Identification of cytogenetic abnormalities is an important clue for the elucidation of carcinogenesis. However, the cytogenetic and clinical significance of adult T-cell leukemia/lymphoma (ATLL) is still unclear. To address this point, cytogenetic findings in 50 cases of ATLL were correlated with clinical characteristics. Karyotypes showed a high degree of diversity and complexity. Aneuploidy and multiple breaks (at least 6) were observed frequently in acute and lymphoma subtypes of ATLL. Breakpoints tended to cluster at specific chromosomal regions, although characteristic cytogenetic subgroups of abnormalities were not found. Of these, aberrations of chromosomes 1p, 1q, 1q10-21, 10p, 10p13, 12q, 14q, and 14q32 correlated with one or more of the following clinical features: hepatosplenomegaly, elevated lactate dehydrogenase, hypercalcemia, and unusual immunophenotype, all indicators of clinical severity of ATLL. Multiple breaks (at least 6); abnormalities of chromosomes 1p, 1p22, 1q, 1q10-21, 2q, 3q, 3q10-12, 3q21, 14q, 14q32, and 17q; and partial loss of chromosomes 2q, 9p, 14p, 14q, and 17q regions correlated with shorter survival. These cytogenetic findings are relevant in predicting clinical outcome and provide useful information to identify chromosomal regions responsible for leukemogenesis. This study also indicates that one model of an oncogenic mechanism, activation of a proto-oncogene by translocation of a T-cell–receptor gene, may not be applicable to the main pathway of development of ATLL and that a multistep process of leukemogenesis is required for the development of ATLL.
Introduction
Adult T-cell leukemia/lymphoma (ATLL) is a distinct clinical entity characterized by a mature T-cell surface-marker profile, the association with human T-cell leukemia virus type-1 (HTLV-1), and abnormal lymphocytosis with markedly deformed pleomorphic nuclei.1-3 Identification of recurring cytogenetic abnormalities may provide important clues to the elucidation of the pathogenesis of ATLL. Cytogenetic findings and the analysis of their clinical significance are still limited in mature T-cell malignancies compared with those of B-cell malignancies.4-6 Although many cytogenetic studies have been performed,7-16 the cytogenetics of ATLL is complicated by clinical heterogeneity and a plethora of secondary abnormalities. To improve the accurate evaluation of karyotypes and to identify specific chromosomal abnormalities in ATLL, a large number of karyotypes from various laboratories were reviewed by the ATL Karyotype Review Committee 1985 in Japan.17 Several recurring abnormalities such as trisomy 3, 7, and 21; monosomy X in the female; loss of a Y in the male; translocations involving 14q11 or 14q32; and deletion in 1p, 3q, 5q, 6q, 7p, 9q, 10p, and 13q have been confirmed.17 However, correlation between these and clinical features, as reported in non-Hodgkin lymphoma (NHL),4-6 has not been attempted in ATLL.
This study reports the detailed cytogenetic findings observed in 50 patients with ATLL and correlates them with clinical characteristics.
Patients, materials, and methods
Patients
A total of 50 newly diagnosed ATLL patients between 1989 and 1996 were studied. These patients were natives of Nagasaki prefecture, one of the clustering areas of ATLL incidence in Japan. Diagnosis and subclassification of ATLL were done according to previously described criteria.18 We classified 3 cases that were in the smoldering or chronic phase before progression to the acute phase as the acute subtype because cytogenetic analysis was performed at the time of progression, and cytogenetic findings exhibited abnormalities possibly of the acute phase. Histologic diagnosis of the lymphoma subtype was based on the findings of biopsied lymph nodes according to the updated Kiel classification.19 Immunophenotypic analysis of tumor cells was performed by direct and indirect immunofluorescence assays, as described previously.20Based on the immunophenotype of ATLL cells, the cases were classified into 2 categories: usual phenotype showing CD4+CD8− phenotype, and unusual phenotype showing CD4+CD8+, CD4−CD8+, or CD4−CD8− phenotypes, because this phenotypic diversity correlates with clinical outcome.20 Patients with acute and lymphoma subtypes were treated by an intensive combination chemotherapy regimen (VEPA, LSG4, or LSG11).21 Patients with the chronic subtype were monitored without chemotherapy until they progressed to the acute or lymphoma subtype, except for one patient who received autologous bone marrow transplantation unsuccessfully.
Cytogenetic analysis
Samples for the cytogenetic studies were obtained from the peripheral blood of patients with acute and chronic subtypes, or from the biopsied lymph nodes of patients with the lymphoma subtype at the time of diagnosis before therapy.
Chromosome analysis was performed using G- and/or Q-banding techniques after culturing without mitogens. Karyotypes were described according to the International System for Human Cytogenetics Nomenclature (1995).22
To identify the specific abnormalities and avoid the secondary changes, the stemline karyotype (ie, the simplest abnormal clone) was used as the representative karyotype for almost all analyses. In patients who showed composite karyotypes (8 cases) or when stemline karyotypes could not be specified (3 cases), we created a “stemline karyotype,” which contained common abnormalities in all analyzed karyotypes.
Numbers of chromosomal breaks from structural abnormalities were calculated as the sum of all breaks whose derivation by band could be identified in the stemline karyotype. Marker chromosomes were not included for analysis of numbers of chromosomal breaks. Breakpoints of isochromosomes 1q, 6p, 7q, 8q, 18q, and 21q were included in breaks on chromosomes 1p10, 6q10, 7p10, 8p10, 18p10, and 21p10, respectively. Multiple rearrangements of the same break site observed in the same patient were recorded separately.
We also assessed recurrent gains or losses of specific chromosomal segments, termed a “chromosomal segmental representation profile (CSRP),” in stemline karyotypes of each patient according to methods described by Thompson et al.23 One patient whose stemline karyotype showed near-triploidy was not included in CSRP analysis.
Statistical analysis
All chromosomal abnormalities occurring in more than 5 cases were correlated with the clinical factors described in Table1. Associations between clinical variables and chromosomal abnormalities were tested using the Fisher exact test as determined by expected frequencies of variables in 2 × 2 tables. P < .05 was used as the criterion for statistical significance.
Generally, survival was calculated from the date of diagnosis. In 3 patients who were in the smoldering or chronic phase before progression to the acute phase, survival was calculated from the date of acute transformation. Overall survival was calculated by the method of Kaplan and Meier.24 Differences between survival curves were assessed using the log-rank test.25 Association between the shorter survival group (less than 6 months) and chromosomal abnormalities was also tested using the Fisher exact test to find specific abnormalities for a shorter survival group.
Results
Clinical characteristics of patients
The clinical characteristics of the 50 patients analyzed are summarized in Table 1. All patients were positive for anti–HTLV-1 antibodies and for monoclonal integration of the HTLV-1 provirus. Among 6 cases of lymphoma type, 5 cases were classified as pleomorphic, medium-sized and large cell type, and one as large cell, anaplastic type.
Peripheral lymphadenopathy and abnormal elevation of serum lactate dehydrogenase (LDH) and calcium levels were significantly more frequent in the combined group of acute and lymphoma subtypes than in the chronic subtype (P < .05, P < .01, andP < .01, respectively). Abnormal elevation of serum LDH and calcium was found significantly more frequently in the shorter-survival group (less than 6 months) than in the longer-survival group (at least 6 months) (P < .05 andP < .01, respectively). Hepatosplenomegaly, central nervous system (CNS) involvement, abnormal elevation of serum LDH and calcium, unusual immunophenotype, and clinical subtype (acute and lymphoma subtypes) were statistically significant clinical factors for overall survival (P < .05, P < .05,P < .01, P < .01, P < .01, and P < .01, respectively, log-rank test). Patients in the combined group of acute and lymphoma subtypes showed shorter survival than patients with the chronic subtype (P < .01, log-rank test). These findings show that clinical factors such as peripheral lymphadenopathy, hepatosplenomegaly, CNS involvement, elevation of serum LDH and calcium, unusual immunophenotype, and clinical subtype (acute and lymphoma subtypes) correlate with clinical severity in this study.
Cytogenetic findings
All 50 patients showed abnormal karyotypes (the table containing the detailed cytogenetic findings can be obtained on request). Of 43 patients with acute and lymphoma subtypes, 11 showed pseudodiploid, 6 hypodiploid, 25 hyperdiploid, and one near-triploid stemline karyotype. Of 7 patients with chronic subtype, 5 showed pseudodiploid and 2 hyperdiploid karyotypes. Aneuploidy was found significantly more frequently in the combined group of acute and lymphoma subtypes than in the chronic subtype (P < .05).
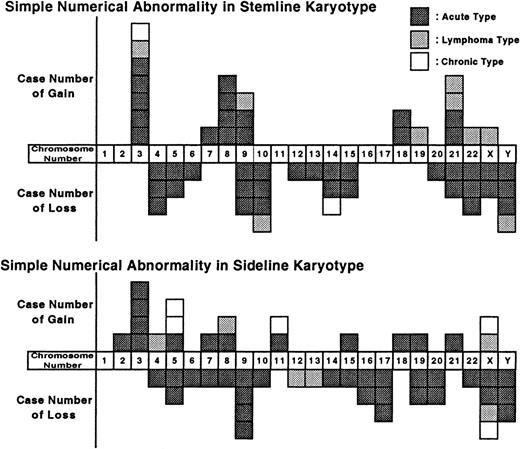
Simple numerical abnormalities (without structural abnormalities) observed in stemline and sideline karyotypes are shown in Figure1. In the stemline karyotype of 49 patients, the most frequently gained chromosomes were chromosome 3 (7 cases, 14%), followed by chromosomes 8 and 21 (4 cases, 8%, each), and chromosome 9 (3 cases, 6%). The most frequently lost chromosomes were chromosomes 10 and Y (4 cases, 8%, each), followed by chromosomes 4, 9, 14, and 22 (3 cases, 6%, each). In the sideline karyotype, gain of chromosomes 3 and 5, and loss of chromosomes 9, 17, X, and Y were also observed.
Numerical chromosomal changes in the stemline and sideline karyotypes (n = 49).
Each box represents one gain or loss.
Numerical chromosomal changes in the stemline and sideline karyotypes (n = 49).
Each box represents one gain or loss.
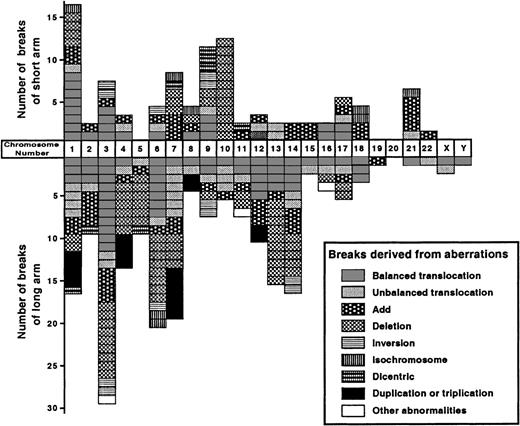
Forty-eight patients had clonal structural abnormalities. Two other patients showed only numerical abnormalities or an extra marker chromosome. A total of 295 breaks were observed (Figures2 and 3). The median number of breaks was highest in the acute subtype (6.0), followed by lymphoma (3.5) and chronic (1.0) subtypes. Multiple breaks (at least 6) were found significantly more frequently in the combined group of acute and lymphoma subtypes than in the chronic subtype (P < .05), which indicates the karyotypic complexity of the acute and lymphoma subtypes compared with the chronic subtype.
Number of breaks observed on each chromosome in the stemline karyotype (total 295 breaks).
Each box represents a single break.
Number of breaks observed on each chromosome in the stemline karyotype (total 295 breaks).
Each box represents a single break.
Schematic representation of cytogenetic abnormalities.
(A) Distribution of breakpoints in the stemline karyotype (n = 50). (B) Chromosomal segmental representation profile in the stemline karyotype (n = 49). Symbols for abnormalities are described in the bottom box of each figure.
Schematic representation of cytogenetic abnormalities.
(A) Distribution of breakpoints in the stemline karyotype (n = 50). (B) Chromosomal segmental representation profile in the stemline karyotype (n = 49). Symbols for abnormalities are described in the bottom box of each figure.
Almost all chromosomes were affected, but with variations in frequency. Chromosomes involved in more than 5 patients (at least 10%) were the short arms of chromosomes 1 (12 cases, 24%), 7, 9, 10 (8 cases, 16%, each), 3 (7 cases, 14%), 21 (6 cases, 12%), and 17 (5 cases, 10%); and the long arms of chromosomes 3 (23 cases, 46%), 6 (18 cases, 36%), 7 (14 cases, 28%), 1 and 14 (13 cases, 26%, each), 13 (11 cases, 22%), 4 and 12 (9 cases, 18%, each), 2 (8 cases, 16%), 9 and 11 (6 cases, 12%, each), and 5, 10, and 17 (5 cases, 10%, each). The types of structural abnormalities were balanced and unbalanced translocations (balanced type, 76 breaks, and unbalanced type, 39 breaks; total 115, 40%), followed by deletion (79, 26.8%), addition (51, 17.3%), duplication or triplication (18, 6.1%), inversion (12, 4.1%), dicentric chromosome (8, 2.7%), isochromosome (8, 2.7%), insertion (2, 0.7%), and translocations with a homogeneously staining region (2, 0.7%). Most abnormalities were not recurrent, with the exception of inv(3)(p25q21) (2 cases), iso(6)(p10) (2 cases), iso(18)(q10) (2 cases), and several deletion abnormalities. Chromosomal breaks tended to cluster at specific bands. The specific bands observed in more than 5 patients (at least 10%) were 3q10-12 (11 cases, 22%), followed by 14q11 (8, 16%), 1q10-21, 3q21, 6q15, 10p11 (7, 14%, each), 3q25, 7q10-11, 13q22, 13q32, 21p11 (6, 12%, each), and 1p22, 4q31, 6q21, 6q23, 7q22, 7q32, 10p13, and 14q32 (5, 10%, each). Deletion frequently affected chromosome 10p (8 cases, 16%), followed by 3q, 6q (7, 14%, each), and 13q (6, 12%).
In the sideline karyotype, isochromosome of 18q (4 cases, 8%) and rearrangement of the centromeric region on chromosome 3 (3 cases, 6%) were observed, which indicates additional changes in the ATLL karyotype (Figure 3A). Del(2)(q), del(3)(p), dup(4)(q), del(6)(p), del(6)(q), del(13)(q), and add(18)(p) were other abnormalities of sideline karyotype observed in 2 patients each.
In many cases, more than one of the structural or numerical abnormalities occurred simultaneously. Associations of structural abnormalities between chromosomes 1p and 3q (10 cases), 1p and 14q (7 cases), 3q and 11q (6 cases), and 7q and 9q, 7q and 13q32, and 9p and 13q (5 cases, each) were found in more than 5 patients and were statistically significant (P < .01).
By CSRP analysis, gains were frequently observed in chromosome 3p (17 cases, 34%), 3q centromeric region (14, 28%), 7q (13, 26%), 1q, 8q (7, 14%, each), 8p, 9q (6, 12%, each), and 21q (5, 10%); and losses were observed in 10p (13 cases, 26%), 6q, 14q telomeric region, 21p (8, 16%, each), 2q telomeric region, 4q (7, 14%, each), 9p telomeric region, 9q telomeric region, 14q centromeric region (6, 12%, each), and 7p telomeric region and 17q telomeric region (5, 10%, each) among 49 stemline karyotypes (Figure 3B). Many of the del(3)(q) and unbalanced translocations of chromosome 1q and 7q resulted in partial trisomy of 3p, 1q, and 7q, respectively.
Correlation between cytogenetic findings and clinical characteristics
Significant correlations noted between cytogenetic findings and clinical characteristics are summarized in Table2. Multiple breaks (at least 6) and aneuploidy were found to be significantly more frequent in the combined group of acute and lymphoma subtypes than in the chronic subtype. There were no significant differences in cytogenetic findings between acute and lymphoma subtypes.
Multiple breaks (at least 6); abnormalities of chromosomes 1p, 1q, 1q10-21, 10p, 10p13, 12q, 14q, and 14q32; and deletion of chromosome 10p correlated with one or more of the following clinical factors: hepatosplenomegaly, elevated serum LDH, hypercalcemia, and unusual immunophenotype. The correlation between these clinical factors and clinical severity suggests that these abnormalities contribute to clinical severity.
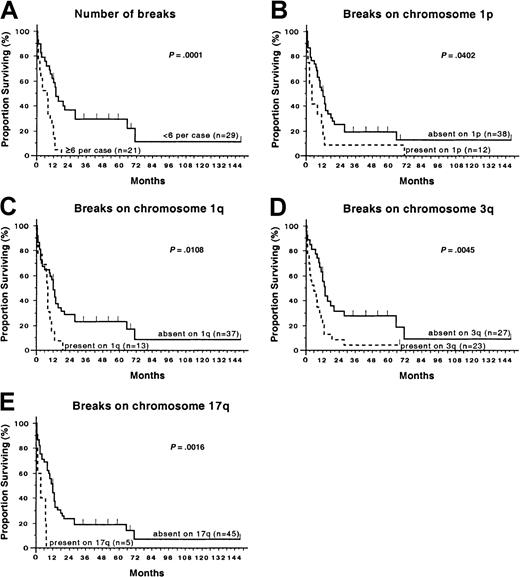
Multiple chromosomal breaks (at least 6) (P < .01, log-rank test) and abnormalities of 1p, 1p22, 1q, 1q10-21, 3q, 3q10-12, 3q21, and 17q (P < .05, P < .01,P < .05, P < .05, P < .01,P < .05, P < .05, andP < .01, respectively, log-rank test) gave statistically significant shorter overall survival (Figure4). Abnormalities of chromosomes 1p, 1p22, 2q, 3q, 14q, and 14q32 (P < .05,P < .05, P < .01, P < .05,P < .05, and P < .05, respectively) were significantly more frequent in the shorter-survival group (less than 6 months), indicating that these abnormalities are specific for the aggressive clinical course of ATLL.
Overall survival curves of 50 patients according to the cytogenetic abnormalities.
(A-E) Statistical significance was assessed using log-rank test.
Overall survival curves of 50 patients according to the cytogenetic abnormalities.
(A-E) Statistical significance was assessed using log-rank test.
Based on the findings of CSRP, we also analyzed the correlation between chromosomal imbalance region and prognosis. Loss of chromosome 9p telomeric region (6 cases) and 17q telomeric region (5 cases) gave significantly shorter overall survival (both P < .05, log-rank test). Loss of chromosome 2q telomeric region (7 cases), 14p11 terminal region (6 cases), and 14q telomeric region (8 cases) were significantly more frequent in the shorter-survival group (P < .01, P < .05, andP < .05, respectively). Other imbalance regions did not correlate with prognosis.
Discussion
Karyotypes of ATLL showed a high degree of diversity and complexity. However, abnormalities were undoubtedly nonrandom, and this study demonstrated clinical significance of some of the cytogenetic abnormalities.
Simple numerical abnormalities observed in this study were trisomy 3, trisomy 21, monosomy X, and loss of Y, reported previously as major numerical changes in ATLL.12,17 Of these, trisomy 3, monosomy X, and loss of Y were also observed in the sideline karyotypes. Trisomy 7 is another common numerical abnormality reported in T-cell malignancies and ATLL.8,12 17 Only one patient showed simple trisomy 7, but partial trisomy of chromosome 7q resulting from unbalanced translocation, duplication, or isochromosome 7q was common in 12 patients. Partial trisomy of 7q was also observed in the sideline karyotype of 2 patients. These findings indicate that trisomy 3, partial trisomy of chromosome 7q, monosomy X, and loss of Y are not primary changes but may be secondary changes of ATLL. Trisomy 3 and partial trisomy of chromosome 7q did not correlate with clinical characteristics.
As reported in other hematologic malignancies,26-28 this study showed that karyotypic complexity, represented by the number of breaks, clearly correlated with poor prognosis (Figure 4A). Interestingly, multiple breaks (at least 6) were observed more frequently in the older age group (50 years or older) (Table 2). This correlation indicates that more genetic changes accumulate in tumor cells of the older age group than in those of the younger age group. This finding is compatible with the hypothesis of multistep carcinogenesis in ATLL.29
This study disclosed that abnormalities of chromosomes 1p, 1q, 2q, 3q, 9p, 10p, 12q, 14p, 14q, and 17q correlated with clinical severity. Of these abnormalities, involvement of chromosomes 1p, 10p, and 14q has been indicated to be specific for T-cell malignancy.4-6 17
Abnormalities of chromosome 1p are indicated to be significantly associated with T-cell NHL,16 but are also observed as primary and secondary abnormalities in B-cell NHL28,30,31and other solid tumors.23,32-35 In this study, abnormality of chromosome 1p, especially 1p22, correlated with aggressive clinical features and poor prognosis (Figure 4B). The presence of loss of heterozygosity (LOH) regions on chromosome 1p, indicating the presence of tumor suppressor genes (TSGs), has been suggested in several malignancies.33 36-40
All abnormalities of chromosome 10p observed in 8 patients were del(10)(p), and the common deleted region was located around band 10p13. In lymphoid malignancies, many del(10)(p) abnormalities have been reported in ATLL cases,17 and less frequently in cutaneous T-cell lymphoma and mycosis fungoides/Sézary syndrome.41,42 Loss of an entire copy of chromosome 10 is common in solid tumors.34 There was a noticeable correlation with unusual phenotype of ATLL cells, resulting in an aggressive clinical course. Recent studies have indicated the presence of several TSGs in the chromosome 10p region.43-45Interestingly, unbalanced translocations of chromosome 10q11 observed in 3 patients resulted in deletion of 10q11-pter.
On chromosome 14, 2 critical breakpoint-clustering regions were reported at bands q11 and q32 in ATLL. We found 14q11 abnormalities in 8 patients and 14q32 abnormalities in 5 patients. These abnormalities were not recurrent, except for del(14)(q11q13). The TCRA/Dis mapped to 14q11,46 and 14q11 is involved frequently in T-cell malignancies.4,5,47,48 Abnormality of chromosome 14q11 in ATLL was first reported by Sadamori et al,11,14and the specificity of this abnormality for ATLL has been confirmed.15-17 Within the 14q32 region, there are 2 loci often involved in B- and T-cell malignancies. One is the IGHlocus and the other is the TCL1 locus, which is often involved in T-cell prolymphocytic leukemia (T-PLL).49Chromosome 14q32 is also involved frequently in ATLL,7,10,17 but it is more commonly involved in B-cell malignancies. Recently, 2 genes (TML150 andTCL651) were cloned within 100 kb around theTCL1 locus. However, the involvement of these genes located at the 14q11 and 14q32 regions has not been confirmed in ATLL. Our recent molecular cytogenetic studies52,53 suggested a lower frequency of involvement of the TCRA than that expected by cytogenetic analysis. More interestingly, our study disclosed the presence of breakpoints clustering or deleted regions different from TCRA/D, TCL1, andIGH loci in the 14q11 and 14q32 regions. The presence of LOH in chromosome 14q has been reported in solid tumors.33,38 54-57 Abnormalities of chromosome 14q correlated with elevated LDH (1100 IU/μL or higher), unusual phenotype, and shorter survival (less than 6 months). These findings suggest the cytogenetic and clinical importance of chromosome 14q rearrangement except for the TCL1, TCRA/D, andIGH loci.
Other abnormalities correlating with clinical severity (1q, 2q, 3q, 9p, 12q, 14p, and 17q) have been reported in various hematologic malignancies and solid tumors.
Abnormalities of chromosome 1q observed in 13 patients correlated with the older age group (50 years or older), elevation of abnormal lymphocytes (15 × 106/L or higher), unusual phenotype, and shorter overall survival (Figure 4C). A correlation between abnormalities around chromosome 1q21-23 region and poor outcome is indicated in B-cell NHL.28,58 CSRP also revealed a high frequency of gain of 1q. Increased copy number of chromosome 1q has been reported by cytogenetic and molecular studies in NHL and solid tumors, and a correlation between gain of 1q and disease progression has been indicated.23,35 59-61
Structural abnormalities of chromosome 3 have been commonly found in various malignancies, and we also observed these abnormalities frequently in this study. Not only deletion, but also various structural abnormalities were found in chromosome 3q, and the breakpoints were clustered at bands q10-12, q21, and q25. Abnormality of 3q (and bands 3q10-12, 3q21) was clearly correlated with poor prognosis in this study (Figure 4D). Involvement of specific regions on chromosome 3q may play an important role in progression of ATLL.
Correlation between chromosome 9p abnormalities and poor outcome has been reported in childhood acute lymphoblastic leukemia.62Deletion of the putative TSGsMTS1/CDK4I/p16INK4A(CDK2) and MTS2/p15INK4B(CDK2B), which are mapped to 9p21-22, was found to correlate with the development and poor outcome of ATLL.63 64 Abnormalities of 9p were observed in 8 patients, and 6 patients with imbalance (loss) of the 9p22-pter region showed shorter overall survival.
Abnormality of chromosome 17 has been shown to correlate with poor prognosis.65-67 The tumor suppressor gene TP53is mapped to chromosome 17p13, and the alteration has been described in lymphoid malignancies, including ATLL.68 LOH of 17q25 region has also been reported in solid tumors.55,69 70 In this study, abnormality of 17p did not correlate with prognosis, but abnormality of 17q (all 5 abnormalities were observed in 17q23-25 region) clearly correlated with shorter survival (Figure 4E), indicating that these abnormalities involve important genes participating in leukemogenesis of ATLL.
Abnormalities of chromosomes 6q, 7, and 13q were also observed frequently in this study and were reported to be major abnormalities in ATLL. In particular, several studies indicated the specificity of del(6)(q) abnormality in ATLL.17 However, the correlation with aggressive clinical course, hypercalcemia, and poor outcome described by Whang-Peng et al13 was not found in this study. Abnormal elevation of white blood cell count correlated with abnormalities of band 7q10-11, but did not correlate with prognosis. The TCRB and TCRG genes have been mapped to chromosome 7q35 and p15, respectively. However, breakpoints were not clustered at these loci in ATLL. Deletion of 13q is reported in progression of B-cell NHL,31,71 and correlation with poor prognosis is indicated.71 In this study, many of the 13q abnormalities were deletion of 13q telomeric region (q22 terminal) different from 13q14, which has been reported to be frequently involved in B-cell chronic lymphocytic leukemia and retinoblastoma.4,5 Abnormality of chromosome 13q did not correlate with clinical characteristics. In addition, abnormalities of chromosomes 6q, 7, and 13 have been frequently observed in various B-cell malignancies and solid tumors.4 5 Their clinical relevance seems to be limited in ATLL.
More than one of the abnormalities described here frequently occurred concomitantly. Interestingly, a similar concomitant correlation between 1p and 14q was observed in advanced stages of malignant meningioma and neuroblastoma by cytogenetic and molecular genetic analyses,32,33 36-38 indicating the possibility that abnormalities of 1p and 14q contribute to disease progression of ATLL.
Abnormalities resulting in chromosomal imbalance regions, such as unbalanced translocation, deletion, duplication, or triplication, were commonly observed in this study. Imbalance patterns observed in some specific chromosomes showed striking similarity to those observed in solid tumors. In addition, characteristic cytogenetic subgroups were not found. These cytogenetic findings are similar to those observed in many solid tumors. ATLL is classified to T-cell malignancies, and involvement of 14q11 region has been reported cytogenetically.11,14-17 The molecular involvement ofTCRA/D, located at 14q11, has been shown repeatedly in T-cell malignancies, most of which are T-cell acute lymphoblastic leukemia (T-ALL) and T-PLL.47,48 By this analogy, the similar mechanism of oncogenesis that translocation ofTCRA/D cause the deregulation of proto-oncogene was speculated in the development of ATLL.47,72However, lack of recurrent balanced rearrangements, presence of many chromosomal imbalance regions, and the molecular complexity of 14q11 and 14q32 regions52 53 indicate that the above paradigm of proto-oncogene deregulation may not be applicable to molecular pathogenesis of ATLL.
Why the same T-cell malignancies show different cytogenetic and clinical characteristics between T-ALL/T-PLL and ATLL is a difficult but interesting question. T-ALL is a prethymic T-cell malignancy and involvement of the TCRA/D by specific translocations occurs frequently in the Dδ-Jδ region,48 and theTCRD locus is not deleted. Many involved genes by translocations of TCR genes in T-ALL are transcription factors, which are closely related to the early lymphocyte differentiation. T-PLL is classified as a postthymic T-cell malignancy, and abnormal T-PLL cells show mature T-cell phenotype.49The Jα region of the TCRA is involved in specific translocations associated with T-PLL.73 Normally, the expression of the TCL1, which is activated by abnormal translocations of TCR genes in T-PLL, is observed in early T- and B-cell progenitors.49 In contrast, we previously reported the biallelic functional deletion of the TCRD locus in all 6 analyzed patients with ATLL.74 Our recent study of 50 cases of ATLL also revealed that the involvement of theTCRA/D locus was less frequent compared with T-ALL and T-PLL, and these rearrangements occurred within the TCRAVregion.52 Actually, HTLV-1 infects mainly CD4+CD8− mature T cells, which is the common phenotype of ATLL cells, and the TCRD locus of mature T cells is functionally deleted biallelically during the T-cell maturation process.75 These findings indicate that acquired abnormal translocations of TCRA/D locus occur at different differentiation stages of T-cell maturation between T-ALL/T-PLL and ATLL. Cytogenetic and molecular genetic differences between T-ALL/T-PLL and ATLL may be based on modified status of chromatin and kinds of activated or inactivated genes corresponding to T-cell differentiation stage when T cells are involved in the malignant process. Lower frequency of involvement of chromosomes 7p15, 7q35, and 14q11 at TCR gene loci in other mature T-cell malignancies may be based on a similar situation of T-cell differentiation. In other words, the mechanism of leukemogenesis speculated for T-PLL in ataxia-telangiectasia47 72 cannot be considered as the main pathway of development of ATLL and other mature T-cell malignancies.
It is now accepted that cancer is a multistep process.70Our cytogenetic findings of ATLL are compatible with this concept. Okamoto et al29 proposed 5 steps of multiple events that are needed for leukemogenesis of ATLL. Actually, alterations of several known TSGs, such as TP53,68Rb1,76MTS1/CDK4I/p16INK4A(CDK2), and MTS2/p15INK4B(CDK2B),63,64 have been found in ATLL. Similar to the role of Epstein-Barr virus in oncogenesis of Burkitt lymphoma, HTLV-1 may not be oncogenic directly.47 Recent studies indicate that HTLV-1 induces DNA instabilities by interference with mitotic checkpoint function77 or defects in DNA repair function.78 The stepwise process and accumulation of chromosomal abnormalities may result in oncogenesis, and variation of these processes and involved genes may show marked clinical diversities of ATLL. We cannot exclude the possibility that unidentified recurrent translocations cause the activation of proto-oncogenes. However, it should be noted that the possible activation of proto-oncogenes is one event in the sequential multistep process of carcinogenesis.
This study revealed that several breakpoints were clustered at regions where LOH was frequently reported in other malignancies and were of clinical relevance. Many efforts have been focused on studying LOH regions. It is still difficult work to narrow the specific regions for oncogenesis. To characterize these breakpoints observed in ATLL may be extremely helpful in identifying the responsible genes, not only for the development of ATLL but also for the oncogenesis of other malignancies.
We thank Dr Gouri Nanjungud for helpful comments on the manuscript and Amelia Panico for artwork assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Takahiro Itoyama, Laboratory of Cancer Genetics, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021; e-mail: itoyamat@mskcc.org.