Abstract
Aberrant DNA methylation is believed to be important in tumorigenesis by causing either transcriptional inactivation of genes or chromosomal instability. Several laboratories have identified promoter hypermethylation of tumor suppressor genes in acute myeloid leukemia (AML). However, these studies do not provide a global assessment of overall methylation changes and do not allow the identification of novel methylated sequences. Previously, nonrandom CpG island methylation was reported in 17 adult de novo AML diagnostic samples when compared with the corresponding remission samples by means of restriction landmark genomic scanning (RLGS). That study has been expanded on by an analysis of a larger set of CpG islands (1740 vs 1184), which now provides details of 33 cloned methylated loci, including 21 known genes or expressed sequence tags. Five of these cloned loci appear to be methylated only in AML and not in the 6 solid tumors studied in this study (more than 98 samples analyzed). Chromosomal location was available for 30 of the 33 loci, and 5 of these 30 (17%) are localized to chromosome 11, suggesting a trend toward overrepresentation of methylation events on this chromosome. These results provide evidence for widespread aberrant methylation in AML, with identification of novel methylation targets, epigenetic changes that appear unique to AML, and apparent preferential methylation on chromosome 11.
Introduction
Acute myeloid leukemia (AML) is a clonal malignancy of hematopoietic cells of the myeloid lineage. It is a heterogeneous disease at the molecular and cytogenetic levels, as evidenced by the mixed clinical response to standard therapy.1 Over half of de novo AML cases exhibit cytogenetic abnormalities, including numerous recurring chromosome translocations and inversions that result in gene fusions.2 Some of the resultant fusion proteins have been shown to be involved in leukemogenesis,3 but it is thought that additional molecular changes are needed for the development of frank leukemia.4-6
DNA methylation is one mechanism that is believed to contribute to cancer initiation and progression by causing the inactivation of gene expression.7,8 This can have important consequences if the inactivated genes are essential for the control of normal cell growth, differentiation, or apoptosis. Aberrant DNA methylation can be responsible for one or both hits of Knudson's 2-hit hypothesis of carcinogenesis.9 The mechanisms that regulate normal and aberrant methylation are not fully understood, nor are the mechanisms by which methylation interferes with transcription. These processes involve complex interactions between methyltransferase and demethylase enzymes, histone acetylases and deacetylases, transcription factors, and chromatin structure (reviewed in Baylin and Herman,10 Cress and Seto,11 and Jones and Wolffe12).
Studies of selected tumor suppressor genes, such as p15 andp16,13,14 as well as candidate tumor suppressor genes, including the estrogen receptor15 and hypermethylated in cancer 1 (HIC1),16 indicate that aberrant DNA methylation is involved in AML. Melki et al17 found that 15 of 20 AML patients showed aberrant methylation in 2 or more cancer-related promoters, suggesting that AML might be characterized by a general deregulation of CpG island methylation.
In contrast to techniques that allow assessment of the methylation status of individual genes, restriction landmark genomic scanning (RLGS) is a tool for the identification of genomewide methylation changes in CpG islands regardless of whether the genomic sequence is known.18 This technique makes use of the rare-cutting methylation-sensitive restriction enzyme NotI (recognition sequence GC↓GGCCGC), which provides the landmarks composing an RLGS profile—a display of up to 2000 CpG islands in a single assay. The majority of these sequences are located in the 5′ regions of genes, thus providing a global portrait of the methylation status of hundreds of promoter sequences within a tumor sample. Some of the methylation changes within a single tumor type are unique to that tumor type, while other changes are common to several different tumor types.19 Therefore, it is possible that the identification of a unique set of methylation changes within a tumor type can establish a methylation signature or fingerprint, allow molecular classifications, and identify novel genes important in pathogenesis and treatment.
Previously, we reported a genomewide study of aberrant methylation in multiple tumor types, including AML. Our results showed that aberrant methylation in adult de novo AML is variable and nonrandom.19 We have expanded our analysis using 16 of the original 17 pairs of diagnostic and remission samples. This expanded analysis includes assessment of a larger number of CpG islands (1740 vs 1184) and a detailed description of 33 methylated loci from diagnostic AML samples. Using this strategy, we have identified novel methylation targets, epigenetic changes that appear specific for AML, and a trend toward overrepresentation of methylation on chromosome 11.
Materials and methods
Restriction landmark genomic scanning
RLGS gel analysis
Gels are analyzed by overlying the tumor sample (AML at diagnosis) and the matching normal control (remission sample), marking the changes on clear acetate, and recording the master profile address of the altered fragments. Previously, we had reported the analysis of 1184 fragments in multiple tumor types.19 Since the AML samples all have matched normal controls (ie, remission samples), this allows us to eliminate the possibility that spot changes are due to polymorphisms, rather than methylation events. Therefore, we were able to analyze 1740 fragments per sample in the current study.
Cloning of methylated loci
RLGS fragments were cloned with the use of a previously constructed arrayed plasmid library derived fromNotI/EcoRV fragments from peripheral blood (PB) genomic DNA from a healthy female donor.21 Confirmation of the clone of interest was accomplished by running a set of 4 RLGS mixing gels in which 5.2, 10.4, 20.8, and 66.3 (or in some cases 83.2) pg of 32P-labeled NotI-EcoRV fragments of the plasmid DNA were added to normal PB DNA for the first dimension gel electrophoresis. The standard RLGS procedure was then followed. Progressive enhancement of the locus of interest on the RLGS profile confirms the validity of the clone (Figure 3).
Determination of CpG island characteristics
RLGS clone sequences (either the entireNotI-EcoRV clone or available sequence from theNotI side) were submitted to the CpG island prediction program at WebGene at: http://www.itba.mi.cnr.it/webgene/ (accessed February 15, 2001). This program uses the CpG island criteria developed by Gardiner-Garden and Frommer.22
Southern hybridization
Southern hybridization was performed as previously described.23
Tissues and cell lines
Patient samples were obtained from the Cancer and Leukemia Group B Tissue Bank (Chicago, IL). The first 16 cases with the same available matched tissue (bone marrow [BM] [n = 3] or PB [n = 13]) from diagnosis and remission with de novo AML were chosen. Samples were collected in anticoagulant (heparin or EDTA) and shipped overnight to the procurement laboratory for processing. Mononuclear cells were separated on a Histopaque 1077 (Sigma, St Louis, MO) gradient by centrifugation at 1500 rpm for 30 minutes. Buffy coats were collected in freezing medium (Sigma) and stored in liquid nitrogen at −80°C until DNA isolation.
Isolation of high molecular weight DNA
High molecular weight DNA (approximately 1 μg/μL) from patient samples and cell lines was isolated according to published protocol.24
Isolation of RNA
Total RNA was extracted by means of Trizol (Gibco BRL, Rockville, MD) according to the manufacturer's directions.
Cell lines
Cell lines (HL-60, Kasumi-1, ML-1, and TI-1) were obtained from the laboratory of one of the authors (M.A.C.). Cells were cultured in RPMI-1640 medium with 20% fetal bovine serum (Gibco BRL), 1% antibiotic-antifungal agent (Gibco BRL), and 1% anti–pleuropneumonia-like organism agent (Gibco BRL) at 37°C in a 5% CO2 humidified atmosphere. We diluted 5-aza-2′-deoxycytidine (Sigma) with sterile water, filtered it, and added it directly to the culture medium in the concentrations and for the durations indicated.
Complementary DNA synthesis
RNA was converted to complementary DNA (cDNA) by means of the Superscript Preamplification System for First Strand cDNA Synthesis (Gibco BRL) following the manufacturer's instructions, and with the use of 2 to 3 μg total RNA and 0.5 μg Oligo(dT)11-17.
Polymerase chain reaction
Polymerase chain reactions (PCRs) were carried out in a total volume of 50 μL. Reaction mixtures contained 2 μL cDNA, 5 μL 10 × PCR buffer (Gibco BRL), 200 μM each dNTP, 1.5 mM MgCl2, 10 pmol each primer, and 2.5 U Platinum TaqDNA polymerase (Gibco BRL), except for the GPIreactions, which used AmpliTaq Gold (Perkin Elmer, Foster City, CA). Some reactions contained 5% dimethyl sulfoxide (DRD4, CYP1B1, and GPI). All reactions were run in a GeneAmp PCR System 9700 (Perkin Elmer Applied Biosystems, Foster City, CA) thermal cycler. For each PCR reaction, 10 μL was loaded onto a nondenaturing 8% polyacrylamide gel, electrophoresed at 250 V for 50 to 60 minutes, stained with ethidium bromide, and visualized under UV light.
Primers for reverse transcription–PCR
DRD4 forward 5′ TCGTCTACTCCGAGGTCCAG and reverse 5′ AGCACACGGACGAGTAGACC; CYP1B1 forward 5′ ACAGCATGATGCGCAACTTC and reverse 5′ TGGTCAGGTCCTTGTTGATG; G-α-Olf forward 5′ CGGAAGACCAGGGCGTCGATGAA and reverse 5′ CCCATTGACGTGCAGGATCCTCA; expressed sequence tag (EST) AI433656 (Genbank accession No.) forward 5′ CCCCAGCGAAGGCGTAG and reverse 5′ CGCGGGGCCACCAGTTTG;WIT1 forward 5′ CCTCACTGCCCTGCTCCTA and reverse 5′ TACCCACCACCCCTCTACC; GPI forward 5′ GACCCCCAGTTCCAGAAGCTGC and reverse 5′ GCATCACGTCCTCCGTCACC. All reactions were performed with an initial denaturing step of 95°C for 10 minutes, followed by 35 cycles of denaturing at 96°C for 30 seconds (GPI, 15 seconds). Annealing was 58°C for 30 seconds for DRD4; 57°C for 30 seconds for CYP1B1; 66°C for 60 seconds forG-α-Olf; 62°C for 60 seconds for EST AI433656; 58°C for 30 seconds for WIT1; and 60°C for 15 seconds forGPI. Extension was performed at 72°C for 60 seconds (G-α-Olf for 30 seconds), and final extension was at 72°C for 10 minutes (GPI, 5 minutes).
Mapping of clone 2D74
This clone was mapped as previously described.25 Primers used for mapping were as follows: No. 1 forward TTAACTCCCTCTGCTGCTCTCG and reverse CAATCCCTTGCACGTTTTGC; No. 2 forward TGGGTTTGCTCTAGGTGTTGGC and reverse TTTAGGGTGTGGTGAAGGGACC.
Statistical analysis
Heterogeneity of fragment losses across samples was determined by a chi-square test comparing the loss distribution with the expected (Poisson) distribution under the homogeneity hypothesis.26Similarly, chi-square goodness-of-fit tests were devised, comparing the observed vs the expected frequencies of fragments exhibiting multiple loss across samples, and comparing the observed vs the expected frequencies of chromosome-specific loss events. For these analyses, standard asymptotic P-value approximations did not apply or were inaccurate. For each analysis, 10 000 simulations from the permutation distribution of the data were performed to obtain empiricalP values. For the fixed observed number of methylation events on each chromosome, we calculated the null expected number of events as proportional to chromosome length or gene number. The variance of the observed number of events per chromosome, however, is sensitive to high-methylation frequencies of individual fragments and may not reflect a propensity of an individual chromosome. We performed a permutation test in which the observed loss at each fragment was fixed, but the fragment was randomly assigned to each chromosome according to the null expectation. To obtain an overall empirical distribution, we calculated, for each of 100 000 random permutations, the maximum chi-square cell contribution: (observed-expected)2/expected. This approach properly considers the varying loss rates of individual fragments and accounts for multiple statistical comparisons across several chromosomes.
Results
Comparison of RLGS profiles from AML diagnostic and remission samples
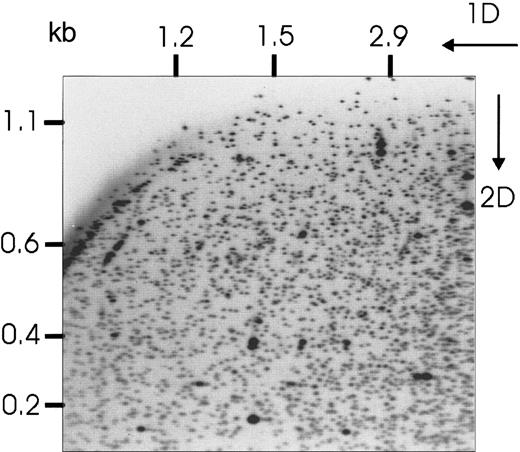
To assess the extent of hypermethylation in AML, RLGS profiles were obtained for 16 pairs of BM (n = 3) or PB (n = 13) samples from adult patients at the time of original diagnosis with AML (Figure1) and again at complete remission (defined according to standard criteria with fewer than 5% blast cells on bone marrow aspirate27) following induction chemotherapy. Profiles were also obtained for 4 AML cell lines: Kasumi-1,28 HL-60,29 ML-1,30 and TI-1.31 Altered RLGS fragments in the patient samples were identified by comparing the diagnostic and remission profiles from the same patient. In each case, BM was compared with BM, and PB with PB. We have previously shown that the absence of a fragment on the diagnostic profile coupled with the presence of the fragment on the remission profile from the same patient is indicative of methylation in the diagnostic tissue19 32 (Figure2). Each fragment on the RLGS profile has a unique address (http://pandora.med.ohio-state.edu/masterRLGS/[accessed February 15, 2001]) to allow for consistent recording of alterations. A total of 1740 fragments were assessed for each pair of diagnostic and remission profiles. The total number of methylation events (ie, loss of a fragment) in the diagnostic profiles when compared with the remission profiles ranged from 0 to 145 (0% to 8.3% of the profile; mean 1.9%), indicating a wide range in the degree of aberrant methylation in these samples (Table1). This range demonstrates heterogeneity across samples, indicating that some samples demonstrated an inherently higher methylation event rate than others (P < .001; see “Statistical analysis”). Of the 1740 loci examined, 208 (16%) were methylated in at least one patient.
RLGS profile prepared from a BM aspirate at initial diagnosis with AML.
Directions of first and second dimension electrophoresis are shown, as well as molecular sizes of the fragments.
RLGS profile prepared from a BM aspirate at initial diagnosis with AML.
Directions of first and second dimension electrophoresis are shown, as well as molecular sizes of the fragments.
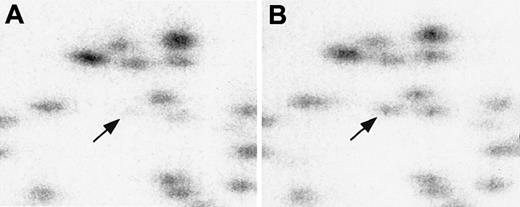
RLGS profiles demonstrating methylation of fragment 3B36 (arrow) in diagnostic sample compared with remission sample from same patient.
Comparison of RLGS profile of AML diagnostic sample (panel A) and remission sample (panel B), showing absence of fragment 3B36, theCPY1B1 gene, in AML diagnostic sample owing to methylation.
RLGS profiles demonstrating methylation of fragment 3B36 (arrow) in diagnostic sample compared with remission sample from same patient.
Comparison of RLGS profile of AML diagnostic sample (panel A) and remission sample (panel B), showing absence of fragment 3B36, theCPY1B1 gene, in AML diagnostic sample owing to methylation.
Cloning, sequencing, and characterization of altered RLGS fragments
In order to further characterize the target sequences that become aberrantly methylated in AML, we selected RLGS loci that had both a high and a low frequency of loss in our diagnostic AML samples and that were available in our cloning library. The cloning was accomplished by using an arrayed NotI-EcoRV plasmid library and RLGS mixing gels as previously described.21,23 Figure3 shows a portion of an RLGS profile demonstrating positive identification of clone 2D70 in a representative mixing gel. A total of 33 RLGS loci that were altered in diagnostic AML samples have been cloned and either partially (single pass from theNotI end) or fully sequenced (Table2). Previously, we reported that certain RLGS fragments are lost exclusively in certain tumor types.19 Here we report the cloning of 5 loci (2C40, 2D14, 3F16, 3F72, and 4E53) methylated exclusively in AML and not in the breast (n = 14), colon (n = 8), head and neck (n = 14), testicular tumors (n = 9), adult brain tumors (n = 14), or pediatric brain tumors (n = 22) in our original analysis (Table 2). All clone sequences were analyzed for CpG island characteristics with the WebGene program (Consiglio Nazionale delle Ricerche, Istituto Tecnologie Biomediche Avanzate, Milan, Italy) (http://www.itba.mi.cnr.it/webgene/). Of the 33 clones, 31 (94%) have CpG island characteristics. BLAST searches show that 11 clones have homology to known genes, 10 to EST sequences, 11 to bacterial artificial chromosomes (BACs) or P1 artificial chromosomes, and 1 has no homology. The results of the database searches are listed in Table2. The data demonstrate the strong bias of RLGS for display of CpG islands and sequences associated with genes, rather than random genomic sequences.
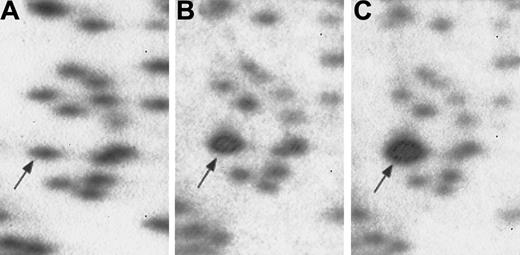
RLGS mixing gels used in cloning procedure.
Normal PB genomic DNA (panel A) with no additionalNotI-EcoRV clone. PB with 5.2 pg (panel B) and 20.8 pg (panel C) of the radiolabeled candidate clone added to the genomic DNA. Progressive enhancement of the RLGS fragment of interest confirms identification of the correct NotI-EcoRV clone.
RLGS mixing gels used in cloning procedure.
Normal PB genomic DNA (panel A) with no additionalNotI-EcoRV clone. PB with 5.2 pg (panel B) and 20.8 pg (panel C) of the radiolabeled candidate clone added to the genomic DNA. Progressive enhancement of the RLGS fragment of interest confirms identification of the correct NotI-EcoRV clone.
To assess the methylation status of selected genes in AML cell lines, Southern blots were prepared and hybridized with 6NotI-EcoRV clones (2C40, 3B36, 3D35, 3C01, 5C08, and 5E34) with the use of either the entire clone or a suitable restriction fragment. Normal donor PB mononuclear cell DNA served as a control. DNA was digested with EcoRV alone (PB) orEcoRV plus the methylation-sensitive enzyme NotI (PB and cell lines). Failure of NotI to cut the cell line DNA was observed in most cases; this is strongly supportive of methylation of these genes in the cell lines (Figure4). Previous work in our laboratory has confirmed that loss of a fragment on the RLGS profile corresponds to methylation in more than 99% of the samples analyzed by Southern hybridization (Costello et al,19 Plass et al,32 and unpublished data).
Southern blots showing methylation of RLGS clones in AML cell lines.
Lane 1: Control normal donor peripheral blood (PB) is digested withEcoRV only (RV only) to show size of unmethylated band. Lanes 2-6: PB and AML cell line DNA is double-digested withEcoRV and methylation-sensitive NotI (RV-NotI). The presence of an uncut band in these lanes is indicative of methylation at the NotI site. Blots are probed with the RLGS clones 2C40 (panel A), 5C08 (panel B), and 3B36 (panel C). The 1.7-kilobase (kb) band in (panel B) is due to partial methylation of an internal NotI site within the larger 3-kbEcoRV-NotI fragment. Kas-1 indicates Kasumi-1.
Southern blots showing methylation of RLGS clones in AML cell lines.
Lane 1: Control normal donor peripheral blood (PB) is digested withEcoRV only (RV only) to show size of unmethylated band. Lanes 2-6: PB and AML cell line DNA is double-digested withEcoRV and methylation-sensitive NotI (RV-NotI). The presence of an uncut band in these lanes is indicative of methylation at the NotI site. Blots are probed with the RLGS clones 2C40 (panel A), 5C08 (panel B), and 3B36 (panel C). The 1.7-kilobase (kb) band in (panel B) is due to partial methylation of an internal NotI site within the larger 3-kbEcoRV-NotI fragment. Kas-1 indicates Kasumi-1.
Reverse transcription–PCR analysis using AML cell lines
As promoter hypermethylation has been linked to transcriptional silencing, we assessed the expression status of 5 sequences (4 genes and 1 EST) that were aberrantly methylated in our diagnostic AML samples. Whereas previously we had examined expression of 3 of these loci (G-α-Olf, WIT1, and 3D41) in brain tumor cell lines, the current studies were carried out with 3 AML cell lines: HL-60, Kasumi, and TI-1. RLGS profiles for each cell line showed all 5 sequences were methylated in the Kasumi-1 line, while 4 of 5 were methylated in the HL-60 (exception: 3B36) and in the TI-1 (exception: 5E34) cell lines (Table 3). Reverse transcription–PCR (RT-PCR) was performed on RNA from these cell lines before and after the cells were treated with the demethylating agent 5-aza-2′-deoxycytidine (Table 3). Expression status of 3 sequences,CYP1B1, G-α-Olf, and EST AI433656, was changed in at least one of the cell lines after treatment. DRD4expression was not detectable regardless of treatment, and expression of WIT1 remained unchanged.
Methylation changes in AML are nonrandom, with a trend toward overrepresentation of chromosome 11 loci
To determine if there were recurrent methylation events among diagnostic AML samples in our expanded set of CpG islands, we compared the analyses of fragment losses across our 16 patients. This comparison revealed that certain fragments (eg, 2C40, Table 2) were altered in the majority of diagnostic AML samples (11 of 16), whereas other fragments were altered much less frequently (eg, 3D41; 3 of 16) or not at all (eg, 4B2; 0 of 16; not listed in Table 2 because not methylated in this sample set). A standard goodness-of-fit test was applied to test the hypothesis of a constant rate of alteration among the fragments. This hypothesis was clearly rejected (P < .001; see “Statistical analysis”), with many more fragments showing repeated alteration than would be expected by chance. The expanded analysis of a larger set of CpG islands reported here upholds our previous observation of nonrandom methylation patterns in AML.
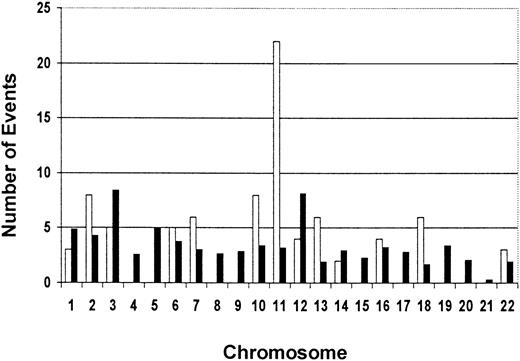
We also examined the BLAST search data for the chromosomal location of our 33 cloned methylated loci. Interestingly, of the 30 clones for which the chromosomal location was available, 5 (17%) map to chromosome 11 (3 to the long arm and 2 to the short arm) (Table 2). We plotted the total number of loss events (ie, the number of patients in which a specific locus was methylated) for each clone with a chromosomal assignment against the chromosome number (Figure5). We calculated an “expected” rate of methylation for the chromosomes using 2 criteria: the relative physical length of the chromosomes33 and the estimated number of genes on each chromosome.34 We compared the observed vs expected frequency of methylation on the chromosomes using a specially designed chi-square test with a permutation-basedP-value (see “Statistical analysis”). While there appeared to be a clear trend for overrepresentation of methylation events on chromosome 11, the test did not reach statistical significance (maximum χ = 110.09; overallP = .056).
Observed vs expected numbers of methylation events per chromosome for 30 RLGS loci for which chromosomal assignment is available.
The total number of methylation events per chromosome was determined by the number of times each RLGS locus with a chromosomal assignment was methylated in the 16 AML diagnostic samples. The data show a trend for an overrepresentation of methylation events on chromosome 11. ■ indicates observed events; ▪, expected events.
Observed vs expected numbers of methylation events per chromosome for 30 RLGS loci for which chromosomal assignment is available.
The total number of methylation events per chromosome was determined by the number of times each RLGS locus with a chromosomal assignment was methylated in the 16 AML diagnostic samples. The data show a trend for an overrepresentation of methylation events on chromosome 11. ■ indicates observed events; ▪, expected events.
Discussion
In this study, we provide a detailed analysis of a genomewide survey of aberrant methylation in adult de novo AML, extending our previous work with characterization of novel methylated CpG islands, identification of leukemia-specific methylation, and the suggestion of preferential methylation events on chromosome 11. There is a remarkable variability in the degree of methylation in our sample set, indicating a much larger degree of methylation in primary patient AML samples than was previously known. Indeed, some AML samples exhibit very little or no aberrant methylation while others exhibit a much larger degree of methylation (up to 8.3% of the RLGS profile, and thus presumably a similar proportion of the 45 000 CpG islands in the genome). The methylation events we have identified do not correlate with specific translocations associated with AML, and in addition, they occur in AMLs with normal cytogenetics. This finding suggests that the aberrant methylation could represent an independent epigenetic change in addition to the genetic changes. The use of complete remission samples for each patient as our normal control eliminates any differences in the profiles due to polymorphisms. Restoration of unmethylated CpG islands in the profiles obtained from remission samples is attributed to eradication of the leukemic blasts and repopulation of the bone marrow with normal hematopoietic cells.
The nonrandom nature of the methylation found in AML is particularly intriguing and can be explained by 2 scenarios. First, aberrant methylation may preferentially target specific genes. Perhaps there are intrinsic features of those sequences, such as repetitive elements, sequence motifs, or chromatin structure, that predispose them to undergo methylation at a high frequency in AML when the regulatory mechanisms of methylation have gone awry. A second explanation is that the methylation itself is completely random but cells with a certain methylation pattern achieve a selective advantage based on the altered gene expression profile that results from the methylation. This explanation is supported by documentation of methylation-induced biallelic inactivation of p15, p16, andMLH1 in various cancers.35 36
In addition to providing a comprehensive assessment of overall methylation changes, RLGS allows the identification of novel methylated genes. As shown in Table 2, one third (11 of 33) of the methylated fragments correspond to known genes, while almost another third (10 of 33) have homology to ESTs. Many of the genes identified in our samples were not previously known to be methylated in cancer. This underscores the utility of RLGS in identifying methylation in novel promoter and gene sequences and provides a rational approach for using these sequences in the study of leukemogenesis.
There are overwhelming data that promoter methylation is associated with transcriptional inactivation.37 In addition, there is evidence that methylation of 3′ CpG islands could have an activating effect on transcription.37 Interestingly, of the 11 genes that we identified, 3 (POU3F1,38TBX18,39,40 and insulin promoter factor41) are known transcription factors. A fourth gene, cold shock domain A, is a member of a family of DNA- and RNA-binding genes involved in transcriptional and translational regulation.42 These observations are particularly relevant in light of the fact that genetic changes in AML frequently involve transcription factors.43 However, since some of the methylated genes that we identified are not expressed in normal blood (eg, DRD4, Table 3), the significance of the methylation of those genes in the pathogenesis of AML is unclear. It is possible, if not likely, that a fraction of the methylated genes are unrelated to the leukemic process and that the methylation is a reflection of a global deregulation of control mechanisms, including methyltransferase44 or demethylase enzymes,45or endogenous stimuli.46
Our RT-PCR analyses showed that methylation of the RLGS landmarkNotI restriction site does not always correlate with complete transcriptional inactivation. This finding agrees with other reports in the literature.19,47 Cameron and colleagues14 have previously shown that the density of methylated CpG dinucleotides is important for inactivation of thep15 gene, and Ganderton and Briggs48 showed that methylation of the 5′ region of PTHrP did not correlate with inactivation. In addition, the location of the CpG island within the gene may play a role in determining the effects of methylation on transcription. In our study, methylation of the CpG island in the middle of CYP1B1 seemed to have a variable effect on transcription.
Importantly, our approach has identified 5 loci that appear to be methylated only in leukemia and not in the other tumor types we have investigated. Thus, we have the means to investigate genes whose effects may be specific to leukemia, as well as genes whose effects may be important in many different types of tumors.19
As shown in Table 2, of the 33 cloned sequences, 30 have been mapped to a specific chromosome, and 5 of these 30 (17%) reside on chromosome 11. As chromosome 11 accounts for only 4.7% of relative length of the autosomes,33 methylation may be overrepresented on this chromosome. However, the trend for overrepresentation does not meet statistical significance. This will need to be further investigated with continuing elucidation of the human genome and a clearer understanding of CpG island and NotI site distribution. In a previous report, de Bustros et al49 postulated that the short arm of chromosome 11 is a “hot spot” for methylation. Our data with AML support these previous results and show that the methylation is not restricted to 11p but also involves 11q.
Chromosome 11 is already known to be involved in AML by virtue of translocations, partial tandem duplications, and trisomy 11.50-54 The association of methylation in these areas of chromosomal instability requires further investigation, but one hypothesis is that methylation occurs in these regions as a cause or consequence of genetic instability. Chromosome 11 is also known to harbor several cancer-related genes, including oncogenes and tumor suppressor genes, such as HRAS,55FGF3,56cyclin D,57MEN1,58ATM,59TSG101,60 andWIT1.32 Aberrant methylation of any of these regions may lead to a selective advantage by activation or inactivation of these genes and an in vivo expansion of a malignant clone. In addition, there are several imprinted genes on chromosome 11, such as IGF2 and H19 at 11p15.5. Altered methylation of these genes is associated with Beckwith-Wiedemann syndrome.61 62 It is possible that the methylation associated with imprinting of these regions becomes deregulated and spreads over a larger region, resulting in the methylation of nearby genes. Together, these data indicate that not only genetic events but also methylation changes on chromosome 11 may play an important role in leukemogenesis.
Our findings provide a genomewide characterization of a highly variable methylation pattern in AML, along with the identification of novel methylated sequences, leukemia-specific methylation, and a trend for hypermethylation on chromosome 11. As AML is known to be a heterogeneous disease at both the cytogenetic and genetic levels,4 this may now also be extended to the epigenetic level since no single fragment is methylated in 100% of the profiles from diagnostic samples. Our data provide new insights into the molecular biology of AML and the role of epigenetic changes in this disease. Further characterization of these events should enhance our understanding of the pathogenesis of AML and could potentially result in the identification of novel cancer-related genes as well as molecular markers for the classification of AML.
We thank Julie Hall, Yue-Zhong Wu, Kellie Archer, Jeff Palatini, Donna Bucci, Mike Ostrowski, and Matthew P. Strout for advice and expert technical assistance; Robert Chadwick, head of the Genotyping and Sequencing Unit at Ohio State University, for assistance with sequencing and clone analysis; and Bo Yuan for bioinformatics assistance.
Supported by grants CA80912 (C.P.), CA16058 (C.D.B.), CA77658 (C.D.B.), CA31946 (C.D.B., M.A.C.), and CA09338 (M.A.C.) from the National Cancer Institute, National Institutes of Health, Bethesda, MD, and the Leukemia Clinical Research Foundation (C.D.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christoph Plass, The Ohio State University, Division of Human Cancer Genetics, Medical Research Facility 494A, 420 West 12th Ave, Columbus, OH 43210; e-mail: Plass-1@medctr.osu.edu.