired and excluded germline mutations, a possibility suggested by Osato et al12 in some of the AML1 mutations they described.
An important issue about mutations described in tumor cells is the demonstration that they affect the function of the protein. Mutations that truncate the AML1 protein within the Runt domain certainly destroy the DNA binding and the heterodimerization activity of the protein. Regarding the missense mutations we observed, R177Q was previously shown by Osato et al12 to abolish transactivation potential and DNA binding but not heterodimerization with CBF beta. R174Q (found in one of our cases) was also reported by Song et al21 in a case of familial platelet disorder (FPD) that evolved to AML and in a case of cleidocranial dysplasia, an autosomal dominant disorder that affects skeletal patterning.26,27In those 2 cases, functional analyses were not performed, but structural analysis with the use of nuclear magnetic resonance and protein-DNA nuclear Overhauser effects26,28 demonstrated that the R174G mutation abolished specific protein-DNA interaction. R174 is part of the last 10 AAs of the Runt domain, and deletion of this region has been shown to abolish DNA binding.29 D171G mutation (found in 2 of our cases) had not been previously reported. D171 is also included in the same region of AML1 protein and could also play a major role in the interaction between DNA and protein. The 2 last missense mutations we observed, R135G and G138D, are located in one of the beta sheets of the AML1 protein. The -E′ beta sheet linked to the beta E′ F loop, which binds DNA. Those 2 mutations are located in the same beta sheet as the R139G mutation described by Song et al21 in a case of FPD. By using structural analysis, it was shown that those AAs are not directly implicated in the DNA binding or heterodimerization but are located near the beta E′ F loop.
In the mutated cases of AML1, we observed in myeloid malignancies with acquired trisomy or tetrasomy 21, one AML1 allele was wild type, suggesting no inactivation of AML1. In similar cases of inactivation of only one AML1 allele in FPD, Song et al21 suggested that haploinsufficiency of AML1 could contribute to the development of leukemia, although this theory would have to be further substantiated. Interestingly, in our patient 12, with the same mutation of 2 AML1 alleles and 1 wild-type allele in chronic phase, loss of this normal allele occurred during progression to MoAML.
In conclusion, we confirmed that AML1 can be altered in leukemias by other mechanisms than translocation. We found in particular a high incidence of AML1 mutations in MoAML and in myeloid malignancies with acquired trisomy 21. The relationship between inactivation of AML1 gene and Mo phenotype in AML will have to be further explored, especially as AML1 protein has been shown to regulate expression of the MPO gene during myeloid differentiation. Mutations of the Runt domain of both AML1 alleles in MoAML could result in loss of the ability of AML1 to interact with the promotor sequences of its target gene and to loss of its transcriptional activity, particularly on the MPO gene, thus contributing to the Mo phenotype.
Acknowledgments
The authors are indebted to D. Lantoine, B. Vaast, R. Coudenis, M. Crepin, and the Institut Fédératif de Recherche for their excellent technical assistance and support in molecular biology and to Dr Helene Cavé and Pr C. Chomienne for helpful discussions and critical review of the manuscript.
Supported by the Centre Hospitalier Universitaire of Lille (PHRC 1997) and the Ligue Nationale Contre le Cancer (Comité du Nord et Aisne).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Pierre Fenaux, Service des Maladies du Sang, CHU, 1 place de Verdun, 59037 LILLE, France; e-mail: pfenaux@chru-lille.fr.