The molecular cloning of the t(5;10)(q33;q22) associated with atypical chronic myeloid leukemia (CML) is reported. Fluorescence in situ hybridization (FISH), Southern blot, and reverse transcriptase– polymerase chain reaction analysis demonstrated that the translocation resulted in an H4/platelet-derived growth factor receptor βR (PDGFβR) fusion transcript that incorporated 5′ sequences from H4 fused in frame to 3′PDGFβR sequences encoding the transmembrane, WW-like, and tyrosine kinase domains. FISH combined with immunophenotype analysis showed that t(5;10)(q33;q22) was present in CD13+ and CD14+ cells but was not observed in CD3+ or CD19+ cells. H4 has previously been implicated in pathogenesis of papillary thyroid carcinoma as a fusion partner of RET. The H4/RET fusion incorporates 101 amino acids of H4, predicted to encode a leucine zipper dimerization domain, whereas the H4/PDGFβR fusion incorporated an additional 267 amino acids of H4. Retroviral transduction of H4/PDGFβR, but not a kinase-inactive mutant, conferred factor-independent growth to Ba/F3 cells and caused a T-cell lymphoblastic lymphoma in a murine bone marrow transplantation assay of transformation. Mutational analysis showed that the amino-terminal H4 leucine zipper domain (amino acids 55-93), as well as H4 amino acids 101 to 386, was required for efficient induction of factor-independent growth of Ba/F3 cells. Tryptophan-to-alanine substitutions in the PDGFβR WW-like domain at positions 566/593, or tyrosine-to-phenylalanine substitutions at PDGFβR positions 579/581 impaired factor-independent growth of Ba/F3 cells. H4/PDGFβR is an oncoprotein expressed in t(5;10)(q33;q22) atypical CML and requires dimerization motifs in the H4 moiety, as well as residues implicated in signal transduction by PDGFβR, for efficient induction of factor-independent growth of Ba/F3 cells.
Introduction
Constitutive activation of tyrosine kinases plays an important role in the pathogenesis of solid tumors and hematological malignancies. Chromosomal translocations and somatic mutations can cause deregulation of tyrosine kinase activity, and an emerging number of chromosomal translocations involving tyrosine kinases have been identified in leukemia.1 An important consequence of these translocations is the expression of fusion proteins with constitutive tyrosine kinase activity. The most studied example is the t(9;22)(q34;q22), resulting in expression of the BCR/ABL fusion, which is present in approximately 95% of patients with chronic myeloid leukemia (CML).2 Distinct genomic breakpoints within theBCR gene result in different BCR/ABL fusion variants (p230, p210, and p190). BCR causes oligomerization and activation of the ABL tyrosine kinase activity and a broad spectrum of downstream effectors, leading to transformation of hematopoietic cells.2 Other fusion genes involving tyrosine kinases associated with hematopoietic disorders include TEL/ABL, TEL/JAK2, and TEL/TRKC fusions involving the FGFR1 gene (ZNF198/FGFR1, CEP110/FGFR1,and FOP/FGFR1) and fusions involving the platelet-derived growth factor receptor β (PDGFβR)gene.3-6 PDGFβR, a member of the PDGF receptor family that includes the PDGFαR, colony-stimulating factor 1 receptor, steel-factor receptor (c-kit) and Flt3/FLK receptor, is characterized structurally by 5 immunoglobulinlike extracellular loops and a split intracellular tyrosine kinase domain.7The first translocation involving the PDGFβR gene was cloned from a chronic myelomonocytic leukemia (CMML) patient with t(5;12)(q33;p13), resulting in fusion of the 5′ terminal region of theTEL(ETV6) gene (a member of the ets family of transcription factors) to the transmembrane and tyrosine kinase domains of the PDGFβR.8 Two other PDGFβR fusion genes have subsequently been cloned. The t(5;14)(q33;q32) and t(5;7)(q33;q11.2) rearrangements give rise to the CEV14/PDGFβR andHuntingtin-interacting protein 1 (HIP1)/PDGFβR fusion genes, respectively.9,10 TheTEL/PDGFβR and HIP1/PDGFβR fusion genes have been associated with CMML, whereas theCEV14/PDGFβR fusion was cloned from a patient with acute monocytic leukemia in relapse. CMML is a subtype of myelodysplastic syndrome characterized by dysplastic monocytosis, variable bone marrow fibrosis, and progression of acute leukemia; its clinical phenotype is similar to CML, but is lacking the Philadelphia chromosome.11 TEL/PDGFβR and HIP1/PDGFβR transform hematopoietic cells in vitro and in vivo.12-15
Constitutive activation of tyrosine kinases due to chromosomal rearrangements or somatic mutations is also observed in solid tumors.16 Overexpression and somatic mutation ofHER-2/p185neu and cMET are observed in breast carcinomas and renal or thyroid carcinoma.17,18Alterations of the RET proto-oncogene, which encodes for a receptor tyrosine kinase, are responsible for the development of multiple endocrine neoplasia types 2A and 2B, Hirschsprung disease, and papillary thyroid cancer.19 Gene rearrangements leading to fusion of its tyrosine kinase domain to the 5′ terminal region of other genes generate the RET/PTC oncogenes, which are associated with human papillary carcinoma.20 Seven different types of RET rearrangement (PTC 1-7) have been molecularly characterized.21RET/PTC1 is the result of a paracentric inversion of the long arm of chromosome 10 inv(10)(q11.2q21), fusing the terminal amino acid 101 of theH4(D10S170) gene to the intracellular split tyrosine kinase domain of RET. This rearrangement is observed in approximately 20% of human papillary carcinomas.22 TheH4(D10S170) gene encodes for a 585–amino acid protein with no significant homology to known genes and has unknown function.23 In vitro studies showed that the leucine zipper region of H4 included in the fusion is responsible for the dimerization of the PTC1 oncoprotein and is essential for tyrosine hyperphosphorylation and transformation in vitro.24
Here we present the molecular characterization of a patient with atypical CML with t(5;10)(q33;q22). Initial classical karyotypic analysis showed a reciprocal translocation t(5;10) in myeloid progenitor cells of the patient.25 Using fluorescence in situ hybridization (FISH), Southern blot, and rapid amplification of complementary DNA (cDNA) ends (RACE)–polymerase chain reaction (PCR) techniques, we show that the translocation is present only in the patient's myeloid cells and results in fusion of theH4(D10S170) gene to the transmembrane and tyrosine kinase domains of the PDGFβR. In vitro experiments show that expression of the H4/PDGFβR fusion gene transforms interleukin-3 (IL-3)–dependent Ba/F3 cells to factor independence. Moreover, in vivo transforming activity of the H4/PDGFβR fusion is demonstrated in a murine bone marrow reconstitution assay. These data show that rearrangements of the H4(D10S170) gene can not only contribute to carcinogenesis by activating RET in papillary carcinomas but also lead to hematopoietic transformation in anH4/PDGFβR fusion gene associated with atypical CML.
Materials and methods
FISH and “fiction” analysis
FISH was performed as previously described.26 The breakpoint on chromosome 5 was investigated by means of the cosmid probe cosmid B (cosB) (kindly provided by Dr W. M. Roberts, M. D. Anderson Cancer Center, Houston, TX),8assigned to the PDGFβR gene at band 5q33. The breakpoint on chromosome 10 was narrowed by applying 2 yeast artificial chromosomes (YACs), 781F5 and 876H2, for bands 10q21.3 and 10q22.1, respectively. In each experiment, the centromeric chromosome 10 probe D10Z1 was added. Dual-color FISH was performed with cosB labeled with digoxigenin, andPAC29F6 for the H4/D10S170 gene,27labeled with biotin.
The “fiction” method was adapted from Weber-Matthiesen et al.28 Cytospins were prepared from mononuclear cells that were obtained from the patient's peripheral blood and stored at −20°C without fixation. After thawing, the slides were fixed in acetone at room temperature for 10 minutes and air-dried. They were then incubated with a monoclonal antibody for 30 minutes at room temperature, and staining was performed in a 3-step technique with the following antibodies: Cy3-conjugated polyclonal goat antimouse, rabbit antigoat, and donkey antirabbit (Jackson Laboratory, Bar Harbor, ME; Dianova, Hamburg, Germany); and the following monoclonal antibodies: anti-CD3, anti-CD13, anti-CD14, anti-CD20, and anti-CD19 (all from Dako, Glostrup, Denmark). After immunostaining, the slides were fixed in Carnoy fixative (methanol-to-acetic acid, 3:1) for 10 minutes, followed by 1% paraformaldehyde for 10 minutes, washed in distilled water; dehydrated in an ethanol series; and hybridized with cosB for the PDGFβR. The biotinylated DNA was detected by incubation with fluorescein isothiocyanate (FITC)–conjugated avidin, followed by biotinylated goat antiavidin antibody, followed by a second incubation with FITC-conjugated avidin. The immunophenotype and hybridization signals were evaluated simultaneously on an Olympus microscope with filter sets for Cy-3 and FITC (Olympus Optical, Japan).
DNA isolation and Southern blot analysis
DNA was isolated from buffy-coat preparations of peripheral blood from the patient and a normal individual after informed consent had been obtained. The patient's clinical history has been described previously.25 DNA was prepared by means of a Purgene DNA isolation kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's protocol. After enzymatic digestion with restriction endonucleases and electrophoretic separation, the genomic DNA was blotted to Hybond-N+ nylon membranes (Amersham, Arlington Heights, IL) by alkaline transfer. The PDGFβR genomic probe was a 1.1-kilobase (kb) HindIII-XhoI fragment prepared from PDGFβR cosB.8 Probes were labeled with 32P by random priming, and Southern hybridizations were performed as described.29
Breakpoint cloning
We isolated 1.6 μg poly(adenylic acid), or poly(A), RNA from 60 × 106 cells from the patient's buffy-coat cells using the Quickprep Micro messenger RNA purification kit (Pharmacia Biotech, Uppsala, Sweden). Anchored PCR was performed to clone the chromosome 10 partner gene by means of a 5′-3 RACE kit (Boehringer Mannheim, Mannheim, Germany). In brief, 500 ng poly(A) RNA was reverse-transcribed by means of avian myeloblastosis virus (AMV) reverse transcriptase and PDGFβRoligonucleotide primer 1873R at 55°C. PDGFβRprimers have been previously described in detail.8 10 A poly-A tail was appended to the purified cDNA by means of terminal transferase and deoxyadenosine triphosphate. The tailed cDNA was amplified (94°C for 2 minutes, 94°C for 15 seconds, annealing at 58°C for 30 seconds, elongation for 40 seconds, and cycle elongation of 20 seconds after 10 cycles of a total of 35 cycles) by means of oligo deoxythymidine anchor primer (5′-GAC CAC GCG TAT GCA TGT CGA CTT TTT TTT TTT TTT TT-3′) and an internal PDGFβRprimer 1848R. We re-amplified 1 μL of the diluted first-round PCR product (1:20) in a nested PCR reaction using the same conditions as the first round with a PCR anchor primer (5′-GAC CAC GCG TAT CGA TGT CGA C-3′) and the PDGFβR internal primer 1829R. Specific bands were detected after 2 rounds of PCR by direct visualization on an agarose gel. Two fragments, 250 base pairs (bp) and 400 bp, appeared in 2 separate experiments and were subcloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and sequenced. The DNA sequence was then compared with sequences in the GenBank by means of the advanced BlastN screening.
Reconstruction of a full-length fusion cDNA for expression experiments
A full-length cDNA clone of the fusion was generated as follows. The H4/PDGFβR breakpoint was amplified by means of a forwardPDGFβR primer with H4-specific anchor that incorporates a unique StuI site 5′ of the breakpoint, together with a PDGFβR reverse primer covering a unique SacII site 250 bp 3′ of the breakpoint by means of Pfu-polymerase (Stratagene, San Diego, CA) according to the manufacturer's recommendations. The product was subcloned into pcDNA3 (Invitrogen) containing an H4(D10S170) full-length cDNA clone in reverse orientation, cloned from pGEM3Z-H4 cDNA,23 and sequenced to confirm that no mutations had been introduced in the PCR step. The 3′ end of the PDGFβR cDNA was added by isolating the fragment from pBluescript.TEL/PDGFβR cDNA8 and cloning it into the unique SacII site. This full-length reverse H4/PDGFβR clone was then subloned inpBluescript, pMSCVneo, and pMSCV-GFP (the pMSCV retroviral expression vectors were kind gifts from R. Hawley, University of Toronto, ON, Canada, and W. Pear, University of Pennsylvania, Philadelphia). Expression of the H4/PDGFβR fusion protein from the full-length H4/PDGFβR cDNA clone was confirmed by means of an in vitro transcription-translation reaction kit (Promega, Madison, WI) (not shown).
Expression of H4/PDGFβR in primary patient cells
Expression of the H4/PDGFβR fusion was studied in peripheral blood cells (buffy-coat preparation) from the patient and in cells of a normal healthy donor. Total RNA was isolated by means of an RNA isolation kit (RNA-STAT-60) (Tel-Test, Friendswood, TX) followed by reverse transcription of 1 μg total RNA by means of AMV-RT (1st Strand cDNA Synthesis Kit) (Boehringer Mannheim) following the manufacturer's instructions. A nested-PCR approach was used as follows: H4.300F (5′CAA GCC AGG GCT GAG CAG GAA GAA TTC 3′) and H4.720F (5′GCT CCA CCA TCG CCT AGA GAT ATC TCC ATG 3′), each withPDGFβR (1848R) for the first cycle; 1 μL of a 1:10 dilution of the first round was used for the second round with the same H4 primers in combination with the PDGFβR (1829R) primer. The PCR cycle parameters were 10 cycles at 94°C for 30 seconds, 65°C (decreasing 1°C per cycle) for 30 seconds, and 72°C for 30 seconds, followed by 30 cycles of 94°C, 55°C, and 72°C for 30 seconds each and a final extension of 7 minutes at 72°C. To detect the reverse PDGFβR/H4 product, the PDGFβR.F1 (5′GGA GAC TAA CGT GAC GTA CTG 3′) primer was used for the first round in combination with H4.R1 (5′CAG GAC TGT TGC TTC TCC GTG 3′) and H4.R2 (5′GCT CCA TTG GAT GAG TCC CAA C 3′) primers; for the second round, the PDGFβR.F2 (5′GAG TTT GAG GTG GTG AGC AC 3′) primer was used with the same H4 primers. For amplification of the H4 gene, primers H4.300F and H4.720F with H4.R2 were used for the first round, followed by a second round with the H4.R1 reverse primer. To assess the quality of the transcribed cDNA, primers for β-actin were used: β-actin.2282F (5′GGG AAA TCG TGC GTG ACA TT 3′) and β-actin.2583R (5′GGA GTT GAA GGT AGT TTC GTG 3′).
Generation of H4/PDGFβR mutants and transformation studies
Mutants were generated by PCR by means ofPfu-polymerase (Stratagene) according to the manufacturer's recommendations and subcloned into the retroviral expression vectorMSCV-PGK-neo and MSCV-IRES-GFP. As shown in Figure 5A, mutations that were generated includedMSCV-H4/PDGFβR-WW (W → A [single-letter amino acid codes] substitutions at positions 566/593 of the PDGFβR moiety); an H4/PDGFβR-F2 mutant analogous to theTEL/PDGFβR-F2 previously described15; a kinase-inactiveMSCV-H4/PDGFβR−634R mutant,12MSCV-H4/PDGFβRΔLZ lacking the H4(D10S170) leucine zipper domain (H4 amino acids 55-93); andMSCV-H4/PDGFβR-ΔEX lacking amino acids 101-368 ofH4(D10S170) between the H4/RET andH4/PDGFβR breakpoints. All PCR-generated fragments were sequenced after subcloning to confirm the correct sequence, and the cDNA clones were translated in vitro to confirm expression of a protein of the expected molecular weight. To test for transforming ability,H4/PDGFβR clones were expressed in the IL-3–dependent hematopoietic cell line Ba/F3, by retroviral transduction with the use of retroviral supernatants generated by transient double transfection of pMSCV and pIk6 into 293T cells as described previously.29 At 48 hours after infection, cells were washed and screened for survival in medium lacking IL-3. The same experiment was also performed by means of stably transfected cells withMSCV-H4/PDGFβR-PGK-neo and the mutants, preselected in G418 (1 mg/mL), for 7 to 10 days.29
To characterize the in vivo transforming activity of the H4/PDGFβR fusion, a murine bone marrow transplantation (BMT) assay was performed as described previously.15,29 Briefly, donor mice were treated with 5-FU (150 mg/kg) 5 days prior to harvest of bone marrow. Bone marrow cells were incubated for 48 hours ex vivo in media that included IL-3, IL-6, and stem cell factor as described15,29 and transduced by spinfection with viral supernatants of the murine ecotropic retrovirus MSCV containing the constructs noted in the text. One million transduced whole bone marrow cells were then injected by tail vein into lethally irradiated syngeneic recipeint mice (Balb/C, 450 cGY × 2) as previously described.15,29 A total of 16 recipient mice were transplanted with bone marrow transduced with MSCV retrovirus containing the full-length H4/PDGFβR fusion gene. Preparation of tissues for histologic examination and flow cytometric analysis was performed as described previously.29
Results
The PDGFβR gene is rearranged in myeloid lineage cells harboring the t(5;10)(q33;q22)
To localize the breakpoint, FISH of metaphase chromosome preparations of bone marrow cells from the patient with atypical CML and t(5;10) as a single karyotypic abnormality was performed. As described previously, the abnormal karyotype, as assessed by conventional cytogenetics, was present in myeloid precursor cells.25
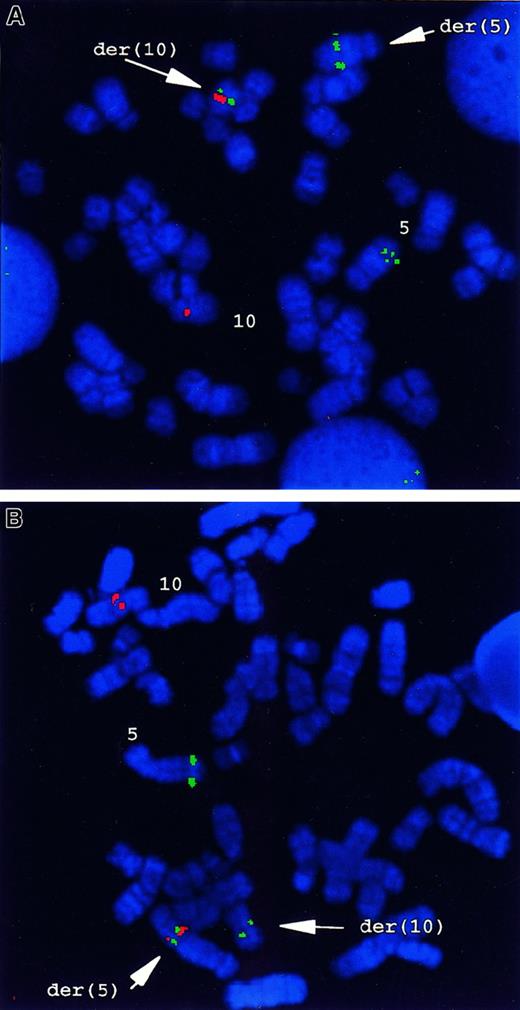
FISH analysis with a chromsome 5q33–specific probe, cosB,which includes a portion of the genomic sequence of the PDGFβRgene, gave a hybridization signal on the normal 5, and a split hybridization signal on the der(5) and the der(10) generated by the t(5;10) (Figure 1A). YACs 781F5 and 876H2, which are specific for 10q21.3 to q22.1, hybridized with the normal chromosome 10 and with der(5). No signals were present on der(10) (Figure 1B). To assess whether t(5;10) was present only in the patient's myeloid cells, a combination of immunostaining and FISH was performed. As shown in Figure 2A-B, hybridization with cosB resulted in 3 hybridization signals in myeloid progenitor cells (CD 13+, CD14+), whereas only 2 signals were observed in T (CD3+) or B lineage (CD19+, CD20+) cells (Figure 2C-D). Within the sensitivity of this assay, these data indicate the presence of the t(5;10) balanced reciprocal in myeloid lineage cells, but not in lymphoid cells of this patient with atypical CML.
FISH analysis of the t(5;10)(q33;q22) in primary patient cells.
(A) FISH analysis using PDGFβR- andH4-locus–specific probes. CosB, a PDGFβR-specific genomic probe, gave 3 hybridization signals on normal 5 and on der(5) and der(10) originating from t(5;10), respectively. Both YACs 781F5 and 876H2, for 10q21.3 to q22.1, hybridized with the normal chromosome 10 and with the der(5). No signals were present on der(10). (B) Dual-color FISH with CosB (red) and PAC29F6 (green) showed the presence of a fusion signal on der(5), whereas a green signal (PAC29F6) was present on the normal 10, and a red signal was seen (CosB) on the normal 5 and the der(10).
FISH analysis of the t(5;10)(q33;q22) in primary patient cells.
(A) FISH analysis using PDGFβR- andH4-locus–specific probes. CosB, a PDGFβR-specific genomic probe, gave 3 hybridization signals on normal 5 and on der(5) and der(10) originating from t(5;10), respectively. Both YACs 781F5 and 876H2, for 10q21.3 to q22.1, hybridized with the normal chromosome 10 and with the der(5). No signals were present on der(10). (B) Dual-color FISH with CosB (red) and PAC29F6 (green) showed the presence of a fusion signal on der(5), whereas a green signal (PAC29F6) was present on the normal 10, and a red signal was seen (CosB) on the normal 5 and the der(10).
Simultaneous phenotypic and genotypic analysis on a cytospin prepared from peripheral blood cells.
Monoclonal antibody (red), PDGFβR probe (green). (A, B) CosB gave 3 hybridization signals on CD13+ or CD14+ cells, respectively, indicating the presence of the t(5;10)(q33;q22) with a split PDGFβR signal in these myeloid lineage cells. (C, D) Only 2 signals were present in the CD3+ or CD19+ cells, respectively, indicating that these lymphoid cells lack the translocation.
Simultaneous phenotypic and genotypic analysis on a cytospin prepared from peripheral blood cells.
Monoclonal antibody (red), PDGFβR probe (green). (A, B) CosB gave 3 hybridization signals on CD13+ or CD14+ cells, respectively, indicating the presence of the t(5;10)(q33;q22) with a split PDGFβR signal in these myeloid lineage cells. (C, D) Only 2 signals were present in the CD3+ or CD19+ cells, respectively, indicating that these lymphoid cells lack the translocation.
The clinical, cytogenetic, and FISH data from this patient were similar to those observed in previously reported cases of CMML with the t(5;12)(q33;p12) TEL/PDGFβR and the t(5;7)(q33;q11.2) HIP1/PDGFβR fusion genes, respectively.8,10 In those previous examples, the PDGFβR cases, the chromosomal breakpoint was identified by means of a 1.1-kbXhoI/HindIII genomic PDGFβR probe derived from cosB.8,10 Southern blot analysis using this genomic PDGFβR fragment as a probe demonstrated rearrangement of the PDGFβR gene in 3 out of 5 restriction-enzyme digests of the patient's DNA, but not in the normal cDNA (Figure 3). These data demonstrated that the chromosome 5 breakpoint was at or near the same intron of thePDGFβR gene as in the t(5;12)(q33;p13) and t(5;7)(q33;q11.2) translocations.8 10
Southern blot analysis of DNA from leukemia cells with t(5;10)(q33;q22).
Genomic DNA was isolated mononuclear cells from Ficoll-Hypaque density gradient sedimentation of peripheral blood from the patient and a normal donor and was digested (10 μg) with 5 different restriction endonucleases. The Southern blot was probed with a 1.1-kb (XhoI/HindIII) genomic PDGFβR probe. Rearrangements were evident in patient DNA when the restriction endonucleases HindIII, PstI, or PvuI were used (lanes 3-8).
Southern blot analysis of DNA from leukemia cells with t(5;10)(q33;q22).
Genomic DNA was isolated mononuclear cells from Ficoll-Hypaque density gradient sedimentation of peripheral blood from the patient and a normal donor and was digested (10 μg) with 5 different restriction endonucleases. The Southern blot was probed with a 1.1-kb (XhoI/HindIII) genomic PDGFβR probe. Rearrangements were evident in patient DNA when the restriction endonucleases HindIII, PstI, or PvuI were used (lanes 3-8).
Identification of the H4(D10S170) gene as fusion partner to the PDGFβR gene in t(5;10)(q33;q22)
The chromosome 10 partner was identified by means of anchored PCR with PDGFβR primers to amplify the fusion transcript from the patient's peripheral blood cell cDNA. Analysis of the amplified cDNA clones showed 2 clones of 250 and 400 bp of non-PDGFβR sequence encoding an open reading frame fused to the transmembrane and tyrosine kinase–encoding regions of thePDGFβR gene (Figure 4A). A database search demonstrated that this sequence was identical to the coding sequence of the H4(D10S170) gene located on chromosome 10q21.23
Structure and RT-PCR of H4/PDGFβR fusion.
(A) Structure of the H4/PDGFβR fusion. H4 is fused at amino acid 381 in frame to the PDGFβR gene beginning at amino acid 528. The breakpoint within PDGFβRis identical to that in the TEL/PDGFβR andHIP1/PDGFβR fusions. (B) RT-PCR of the H4/PDGFβRfusion. RNA was extracted from leukemic cells containing the t(5;10)(q33;q22) and from a normal donor and was used as a template for RT-PCR as described in “Materials and methods.” Two sets ofH4/PDGFβR primers, denoted 300F and 720F, were used.H4/PDGFβR-specific transcripts were detected in the leukemic sample but not in the normal control. (C) RT-PCR ofH4(D10S170) was also performed to demonstrateH4(D10S170) expression in both normal and patient samples. As expected, signal was seen in patient, donor, and H4containing plasmid with the use of the 720F primer set, but not in theH4/PDGFβR plasmid.
Structure and RT-PCR of H4/PDGFβR fusion.
(A) Structure of the H4/PDGFβR fusion. H4 is fused at amino acid 381 in frame to the PDGFβR gene beginning at amino acid 528. The breakpoint within PDGFβRis identical to that in the TEL/PDGFβR andHIP1/PDGFβR fusions. (B) RT-PCR of the H4/PDGFβRfusion. RNA was extracted from leukemic cells containing the t(5;10)(q33;q22) and from a normal donor and was used as a template for RT-PCR as described in “Materials and methods.” Two sets ofH4/PDGFβR primers, denoted 300F and 720F, were used.H4/PDGFβR-specific transcripts were detected in the leukemic sample but not in the normal control. (C) RT-PCR ofH4(D10S170) was also performed to demonstrateH4(D10S170) expression in both normal and patient samples. As expected, signal was seen in patient, donor, and H4containing plasmid with the use of the 720F primer set, but not in theH4/PDGFβR plasmid.
The H4(D10S170) cDNA is ubiquitously expressed and encodes a protein of unknown function and no significant homology to any mammalian gene. The protein has a predicted alpha helical conformation similar to the myosin heavy-chain tail, 2 putative leucine zipper domains, and a putative SH3 domain at the C-terminus, and it has been suggested to be a cytoskeletal protein.23H4(D10S170) is involved in RET rearrangements as a result of a paracentric inversion of chromosome 10q inv(10)(q11.2q21) associated with about 25% of human papillary thyroid carcinomas.20,23,30 In these carcinomas, the genomic breakpoint in H4(D10S170) occurs in intron 1.31,32 The resulting oncogene RET/PTC1(papillary thyroid carcinoma gene 1) is formed by in-frame fusion of 5′ sequences from the H4(D10S170) gene (amino acids 1-101) to the RET gene, 39 bp 3′ of RET tyrosine kinase–coding sequences.20 In contrast toRET/PTC1 (Figure 4A), the breakpoint within theH4 gene in t(5;10)(q33;q21) lies 3′ of the breakpoint in theH4/RET fusion and fuses the first 368 amino acids of H4(D10S170) to the transmembrane and tyrosine kinase domains of thePDGFβR. To confirm expression of theH4/PDGFβR fusion transcript in primary leukemic cells from the patient, reverse transcriptase (RT) PCR analysis was performed with 2 different pairs of H4/PDGFβR primers.H4/PDGFβR fusion transcripts were detected in the patient's peripheral blood cells but not in the normal control (Figure4B). Expression of the reciprocal PDGFβR/H4 fusion was not detected (data not shown), which is similar to results obtained in patient samples harboring the TEL/PDGFβR andHIP1/PDGFβR gene rearrangements.8,10 The normal H4(D10S170) allele is deleted in a papillary thyroid cancer cell line (TPC-1) carrying a RET/PTC1 fusion gene.27 However, FISH analysis of t(5;10)(q33;q21) cells showed no evidence of loss of the residual allele, and theH4(D10S170) transcript was detectable in leukemic cells containing the translocation, indicating the presence of a functional residual H4(D10S170) allele (Figure 4C). Collectively, these data demonstrate that t(5;10)(q33;q21) results in fusion ofH4(D10S170) to the PDGFβR and that theH4/PDGFβR fusion transcript, but not the reciprocal, is expressed in the patient's myeloid lineage cells.
The H4/PDGFβR fusion gene has transforming activity in vitro and in vivo
To characterize the biological properties of the H4/PDGFβR fusion, a full-length cDNA clone was constructed and subcloned intoMSCV-PGK-neo and MSCV-IRES-GFP retroviral expression vectors (Figure 5A). Murine IL-3–dependent hematopoietic Ba/F3 cells were retrovirally transduced with MSCV-H4/PDGFβR and assayed for growth-factor independence. Ba/F3 cells expressing H4/PDGFβR were generated either by retroviral transduction with MSCV-H4/PDGFβR-neo and selection with G418 or by transduction withMSCV-H4/PDGFβR-GFP without selection (data not shown). These cells were able to sustain log-phase growth in the absence of IL-3, whereas Ba/F3 cells transduced with empty vector died rapidly after IL-3 depletion (Figure 5B). In vitro transforming activity of the H4/RET and the TEL/PDGFβR is dependent on the presence of the leucine zipper and pointed dimerization domains in H4 and TEL, respectively.12 24 To determine which structural components of the H4/PDGFβR fusion were required for transformation of Ba/F3 cells to factor-independent growth, several mutants were constructed (Figure 5A). MSCV-H4/PDGFβRΔLZ lacks the 5′-leucine zipper domain (amino acids 55-93) andMSCV-H4/PDGFβR-ΔEX lacks the portion ofH4(D10S170) gene that is present in theH4/PDGFβR fusion but absent in the correspondingH4/RET fusion (H4 amino acids 101-368). Thus, transformation of Ba/F3 cells to factor-independent growth by H4/PDGFβR, in a 4-day assay after IL-3 deprivation, required the presence of the amino-terminal leucine zipper domain. In addition, H4 coding sequences that are present in H4/PDGFβR but absent in H4/RET were required for full transforming activity (Figure 5B).
Structure and transforming properties ofH4/PDGFβR and related mutants.
(A) Schematic diagram of H4/PDGFβR fusion and related mutants. H4/PDGFβR (wt) is the wild-type full-length cDNA. H4/PDGFβR-ΔEX lacks amino acids 101 to 368 of H4; H4/PDGFβR-ΔLZ lacks the leucine zipper domain from amino acids 55 to 93 of H4 moiety;H4/PDGFβR-WW contains 2 W → A substitutions at positions 566/593 of the PDGFβR gene;H4/PDGFβR-634R is a kinase-inactive mutant; andH4/PDGFβR-F2 contains 2 Y → F mutations of residues 579/581 of the PDGFβR portion of the fusion. (B) H4/PDGFβR transformation of Ba/F3 cells. H4/PDGFβR was retrovirally transduced by retrovirus into Ba/F3 cells, followed by selection with G418 (1 mg/mL) in the presence of IL-3. After 7 days, the cells were washed and IL-3 was withdrawn. Cells were counted every 24 hours with the use of trypan blue solution (0.4%) to distinguish between viable and nonviable cells. None of the H4/PDGFβR mutants tested were able to efficiently induce IL-3–independent growth, although longer term culture allowed for eventual outgrowth of IL-3–independent clones (data not shown). Control experiments demonstrated that TEL/JAK2, but not a PGK-neo empty vector, conferred factor independence to Ba/F3 cells. (C) Expression of H4/PDGFβR and related mutants. Expression of H4/PDGFβR and related mutants was confirmed by Western blot analysis of Ba/F3 whole cell lysates by means of anti-PDGFβR antibody as described in “Materials and methods.” Expected band sizes are H4/PDGFβR, 110 kd; H4/PDGFβR ΔEX, 85 kd; and H4/PDGFβR ΔLZ, 105 kd.
Structure and transforming properties ofH4/PDGFβR and related mutants.
(A) Schematic diagram of H4/PDGFβR fusion and related mutants. H4/PDGFβR (wt) is the wild-type full-length cDNA. H4/PDGFβR-ΔEX lacks amino acids 101 to 368 of H4; H4/PDGFβR-ΔLZ lacks the leucine zipper domain from amino acids 55 to 93 of H4 moiety;H4/PDGFβR-WW contains 2 W → A substitutions at positions 566/593 of the PDGFβR gene;H4/PDGFβR-634R is a kinase-inactive mutant; andH4/PDGFβR-F2 contains 2 Y → F mutations of residues 579/581 of the PDGFβR portion of the fusion. (B) H4/PDGFβR transformation of Ba/F3 cells. H4/PDGFβR was retrovirally transduced by retrovirus into Ba/F3 cells, followed by selection with G418 (1 mg/mL) in the presence of IL-3. After 7 days, the cells were washed and IL-3 was withdrawn. Cells were counted every 24 hours with the use of trypan blue solution (0.4%) to distinguish between viable and nonviable cells. None of the H4/PDGFβR mutants tested were able to efficiently induce IL-3–independent growth, although longer term culture allowed for eventual outgrowth of IL-3–independent clones (data not shown). Control experiments demonstrated that TEL/JAK2, but not a PGK-neo empty vector, conferred factor independence to Ba/F3 cells. (C) Expression of H4/PDGFβR and related mutants. Expression of H4/PDGFβR and related mutants was confirmed by Western blot analysis of Ba/F3 whole cell lysates by means of anti-PDGFβR antibody as described in “Materials and methods.” Expected band sizes are H4/PDGFβR, 110 kd; H4/PDGFβR ΔEX, 85 kd; and H4/PDGFβR ΔLZ, 105 kd.
A recent report has proposed an important role for the WW-like domain (amino acids 566/593), which is C-terminal to the PDGFβR transmembrane region, for normal signal transduction mediated by the native PDGFβR.33 We found that a WW mutation (Figure 5A) also impaired the ability of the H4/PDGFβR fusion to confer factor-independent growth to Ba/F3 cells (Figure 5B). In addition, an H4/PDGFβR F2 mutant (Y → F mutants of the tyrosines 579 and 581, near the PDGFβR transmembrane domain) was generated34 (Figure 5A). These tyrosines are important for PDGFβR signal transduction mediated by interaction with the SH2-containing SRC protein and STAT5 family members,35 and an analogous TEL/PDGFβR-F2 mutant cannot induce myeloproliferative disease in murine BMT models.15 TheH4/PDGFβR-F2 mutant also had impaired ability to induce factor-independent growth in Ba/F3 cells (Figure 5B). Western blot analysis of whole cell lysates using an anti-PDGFβR antibody demonstrated expression of H4/PDGFβR and related mutants at the expected molecular weights. (Figure 5C). No differences in tyrosine kinase activity could be detected as a consequence of these mutations as assessed by phosphotyrosine blots to detect autophosphorylation.
We tested the transforming activity of the H4/PDGFβR fusion in vivo using a murine BMT assay. In previous studies, we demonstrated that transplantation of primary bone marrow cells that have been transduced with a retrovirus encoding a tyrosine-kinase fusion gene results in induction of a lethal hematological disease in syngeneic recipient animals.15,29,36,37 Murine bone marrow was transduced with retroviral supernatants containing MSCV-H4/PDGFβR-GFP (16 animals), MSCV-H4/PDGFβR-634R-GFP (8 animals), andMSCV-TEL/PDGFβR-GFP (8 animals), and one million cells were transplanted by tail-vein injection into lethally irradiated syngeneic hosts. Control animals that received bone marrow transduced with the TEL/PDGFβR retrovirus developed a rapidly lethal myeloproliferative disease with a latency of 25 to 30 days, as previously reported,15 whereas the kinase-inactiveMSCV-H4/PDGFβR-634R-GFP caused no disease with 8 months of follow-up. In contrast, H4/PDGFβR induced an aggressive T-cell lymphoblastic lymphoma/leukemia with prolonged latency (median, 150 days post-transplant), as assessed by morphologic (Figure6A-C) and flow-cytometric analysis (Figure 6D). Retroviral integration was confirmed by Southern blot analysis (Figure 7A), and the neoplastic cells were monoclonal in spleen and bone marrow (Figure 7B). An additional, less-intense band indicative of biclonal disease was identified in lymph node and thymus. These data demonstrate that the H4/PDGFβR fusion resulting from t(5;10)(q33;q22) has transforming potential in vitro and in vivo.
Histopathologic and flow-cytometric analysis of mice transplanted with bone marrow transduced by H4/PDGFβR retrovirus.
Mice transplanted with bone marrow transduced with H4/PDGFβRretrovirus develop a lethal hematopoietic malignancy after a latency of approximately 150 days. The disease is characterized by proliferation of immature T cells (CD4+/+ and CD8+/+ cells), with infiltration of spleen (A, high-power view), bone marrow (B, low-power view), and lymph node (C, high-power view). Flow cytometry of the lymph node (D) demonstrates a predominant CD4+/+CD8+/+ T-cell population.
Histopathologic and flow-cytometric analysis of mice transplanted with bone marrow transduced by H4/PDGFβR retrovirus.
Mice transplanted with bone marrow transduced with H4/PDGFβRretrovirus develop a lethal hematopoietic malignancy after a latency of approximately 150 days. The disease is characterized by proliferation of immature T cells (CD4+/+ and CD8+/+ cells), with infiltration of spleen (A, high-power view), bone marrow (B, low-power view), and lymph node (C, high-power view). Flow cytometry of the lymph node (D) demonstrates a predominant CD4+/+CD8+/+ T-cell population.
Analysis of proviral integration in disease tissue in transplanted mice.
Southern blot analysis was performed with 10 μg DNA extracted from murine tissues. (A) DNA was digested with the restriction enzymeNheI, which cleaves in the viral long-terminal repeat sequences and demonstrates viral integration. The expected DNA band size is 5.8 kb. (B) Clonality of the disease was assessed by digestion with EcoRI, which cleaves once within the provirus. There is clonal disease in spleen and bone marrow, and the presence of a second clone was detected in lymph node and thymus. (C) Restriction sites within the proviral sequence.
Analysis of proviral integration in disease tissue in transplanted mice.
Southern blot analysis was performed with 10 μg DNA extracted from murine tissues. (A) DNA was digested with the restriction enzymeNheI, which cleaves in the viral long-terminal repeat sequences and demonstrates viral integration. The expected DNA band size is 5.8 kb. (B) Clonality of the disease was assessed by digestion with EcoRI, which cleaves once within the provirus. There is clonal disease in spleen and bone marrow, and the presence of a second clone was detected in lymph node and thymus. (C) Restriction sites within the proviral sequence.
Discussion
We have cloned a novel fusion between theH4(D10S170) gene on chromosome 10q and PDGFβRon 5q, the result of a reciprocal t(5;10)(q33;q22) in a patient with atypical CML. H4(D10S170) encodes a ubiquitously expressed protein of unknown function and has been previously characterized as fusion partner of the RET receptor tyrosine kinase,20,22,23 whose ligand is the glial cell line–derived neurotrophic factor.38,39 TheH4/RET fusion is one of 6 known RET rearrangements associated with human papillary thyroid carcinoma and is found in approximately 25% of the cases.21
Both fusion genes involving the H4(D10S170) gene,H4/PDGFβR and H4/RET, are associated with a specific disease phenotype. Rearrangements of RET occur exclusively in papillary thyroid carcinomas, whereasPDGFβR fusions have been associated with acute myeloid leukemia and chronic myeloproliferative syndromes. Both H4 fusion genes(H4/RET and H4/PDGFβR) have transforming activity in vitro and in vivo. In vitro transforming activity of H4/RET depends on the presence of the leucine zipper domain at the 5′ end of H4(D10S170), which acts as a dimerization/oligomerization motif leading to activation of the fused kinase domain.24 In addition, targeted expression of the RET/PTC1 oncogene induces papillary thyroid carcinomas in mice.40 41
There are several differences between H4/PDGFβR and H4/RET. TheH4/RET genomic breakpoint occurs in H4 intron 1 and thus incorporates only the first exon of the H4(D10S170)gene. In contrast, the H4/PDGFβR contains an additional 801 bp of H4(D10S170) coding sequence. The function of this domain is unknown, although a protein sequence analysis reveals a series of 5 × 29 tandem repeats (positions 106-235, SWISS-PROT Q16204) and a small second putative leucine zipper domain in this region (positions 265-278, Pfam 4.2, HMM search). H4/PDGFβR conferred factor-independent growth to the IL-3–dependent hematopoietic cell line Ba/F3 cells. Transformation activity was significantly impaired in H4/PDGFβR-ΔLZ mutant lacking the amino-terminal leucine zipper domain, as well as theH4/PDGFβR-ΔEX mutant in which H4 coding sequences unique to the H4/PDGFβR fusion were deleted. These data suggest that there are functional components in each of these domains that are required for full transforming activity in the Ba/F3 assay. Additional mutational analysis will be required to further delineate these critical regions.
Mutations of the juxtamembrane tyrosine residues or in the WW-like domain of the H4/PDGFβR also impaired factor-independent growth of Ba/F3 cells. First, the H4/PDGFβR-F2 mutant contains Y → F substitutions at PDGFβR residues 579/581 and participates in signal transduction to SRC and STAT family members. In the context of the H4/PDGFβR fusion, phenylalanine substitutions at positions 579/581 abrogate induction of IL-3–independent growth of Ba/F3 cells. In addition, an analogous TEL/PDGFβR F2 mutant also has impaired ability to confer factor-independent growth to Ba/F3 cells (D.W.S. and D.G.G., unpublished observation, 2000) and is incapable of causing a myeloproliferative disease in a murine BMT model.15 Taken together, these data suggest that these juxtamembrane residues play an important role in the myeloproliferative phenotype associated with the H4/PDGFβR and TEL/PDGFβR fusion proteins. Second, the W → A point mutations in the WW-like domain of H4/PDGFβR impaired IL-3–independent survival of Ba/F3 cells. These 2 tryptophan residues are characteristic of WW-domains,42 and a single amino acid substitution in the WW-like domain of the native PDGFβR causes constitutive activation of the receptor.43 WW domains have been shown to associate with proline-rich motifs44-46 and with protein-phosphoserine and phosphothreonine motifs.47 We speculate that the WW-like domain of the H4/PDGFβR fusion mediates direct interaction with downstream effector molecules and that disruption of these interactions through mutation impairs transformation of hematopoietic cells. Further analysis of the signal transduction and transforming properties of these mutants in vivo will be necessary to identify the relevant downstream effector molecules.
We next tested in vivo transforming activity of H4/PDGFβR in a murine BMT model.15,29 In this assay, expression of the tyrosine kinase fusion genes BCR/ABL, TEL/JAK2, andTEL/PDGFβR induces a lethal myeloproliferative syndrome with an average latency of 25 to 30 days.15,29,36 37 However, mice transplanted with bone marrow transduced with a retrovirus encoding H4/PDGFβRdeveloped a long-latency T-cell lymphoblastic lymphoma/leukemia. These data demonstrate that H4/PDGFβR transforms primary hematopoietic cells in the murine BMT assay. In addition, these data show that, although there is myeloid-lineage restriction of the t(5;10)(q33;q22) translocation in human atypical CML, the H4/PDGFβR fusion is capable of transformation of lymphoid cells. In a control experiment, mice transplanted with TEL/PDGFβR died of a myeloproliferative disease within 30 days, as expected. There are several possible explanations for the disparity in latency and phenotype between the TEL/PDGFβR and H4/PDGFβR fusions, including variable contribution to transformation by the H4 and TEL moieties, respectively. However, the most likely explanation is that H4/PDGFβR viral supernatants had a substantially lower titer than the TEL/PDGFβR supernatants (10−5 vs 10−6/mL, respectively), despite repeated efforts to generate higher titer H4/PDGFβR retrovirus. TEL/PDGFβR BMT performed with viral supernatant titers of 10−5/mL also gives long-latency, clonally derived lymphoblastic lymphoma (D.W.S. and D.G.G., unpublished observation, 2000).
Another case of myeloproliferative disease associated with t(5;10)(q33;q22) and an H4/PDGFβR fusion has recently been reported by Cross and colleagues, indicating that this is a recurring gene rearrangement.48
In summary, the H4/PDGFβR fusion has transforming activity both in vitro and in vivo. Mutational analysis shows that transformation is dependent on the PDGFβR-kinase domain. In addition, induction of factor-independent growth is impaired by deletion of H4sequences encoding the amino-terminal leucine zipper domain. Induction of factor-independent growth is also impaired by deletion ofH4 sequences that are present in the H4/PDGFβR fusion, but absent in the H4/RET fusion associated with papillary thyroid carcinoma. In addition, mutation of PDGFβR juxtamembrane tyrosine residues 579/581 to phenylalanine, as well as W → A mutations of the WW-like domain, significantly impairs transformation in this assay. Further analysis of related mutations and assays in the murine BMT assay should provide additional insights into the mechanisms of transformation by the increasing number of PDGFβR gene rearrangements associated with human leukemias.
The authors thank Francesca Garcia for administrative assistance and members of the Gilliland laboratory for valuable discussion. J.S. is a recipient of a Special Fellowship from the Leukemia Society of America, and D.G.G. is an Associate Investigator in the Howard Hughes Medical Institute.
Supported in part by a grant from the Swiss National Science Foundation (3100-056984.99) (J.S.); by the Associazione Italiana per la Richerca sul Cancro (C.M.); and by grants from the National Institutes of Health (P01CA66996-01 and P01OK50654) and the MarJo Foundation (D.G.G.).
J.S. and E.A. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Juerg Schwaller, Institute de pathologie clinique, Hôpitaux univérsitaire de Genève, CMU, 1 Rue Michel Servet, Genéve, CH-1211, Switzerland; or Gary Gilliland, Division of Hematology/Oncology, Brigham and Women's Hospital, Harvard Medical School, 4 Blackfan Circle, Boston, MA, 02115.