It was previously shown that plasmin activates human peripheral monocytes in terms of lipid mediator release and chemotactic migration. Here it is demonstrated that plasmin induces proinflammatory cytokine release and tissue factor (TF) expression by monocytes. Plasmin 0.043 to 1.43 CTA U/mL, but not active site-blocked plasmin, triggered concentration-dependent expression of mRNA for interleukin-1α (IL-1α), IL-1β, tumor necrosis factor-α (TNF-α), and TF with maximum responses after 4 hours. Plasmin-mediated mRNA expression was inhibited in a concentration-dependent manner by the lysine analoguetrans-4-(aminomethyl)cyclohexane-1-carboxylic acid (t-AMCA). Increases in mRNA levels were followed by concentration- and time-dependent release of IL-1α, IL-1β and TNF-α and by TF expression on monocyte surfaces. Neither cytokines nor TF could be detected when monocytes were preincubated with actinomycin D or cycloheximide. Electrophoretic mobility shift assays indicated plasmin-induced activation of NF-κB; DNA-binding complexes were composed of p50, p65, and c-Rel, as shown by supershift experiments. Nuclear translocation of NF-κB/Rel proteins coincided with IκBα degradation. At variance with endotoxic lipopolysaccharide, plasmin elicited the rapid degradation of another cytoplasmic NF-κB inhibitor, p105. Proteolysis of NF-κB inhibitors was apparently due to transient activation of IκB kinase (IKK) β that reached maximum activity at 1 hour after plasmin stimulation. In addition, AP-1 binding was increased in plasmin-treated monocytes, with most complexes composed of JunD, c-Fos, and FosB. These findings further substantiate the role of plasmin as a proinflammatory activator of human monocytes and reveal an important new link between the plasminogen-plasmin system and inflammation.
Introduction
Most blood cells, including monocytes, bind plasmin and plasminogen through low-affinity binding sites, a fact that has usually been regarded in terms of fibrinolytic activity.1However, several studies2-5 suggest that the serine protease plasmin might have physiological functions beyond fibrinolysis. Thus, plasmin induces neutrophil aggregation, platelet degranulation, and arachidonate release from endothelial cells, implying activity as a proinflammatory agonist.6-8Moreover, plasmin was found to be a potent and selective stimulus for human peripheral monocytes.2-4 In monocytes, plasmin triggered the release of lipid mediators, such as the chemotactic leukotriene B4, and a chemotactic response equipotent to that of N-formyl-methionine-leucine-phenylalanine.2-4Studies with transgenic mice revealed that plasmin is important for monocyte recruitment to sites of inflammation and for the development of atherosclerotic lesions.5,9 Additional support for an extended pathophysiological function of plasmin in vivo comes from clinical studies showing elevated levels of plasmin in synovial fluid from arthritic joints and an increased expression of fibrinolytic genes in atherosclerotic lesions.9-11
Nuclear factor–κB (NF-κB) is a ubiquitous transcription factor that regulates the expression of numerous genes, including early-response genes encoding cytokines, adhesion molecules, and tissue factor.12-14 Disorders in NF-κB activation have been linked to inflammatory reactions associated with arthritis, atherosclerosis, septic shock, and various other diseases.12-14 In resting cells, the NF-κB/Rel nuclear factors reside in the cytoplasm bound to inhibitory proteins.12 Five members of the NF-κB family have been identified. The most abundant form of NF-κB is a heterodimer composed of a p65 subunit and a p50 or p52 subunit. Other complexes, such as p65/c-Rel, have also been detected.12-14
Although NF-κB is considered a genetic switch for early-response genes, the synergistic interaction of NF-κB with other transcription factors is essential for the optimal induction of distinct gene expression.15-17 Cooperative and coordinate binding of NF-κB and AP-1 transcription factors enhances the activity of cytokine and TF gene promoters.15-18 Transcription factor AP-1 is composed of Jun homodimers or Jun/Fos heterodimers. AP-1 is activated by a wide array of stimuli and is implicated in such different processes as cell cycle progression, cell cycle arrest, and apoptosis19 and in leukocyte activation and differentiation.17
In the present study, we have investigated the plasmin-induced activation of human peripheral monocytes in terms of pro-inflammatory gene expression. We demonstrate that in human peripheral monocytes, plasmin stimulates proinflammatory cytokines and TF expression associated with proinflammatory activation of macrophages,20 indicating a new link between the plasminogen/plasmin system and inflammation. We further show that plasmin-induced signal transduction entails the activation of transcription factors NF-κB and AP-1.
Materials and methods
Materials
Purified human plasmin (lots 354915 and 352817; specific activity, 14.83 CTA U/mg) was from Fluka (Deisenhofen, Germany). Antibodies against nuclear factors, IκBα and IκBβ, IKKα and IKKβ, and tagged IκBα fusion protein were from Santa Cruz Biotechnology (Santa Cruz, CA). Double-stranded oligonucleotides containing the NF-κB, AP-1, AP-2, and SP-1 binding sites and T4 polynucleotide kinase were purchased from Promega (Madison, WI). Anti-CD41 and anti-CD14 monoclonal antibodies were obtained from Immunotech (Marseilles, France). Mouse monoclonal antibodies against human TF and control MOPC21 mouse IgG1 were from American Diagnostica (Greenwich, CT) and Sigma (St Louis, MO), respectively. Lysine-free RPMI 1640, the Limulus amebocyte lysate assay cycloheximide, trans-4-(aminomethyl)cyclohexane-1-carboxylic acid (t-AMCA), and lipopolysaccharide (LPS; Escherichia coliserotype 055:B5) were also from Sigma. Actinomycin D and D-valyl-L-phenylalanyl-L-lysine chloromethyl ketone were from Calbiochem (San Diego, CA). Percoll was from Pharmacia Biotech (Uppsala, Sweden). The plasmin substrate S-2251 (H-D-valyl-leucyl-L-lysine-P-nitroanilide dihydrochloride) was supplied by Chromogenix (Mölndal, Sweden). Oligo(dT)25 magnetic beads were from Dynal (Oslo, Norway). Other chemicals were of analytical grade.
Monocyte preparation and incubation
Peripheral monocytes were isolated by Percoll gradient centrifugation.2-4 Preparations with 94% or more CD14+ cells were used. Contaminating cells were lymphocytes (2%-6%). Flow cytometry of cells stained additionally with anti-CD41 antibodies did not reveal any platelets associated with monocytes.
Monocytes were generally incubated in lysine-free RPMI 1640 in the presence or absence of LPS (1 μg/mL), a concentration that was found to yield maximum responses in terms of tumor necrosis factor-α (TNF-α) and TF biosynthesis, or LPS-free human plasmin (0.043-1.43 CTA U/mL). All plasmin batches were regularly checked for LPS contamination with the Limulus amebocyte lysate assay. Occasionally, plasmin was added in the presence of the lysine analogue t-AMCA.
In some experiments, monocytes were treated for 4 hours with active site-blocked plasmin, equivalent to 0.43 CTA U/mL native plasmin. D-Val-Phe-Lys chloromethyl ketone (VPLCK) was used to block the catalytic center of plasmin.3 4 Active site-blocked plasmin had no detectable residual plasmin activity, as tested with S-2251 as a substrate. Controls and LPS-stimulated samples received the appropriate amounts of VPLCK.
Semiquantitative reverse transcription–polymerase chain reaction analysis
mRNA isolated from monocytes (0.5 × 106cells/assay) with oligo(dT)25 magnetic beads was analyzed by reverse transcription–polymerase chain reaction (RT-PCR) with primers specific for interleukin-1α (IL-1α), IL-1β, TNF-α, and TF.21 22 Conditions were such that the PCR reactions did not reach the saturation phase.
Control experiments showed no DNA contaminations. Normalization of semiquantitative PCR was carried out using HLA(B) as an internal standard.23 The identity of the PCR products was confirmed by direct sequencing (Abi Prism 310; Applied Biosystems, Foster City, CA).
For determination of mRNA stability, monocytes were stimulated with plasmin (1.43 CTA U/mL) or LPS (1 μg/mL) for 4 hours before the addition of actinomycin D (5 μg/mL). Levels of corresponding cytokines or TF mRNA at the beginning of the actinomycin D chase (time point, 0 hour) were set to 100%. Curves fitted by least-squares regression were used for the calculation of the half-life of each mRNA.
Cytokine and TF biosynthesis
Cytokines were assayed in cell-free supernatants using enzyme-linked immunosorbent assay (ELISA) specific for IL-1α (Cytimmune; College Park, MD), IL-1β, interferon (IFN)-γ (Biosource, Camarillo, CA), and TNF-α (R&D Systems, Minneapolis, MN). TF was analyzed in whole cell extracts using a TF-specific ELISA from American Diagnostica. In some experiments, monocytes were preincubated for 15 minutes with cycloheximide (10 μg/mL) or actinomycin D (5 μg/mL) before stimulation with 1.43 CTA U/mL plasmin for 8 hours.
For flow cytometry of TF expression, monocytes plated on hydrophobic Petriperm dishes (In Vitro Systems, Osterode, Germany) were stimulated with plasmin (0.43 CTA U/mL) or LPS (1 μg/mL) for 8 hours. Cells were carefully detached by scraping. Then they were washed with 15 mM 6-aminohexanoic acid and phosphate-buffered saline (PBS) to remove bound plasmin, stained with either monoclonal antihuman TF or control MOPC21 and anti-CD14 monoclonal mouse antibodies, and analyzed by FACScan (Becton Dickinson, San Jose, CA).
Electrophoretic mobility shift assays
Monocytes (5 × 106 cells/assay) were stimulated with plasmin (0.43 CTA U/mL) or LPS (1 μg/mL). Nuclear extracts for NF-κB and AP-1 electrophoretic mobility shift assay (EMSA) were prepared as described.24,25 DNA–protein interactions were assayed by incubating 5 μg nuclear extract with 50 000 cpm32P-end labeled double-stranded NF-κB site-specific probe in the presence of 1 μg poly [dI-dC] (Pharmacia Biotech) in 20 μL binding buffer, pH 5.0 (10 mM Tris-HCl, 10% glycerol, 1.0 mM EDTA, 40 mM NaCl, 1.0 mM dithiothreitol, and 4.0 mM MgCl2) at 24°C for 30 minutes. AP-1 EMSA was assayed as described25,26except for 10% glycerol in the buffer. In supershift experiments, nuclear extracts were incubated with the corresponding antibodies (2 μg) for 1 hour at 4°C after the addition of 32P-end labeled NF-κB DNA or for 12 hours at 4°C before the addition of the32P-labeled AP-1 probe.25 26
Western blot analysis and immunostaining
Monocytes (5 × 106 cells/assay) were cultured on hydrophobic Petriperm membranes in the presence of plasmin (0.43 CTA U/mL) or LPS (1 μg/mL). Cells were lysed in 30 μL PBS, pH 7.4, containing 1% Igepal CA-630 (Sigma), 0.5% Na-deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and 0.3 μL protease inhibitor cocktail set III (Calbiochem). Samples containing equal amounts of protein were resolved by 10% SDS polyacrylamide gel electrophoresis (PAGE) and electroblotted onto nitrocellulose membranes. Membranes were incubated with appropriate antibodies (1 μg/mL) and subsequently with secondary antibodies conjugated with horseradish peroxidase (1:1000). Antibody complexes were visualized using the enhanced chemiluminescence Western blotting detection reagent system (ECL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) and subsequent exposure to Hyperfilm ECL (Amersham).
Immunoprecipitation and kinase assay
Monocytes (5 × 106 cells/assay) were lysed with 500 μL buffer, pH 8.0 (25 mM Tris-HCl, 100 mM NaCl, 25 mM β-glycerophosphate, 100 μM Na-orthovanadate, 2 mM EDTA, 2 mM EGTA, 10% glycerol, 1% Triton X-100, 5 μL protease inhibitor cocktail set III [Calbiochem]), and supernatants were precleared by incubation with 1 μg normal rabbit IgG and 10 μL protein A–agarose. For immunoprecipitation of IKKα and IKKβ, precleared cell lysates were further incubated for 1 hour at 4°C with 1 μg anti-IKKα or anti-IKKβ rabbit polyclonal antibodies. Afterward, 10 μL protein A–agarose was added, and incubations were continued for 1 hour. IKKs attached to the agarose beads were extensively washed and finally resuspended in 15 μL kinase buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, 20 mM β-glycerophosphate, 100 μM Na-orthovanadate, 1 mM dithiothreitol). Fifteen microliters IKK, 10 μM adenosine triphosphate (ATP), and 5 μCi [γ-32P]-ATP (6000 Ci/mmol, 10 mCi/mL; Amersham) were incubated with substrate IκBα (1 μg) tagged fusion protein corresponding to the full-length IκBα (amino acids 1-317) of human origin (Santa Cruz Biotechnology) at 30°C for 20 minutes. Samples were resolved by 10% SDS-PAGE, blotted on nitrocellulose, visualized by autoradiography, and quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). To check loading and for confirmation of IKK immunoprecipitation, blots were immunostained with 1:500 dilutions of IKKα or IKKβ mouse monoclonal antibodies, respectively.
Statistical analysis
Values shown represent mean ± SEM where applicable. Statistical significance was calculated with the Newman-Keuls test. Differences were considered significant for P < .05.
Results
Expression of cytokine and TF mRNA
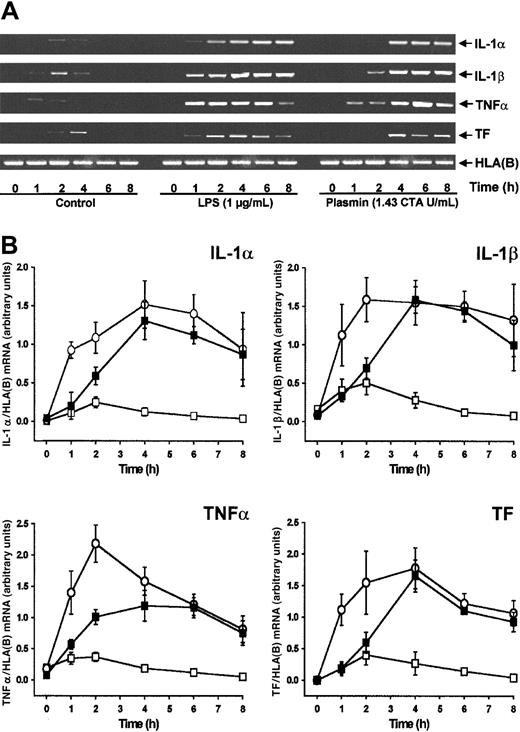
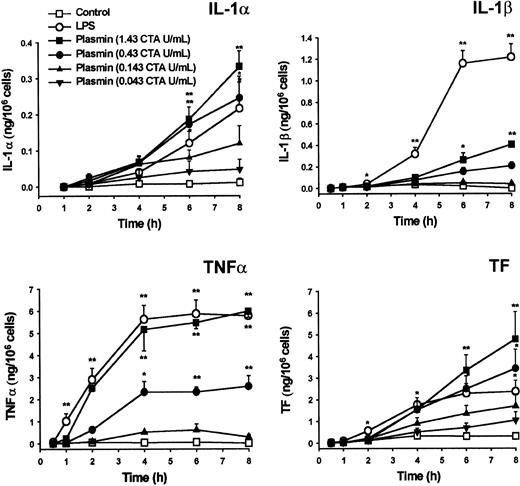
By semiquantitative RT-PCR analysis, human monocytes did not express detectable cytokine or TF mRNA at 0 hour (Figure1A). Compared with controls, stimulation of monocytes with plasmin (1.43 CTA U/mL) induced the time-dependent expression of mRNA for IL-1α, IL-1β, and TNF-α and for TF (Figure 1A-B). All 4 mRNAs investigated followed similar kinetics, with maximum expression 4 hours after stimulation and a slight decrease at 8 hours. IL-1α, IL-1β, and TF mRNA expression was comparable in monocytes stimulated with either plasmin or LPS, though the kinetics differed. Within 1 hour of 1 μg/mL LPS stimulation, a rapid increase of mRNA expression occurred, whereas mRNA expression in the presence of plasmin was delayed. In accordance with the existing literature,26,27 culture of the monocytes led to weak adherence-induced gene expression (Figure 1B). Adhesion of monocytes28 was, however, not promoted by plasmin (0.43 to 1.43 CTA U/mL), nor did it induce homotypic aggregation as measured by either spectrophotometric aggregometry29 or flow cytometric doublet discrimination (data not shown). Therefore, plasmin-induced changes in mRNA expression are not secondary to any such events.
Plasmin stimulates time-dependent expression of IL-1α, IL-1β, TNF-α, and TF mRNA in human monocytes.
Monocytes (0.5 × 106/mL) were cultured in the presence of plasmin 1.43 CTA U/mL (▪) or LPS 1 μg/mL (○) or in the absence of any stimulus (■). At indicated times cells were harvested, and mRNA was extracted and subjected to RT-PCR. HLA(B) was used for normalization. (A) Representative gels are shown. (B) Semiquantitative analysis of the mRNA expression. Results are the mean ± SEM of 5 independent experiments.
Plasmin stimulates time-dependent expression of IL-1α, IL-1β, TNF-α, and TF mRNA in human monocytes.
Monocytes (0.5 × 106/mL) were cultured in the presence of plasmin 1.43 CTA U/mL (▪) or LPS 1 μg/mL (○) or in the absence of any stimulus (■). At indicated times cells were harvested, and mRNA was extracted and subjected to RT-PCR. HLA(B) was used for normalization. (A) Representative gels are shown. (B) Semiquantitative analysis of the mRNA expression. Results are the mean ± SEM of 5 independent experiments.
RT-PCR products had the expected molecular weights and were identified by sequencing. Each PCR band represented a single PCR product, with more than 99% sequence identity with the investigated cytokine or TF (data not shown).
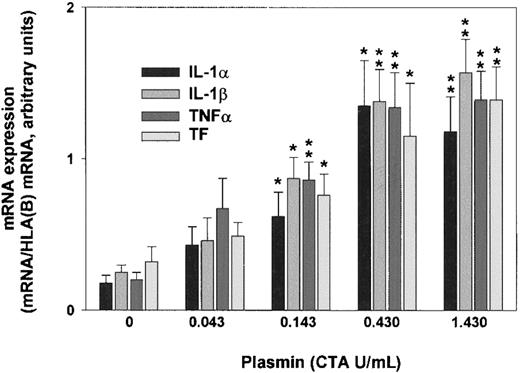
Plasmin triggered a concentration-dependent expression of IL-1α, IL-1β, TNF-α, and TF mRNA in monocytes incubated for 4 hours (Figure 2). By contrast, unstimulated controls expressed only small amounts of cytokine and TF mRNA. Although 0.043 CTA U/mL plasmin already increased IL-1α, IL-1β, TNF-α, and TF mRNA expression, increases were significant at 0.143 CTA U/mL plasmin (n = 5; P < .05 vs unstimulated controls). The concentration of 0.43 CTA U/mL plasmin appeared to be optimal for cytokine expression because there was almost no further mRNA augmentation with plasmin 1.43 CTA U/mL. Consistent with previous studies, up to 1.43 CTA U/mL plasmin for 8 hours did not alter monocyte viability, as assessed by trypan blue dye exclusion, nor did it induce apoptosis in terms of annexin V binding to externalized phosphatidylserine.30 By contrast, UV (254 nm, 150 J/m2)-irradiated monocytes stained positive (data not shown).
Plasmin triggers concentration-dependent expression of IL-1α, IL-1β, TNF-α, and TF mRNA in human monocytes.
Monocytes were incubated with the indicated concentrations of LPS-free plasmin for 4 hours. Expression of mRNA was measured by semiquantitative RT-PCR. HLA(B) was used for normalization. Results are the mean ± SEM of 5 independent experiments. *P < .05 and **P < .01. versus unstimulated controls.
Plasmin triggers concentration-dependent expression of IL-1α, IL-1β, TNF-α, and TF mRNA in human monocytes.
Monocytes were incubated with the indicated concentrations of LPS-free plasmin for 4 hours. Expression of mRNA was measured by semiquantitative RT-PCR. HLA(B) was used for normalization. Results are the mean ± SEM of 5 independent experiments. *P < .05 and **P < .01. versus unstimulated controls.
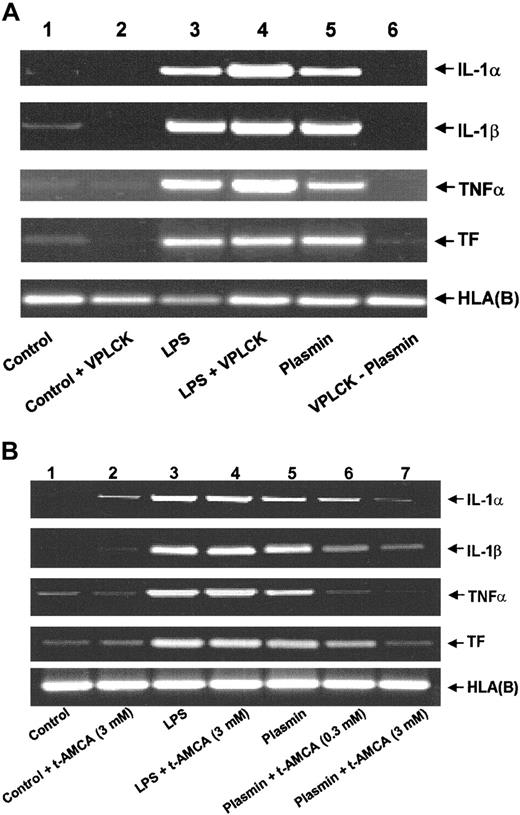
An intact plasmin catalytic center is essential for the effects observed because active site-blocked plasmin was unable to trigger either mRNA expression (Figure 3A) or corresponding protein release (data not shown). The plasmin inhibitor VPLCK had no effect on mRNA expression in untreated or LPS-stimulated monocytes (Figure 3A). Similarly, the lysine analogue t-AMCA (3.0 mM) had no effect on LPS-stimulated mRNA expression, nor did it modulate basal control expression (Figure 3B). By contrast, t-AMCA at 0.3 and 3.0 mM induced a concentration-dependent inhibition of plasmin-induced mRNA expression (Figure 3B), implying the necessity of the plasmin molecule to bind through its lysine-binding sites.
Role of the intact catalytic center and lysine binding sites for the plasmin-induced expression of IL-1α, IL-1β, TNF-α, and TF mRNA in human monocytes.
(A) Effect of active site-blocked plasmin. The catalytic center of plasmin was blocked by preincubation with the irreversible inhibitor VPLCK. Monocytes were cultured in the absence of any stimulus (lane 1), VPLCK 25 μM (lane 2), LPS 1 μg/mL (lane 3), LPS 1 μg/mL, and VPLCK 25 μM (lane 4), in the presence of plasmin 0.43 CTA U/mL (lane 5) or with 0.43 CTA U/mL active site-blocked plasmin (VPLCK-plasmin, lane 6). (B) Effects of t-AMCA. Monocytes were cultured in the absence of any stimulus (lane 1), t-AMCA 3 mM (lane 2), LPS 1 μg/mL (lane 3), LPS 1 μg/mL, and t-AMCA 3.0 mM (lane 4), in the presence of plasmin 0.43 CTA U/mL (lane 5) and with plasmin 0.43 CTA U/mL and t-AMCA 0.3 mM (lane 6) or 3.0 mM (lane 7), respectively. In both experiments, cells were harvested after 4 hours, and mRNA was extracted and subjected to RT-PCR. HLA(B) was used for normalization. Results of 1 of 3 experiments are shown in each case.
Role of the intact catalytic center and lysine binding sites for the plasmin-induced expression of IL-1α, IL-1β, TNF-α, and TF mRNA in human monocytes.
(A) Effect of active site-blocked plasmin. The catalytic center of plasmin was blocked by preincubation with the irreversible inhibitor VPLCK. Monocytes were cultured in the absence of any stimulus (lane 1), VPLCK 25 μM (lane 2), LPS 1 μg/mL (lane 3), LPS 1 μg/mL, and VPLCK 25 μM (lane 4), in the presence of plasmin 0.43 CTA U/mL (lane 5) or with 0.43 CTA U/mL active site-blocked plasmin (VPLCK-plasmin, lane 6). (B) Effects of t-AMCA. Monocytes were cultured in the absence of any stimulus (lane 1), t-AMCA 3 mM (lane 2), LPS 1 μg/mL (lane 3), LPS 1 μg/mL, and t-AMCA 3.0 mM (lane 4), in the presence of plasmin 0.43 CTA U/mL (lane 5) and with plasmin 0.43 CTA U/mL and t-AMCA 0.3 mM (lane 6) or 3.0 mM (lane 7), respectively. In both experiments, cells were harvested after 4 hours, and mRNA was extracted and subjected to RT-PCR. HLA(B) was used for normalization. Results of 1 of 3 experiments are shown in each case.
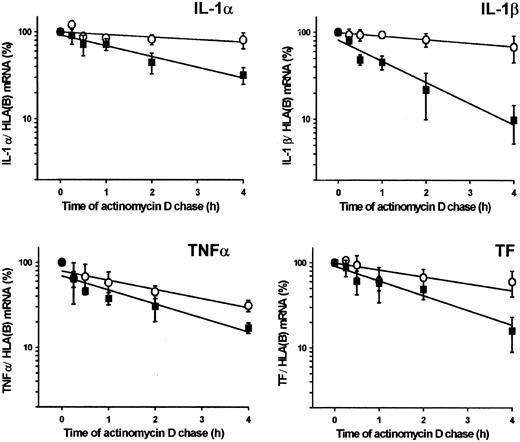
Effects of plasmin on mRNA stability were investigated by monitoring the decay of cytokine and TF mRNA in monocytes stimulated with plasmin (1.43 CTA U/mL) or LPS (1 μg/mL). Exponential regression analysis of the data indicated that stimulation with plasmin unexpectedly reduced the stability of cytokine and TF mRNA compared with LPS (Figure4). The mRNA half-life in plasmin-stimulated monocytes ranged from 0.9 hours for IL-1β to 2.2 hours for IL-1α. In LPS-stimulated cells, mRNA half-life varied between 1.9 hours for TNF-α and 10.4 hours for IL-1α.
Stability of IL-1α, IL-1β, TNF-α, and TF mRNA in plasmin-treated monocytes.
Monocytes were stimulated for 4 hours with plasmin 1.43 CTA U/mL (▪) or LPS 1 μg/mL (○). The level of the corresponding cytokine or TF mRNA at this time point was 100%. Actinomycin D (5 μg/mL) was added, and incubation continued for the indicated time. Poly(A)+RNA was isolated and subjected to RT-PCR. HLA(B) was used for normalization; its stability over 4 hours was not significantly different in controls than in plasmin- or LPS-treated cells. Results are the mean ± SEM of 3 independent experiments. Curves were fitted by least-squares regression analysis and were used to calculate the half-life of each mRNA species.
Stability of IL-1α, IL-1β, TNF-α, and TF mRNA in plasmin-treated monocytes.
Monocytes were stimulated for 4 hours with plasmin 1.43 CTA U/mL (▪) or LPS 1 μg/mL (○). The level of the corresponding cytokine or TF mRNA at this time point was 100%. Actinomycin D (5 μg/mL) was added, and incubation continued for the indicated time. Poly(A)+RNA was isolated and subjected to RT-PCR. HLA(B) was used for normalization; its stability over 4 hours was not significantly different in controls than in plasmin- or LPS-treated cells. Results are the mean ± SEM of 3 independent experiments. Curves were fitted by least-squares regression analysis and were used to calculate the half-life of each mRNA species.
Plasmin stimulates cytokine release and TF biosynthesis
Cytokine mRNA expression does not inevitably lead to translation.27,31 32 Similar to LPS, plasmin triggers time- and concentration-dependent release of IL-1α, IL-1β, and TNF-α from monocytes (Figure 5). Cytokines and TF were detectable as early as 2 to 4 hours after stimulation. Plasmin at 0.43 and 1.43 CTA U/mL was at least as potent as 1 μg/mL LPS with respect to IL-1α release. However, LPS triggered a more rapid and extensive release of IL-1β, indicating clear differences between LPS- and plasmin-mediated activation of monocytes. Although the release of TNF-α was delayed in plasmin-stimulated monocytes, at 2 hours after exposure there was no significant difference in the amount of TNF-α released by cells stimulated with either 1.43 CTA U/mL plasmin or LPS 1 μg/mL. Similarly, TF production analyzed in whole-cell extracts commenced with some delay in plasmin-treated monocytes; however, at 6 to 8 hours after the onset of stimulation with 0.43 or 1.43 CTA U/mL plasmin, monocytes generated even more TF than LPS-stimulated cells (Figure 5). Monocytes pretreated with inhibitors of transcription (5 μg/mL actinomycin D) or translation (10 μg/mL cycloheximide) produced no detectable amounts of cytokines or TF after stimulation with 1.43 CTA U/mL plasmin (data not shown). There was no detectable release of IFN-γ by cultured cells at any of the time points investigated (data not shown).
Generation of IL-1α, IL-1β, TNF-α, and TF by plasmin-stimulated human monocytes.
Monocytes were incubated with various concentrations of plasmin for the times indicated. Monocytes treated with LPS 1 μg/mL served as positive controls. Supernatants were collected, and the release of IL-1α, IL-1β, and TNF-α was analyzed by ELISA. Cell-associated TF was measured in whole-cell lysates. Results are the mean ± SEM of 4 to 6 independent experiments. *P < .05 and **P < .01. versus unstimulated controls.
Generation of IL-1α, IL-1β, TNF-α, and TF by plasmin-stimulated human monocytes.
Monocytes were incubated with various concentrations of plasmin for the times indicated. Monocytes treated with LPS 1 μg/mL served as positive controls. Supernatants were collected, and the release of IL-1α, IL-1β, and TNF-α was analyzed by ELISA. Cell-associated TF was measured in whole-cell lysates. Results are the mean ± SEM of 4 to 6 independent experiments. *P < .05 and **P < .01. versus unstimulated controls.
TF expression is up-regulated in plasmin-stimulated monocytes
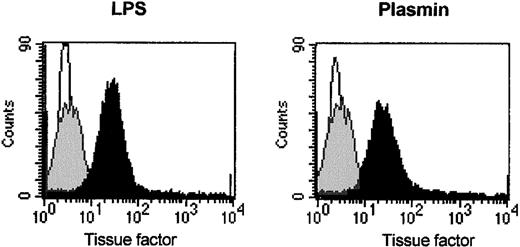
Flow cytometric analysis of CD14+ cells revealed TF expression on the surfaces of plasmin-stimulated monocytes. In agreement with published data,33 unstimulated monocytes expressed only low levels of TF. Stimulation with 0.43 CTA U/mL plasmin for 8 hours led to significantly increased TF expression, comparable to that of cells stimulated with LPS 1 μg/mL (Figure6). TF could not be detected in supernatants of plasmin-stimulated monocytes (data not shown), indicating that membrane-bound TF was not proteolytically released by plasmin. LPS-stimulated cells did not release TF either (data not shown).
Induction of TF expression on the monocyte membrane by plasmin.
Flow cytometric analysis of TF on human monocytes incubated for 8 hours with plasmin 0.43 CTA U/mL (black), LPS 1 μg/mL (black), or unstimulated controls (gray). Cells were stained with unspecific IgG1 (MOPC21, negative control, white) and antihuman TF antibodies. Binding of primary antibodies was visualized by anti–mouse IgG PE-conjugated secondary antibodies. Results of 1 of 3 experiments are shown.
Induction of TF expression on the monocyte membrane by plasmin.
Flow cytometric analysis of TF on human monocytes incubated for 8 hours with plasmin 0.43 CTA U/mL (black), LPS 1 μg/mL (black), or unstimulated controls (gray). Cells were stained with unspecific IgG1 (MOPC21, negative control, white) and antihuman TF antibodies. Binding of primary antibodies was visualized by anti–mouse IgG PE-conjugated secondary antibodies. Results of 1 of 3 experiments are shown.
Plasmin triggers nuclear translocation of the NF-κB complex
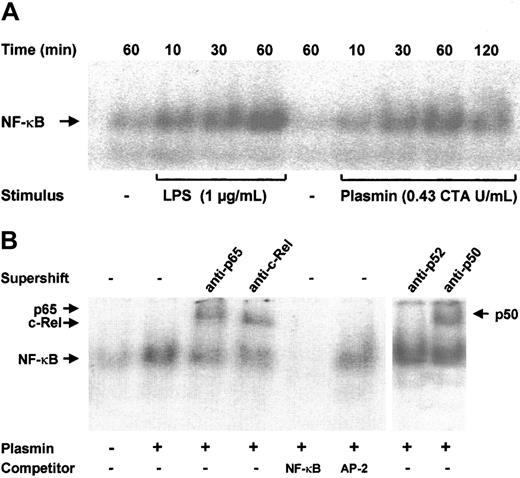
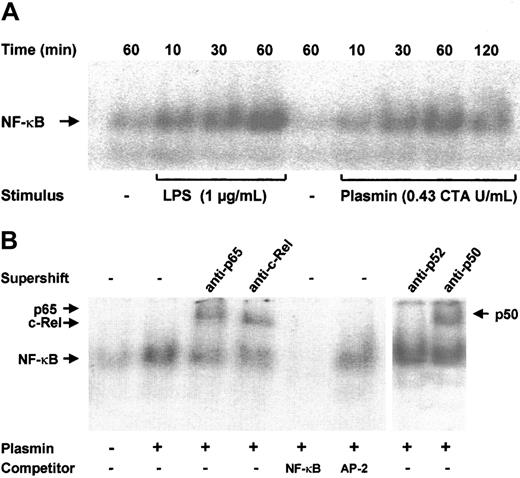
NF-κB–binding sites are present in the promoter regions of many cytokine genes.13 Transcription of the TF gene is also strongly dependent on the activation of NF-κB/Rel proteins.18 Increased binding of nuclear extracts to an NF-κB DNA probe was observed as early as 10 minutes after stimulation of monocytes with 0.43 CTA U/mL plasmin; it reached a maximum at 1 hour and declined after 2 hours (Figure 7A). A similar time-course of NF-κB activation was found in LPS-stimulated monocytes, though the DNA-binding activity was stronger. The specificity of NF-κB binding was confirmed in competition experiments; a 100-fold molar excess of unlabeled NF-κB, but not of AP-2 consensus oligonucleotides, abolished binding of the nuclear extracts to the labeled NF-κB–binding site sequence (Figure 7B). The protein composition of the DNA-protein band was further investigated through EMSA. Of the 5 known mammalian NF-κB/Rel proteins, p50, p52, c-Rel, and p65 are highly expressed in monocytes and macrophages; a high expression of RelB is confined to maturing and differentiated dendritic cells.14 34 Stimulation of monocytes with 0.43 CTA U/mL plasmin resulted in the nuclear translocation of p50, p65, and c-Rel but not of p52 (Figure 7B).
NF-κB activation by plasmin.
(A) Kinetic study. Monocytes were stimulated with plasmin 0.43 CTA U/mL or LPS 1 μg/mL (positive control) for the times indicated. Cells were lysed, and nuclei were isolated. Nuclear extracts (5 μg) were subjected to EMSA with a 32P-labeled DNA probe containing the NF-κB binding site. (B) Supershift and competition study. Monocytes were stimulated with plasmin 0.43 CTA U/mL for 1 hour. Nuclear extracts were incubated for 1 hour with anti-p65, anti-c-Rel, anti-p50, and anti-p52 antibodies or with unlabeled NF-κB or AP-2 specific oligonucleotides (100-fold molar excess). Supershift assay revealed binding of p65, c-Rel, and p50, but not of p52 nuclear factor. Preincubation with NF-κB–specific oligonucleotide sequence abolished formation of the NF-κB complex. AP-2–specific oligonucleotides had no effect on the NF-κB/DNA complex formation. Results of 1 of 3 experiments are shown.
NF-κB activation by plasmin.
(A) Kinetic study. Monocytes were stimulated with plasmin 0.43 CTA U/mL or LPS 1 μg/mL (positive control) for the times indicated. Cells were lysed, and nuclei were isolated. Nuclear extracts (5 μg) were subjected to EMSA with a 32P-labeled DNA probe containing the NF-κB binding site. (B) Supershift and competition study. Monocytes were stimulated with plasmin 0.43 CTA U/mL for 1 hour. Nuclear extracts were incubated for 1 hour with anti-p65, anti-c-Rel, anti-p50, and anti-p52 antibodies or with unlabeled NF-κB or AP-2 specific oligonucleotides (100-fold molar excess). Supershift assay revealed binding of p65, c-Rel, and p50, but not of p52 nuclear factor. Preincubation with NF-κB–specific oligonucleotide sequence abolished formation of the NF-κB complex. AP-2–specific oligonucleotides had no effect on the NF-κB/DNA complex formation. Results of 1 of 3 experiments are shown.
IκBα and p105 are differentially degraded in plasmin- and LPS-stimulated monocytes
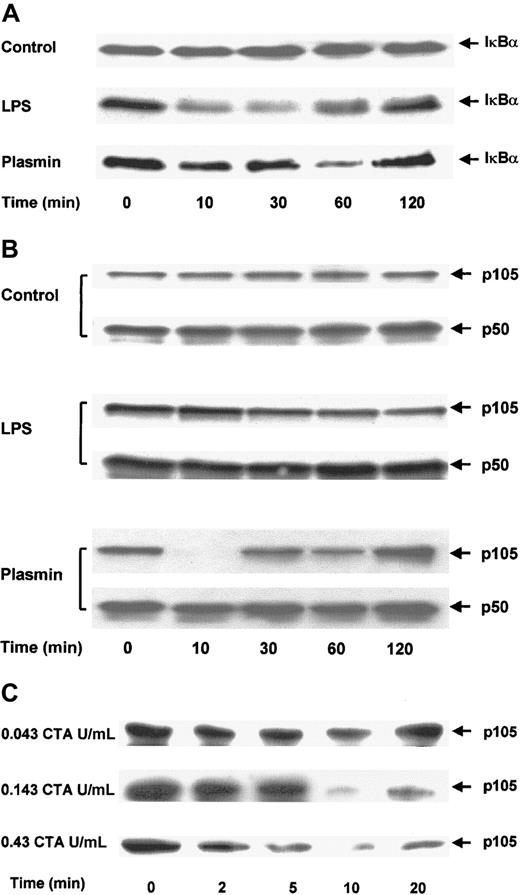
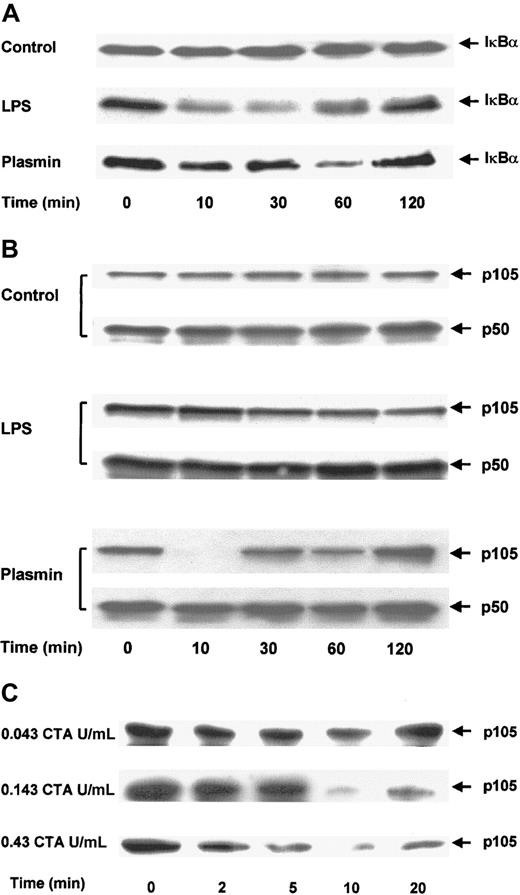
The effect of plasmin on the degradation of IκBα was analyzed in immunoblots of cell extracts. Control cells showed no significant changes in IκBα protein during 2 hours (Figure8A). Stimulation of monocytes with 1 μg/mL LPS for 10 minutes was sufficient for the rapid degradation of IκBα. By 30 minutes, only 30% of the initially present IκBα was detectable. However, 1 hour after stimulation, the level of IκBα started to increase and reached almost the initial level by 2 hours. The degradation of IκBα in monocytes stimulated with 0.43 CTA U/mL plasmin was delayed. Proteolysis of IκBα was detectable after 10 minutes, but almost complete (80%) degradation was not observed until after 1 hour. As in LPS-stimulated cells, 2 hours after plasmin stimulation IκBα returned almost to its initial level. This normalization of IκBα coincided with the decrease of NF-κB binding, as measured by EMSA (Figure 7A).
Proteolytic degradation of IκBα and p105 in plasmin-stimulated monocytes.
(A,B) Kinetic study. Cells were either unstimulated (control) or treated with LPS 1 μg/mL (positive control) or plasmin 0.43 CTA U/mL for the times indicated. (C) Detailed time- and concentration-dependent effects of plasmin from 0.043 to 0.43 CTA U/mL on p105 degradation. Cell extracts were prepared and used for immunoblots with specific anti-IκBα (A) and anti-p50 (B,C) antibodies. Because p105 contains the complete sequence of p50, it is also recognized by anti-p50 antibodies. Positions of IκBα (37 kd), p105 and p50 (105 kd and 50 kd, respectively) are indicated. In each case, a representative blot of 3 independent experiments is shown.
Proteolytic degradation of IκBα and p105 in plasmin-stimulated monocytes.
(A,B) Kinetic study. Cells were either unstimulated (control) or treated with LPS 1 μg/mL (positive control) or plasmin 0.43 CTA U/mL for the times indicated. (C) Detailed time- and concentration-dependent effects of plasmin from 0.043 to 0.43 CTA U/mL on p105 degradation. Cell extracts were prepared and used for immunoblots with specific anti-IκBα (A) and anti-p50 (B,C) antibodies. Because p105 contains the complete sequence of p50, it is also recognized by anti-p50 antibodies. Positions of IκBα (37 kd), p105 and p50 (105 kd and 50 kd, respectively) are indicated. In each case, a representative blot of 3 independent experiments is shown.
Proteolytic degradation of p105, a cytoplasmic inhibitor of NF-κB/Rel nuclear factors and a putative precursor of p50, has been implicated in cell activation.35 36 Levels of p50 and p105 were analyzed by immunoblotting. Antibodies directed against p50 also recognized p105. There were no changes in the levels of p50 or p105 in control cells (Figure 8B). Stimulation with 1 μg/mL LPS caused partial degradation of p105, with approximately 50% reduction at 2 hours. In contrast, stimulation of monocytes with 0.43 CTA U/mL plasmin almost resulted in the disappearance of the p105 band 10 minutes after stimulation (Figure 8B). However, this degradation did not lead to an accumulation of the p50 product, indicating that plasmin induces the proteolysis of p105 but not of its processing to p50. The profound effect of plasmin stimulation on p105 degradation was studied in greater detail (Figure 8C). Plasmin 0.043 to 0.43 CTA U/mL induced a concentration-dependent proteolysis of p105. Analysis of the time dependence between 0 and 20 minutes revealed proteolytic degradation as early as 2 minutes after plasmin stimulation and confirmed the maximum effect at 10 minutes.
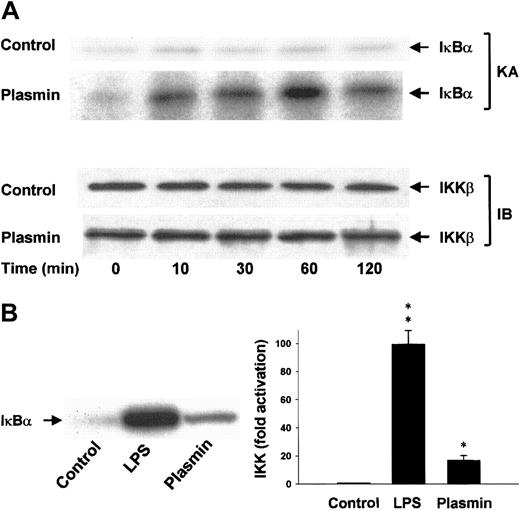
Differential activation of IKKα and IKKβ
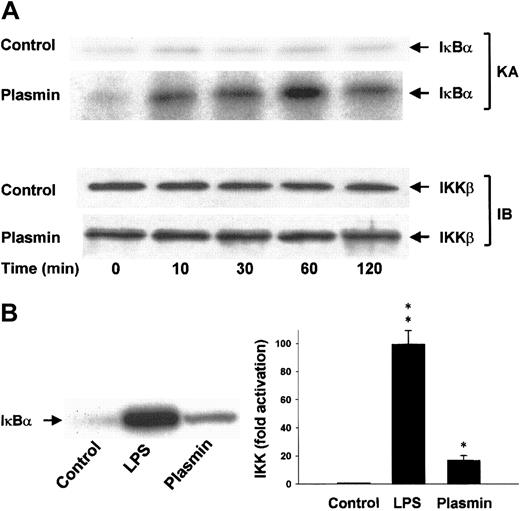
Lysis of the monocytes in the presence of 1% Triton X-100 disrupts the integrity of the signalsome complex37and thus allows separate analysis of IKKα and IKKβ activities after immunoprecipitation of the IκB kinases from the cellular extracts. The protein composition of the precipitates was analyzed, and homogeneity and equal sample distribution were confirmed by immunoblot analysis with anti-IKKα (data not shown) and anti-IKKβ antibodies (Figure 9A). IKK assays revealed that, in contrast to unstimulated cells, monocytes stimulated with 0.43 CTA U/mL plasmin elicited rapid activation of IKKβ detectable within 10 minutes. IKKβ activity peaked at 1 hour and decreased by 2 hours, indicating the transient character of the activation (Figure 9A). LPS (1 μg/mL) was a more potent activator of IKKβ and led to approximately 100-fold activation within 10 minutes (Figure 9B). Plasmin did not affect IKKα activity (data not shown).
Induction of IKKβ activation by plasmin.
(A) Kinetic study of IKKβ activation. Monocytes were either unstimulated or stimulated with plasmin 0.43 CTA U/mL for the indicated times. IKKβ was precipitated from the cell extracts with a specific rabbit anti-IKKβ antibody and protein A–agarose. In vitro kinase assays (KA) were performed using IκBα-tagged fusion protein corresponding to the full-length human IκBα (amino acids 1-317) as a substrate. Composition of the immunoprecipitates was analyzed by immunoblot with mouse anti-IKKβ (IB). Results of 1 of 3 experiments are shown. (B) Monocytes were incubated with or without plasmin 0.43 CTA U/mL or LPS 1 μg/mL for 10 minutes. IKKβ was immunoprecipitated with a specific rabbit anti-IKKβ antibody. Precipitates were analyzed for kinase activity (KA). Phosphorylated substrate was visualized by autoradiography and quantified densitometrically. *P < .05 and **P < .01 versus unstimulated controls. Results are the mean ± SEM of 3 independent experiments.
Induction of IKKβ activation by plasmin.
(A) Kinetic study of IKKβ activation. Monocytes were either unstimulated or stimulated with plasmin 0.43 CTA U/mL for the indicated times. IKKβ was precipitated from the cell extracts with a specific rabbit anti-IKKβ antibody and protein A–agarose. In vitro kinase assays (KA) were performed using IκBα-tagged fusion protein corresponding to the full-length human IκBα (amino acids 1-317) as a substrate. Composition of the immunoprecipitates was analyzed by immunoblot with mouse anti-IKKβ (IB). Results of 1 of 3 experiments are shown. (B) Monocytes were incubated with or without plasmin 0.43 CTA U/mL or LPS 1 μg/mL for 10 minutes. IKKβ was immunoprecipitated with a specific rabbit anti-IKKβ antibody. Precipitates were analyzed for kinase activity (KA). Phosphorylated substrate was visualized by autoradiography and quantified densitometrically. *P < .05 and **P < .01 versus unstimulated controls. Results are the mean ± SEM of 3 independent experiments.
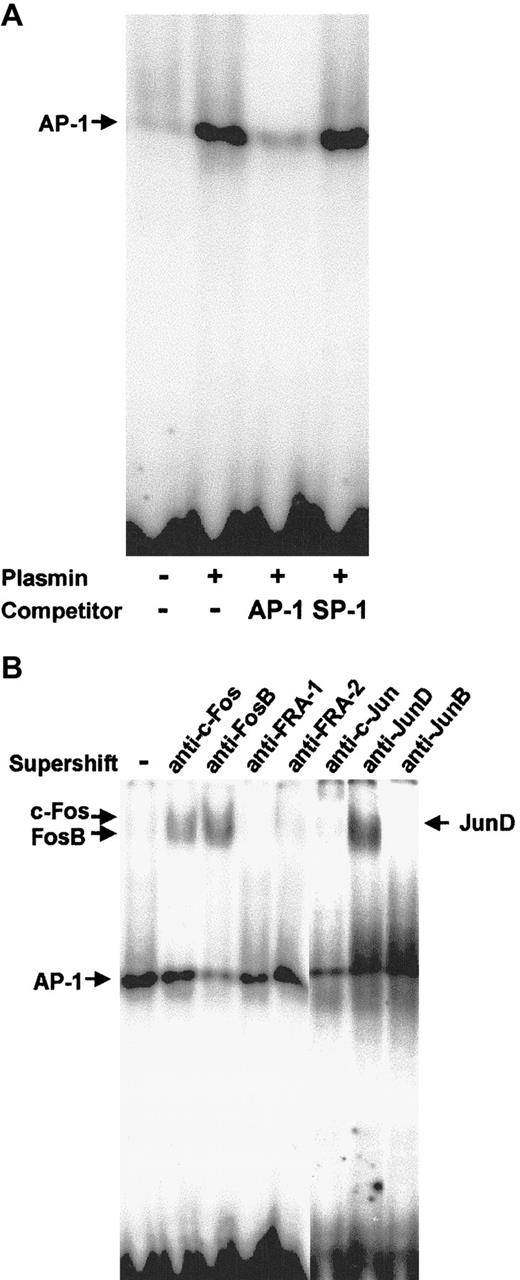
Plasmin triggers activation of AP-1
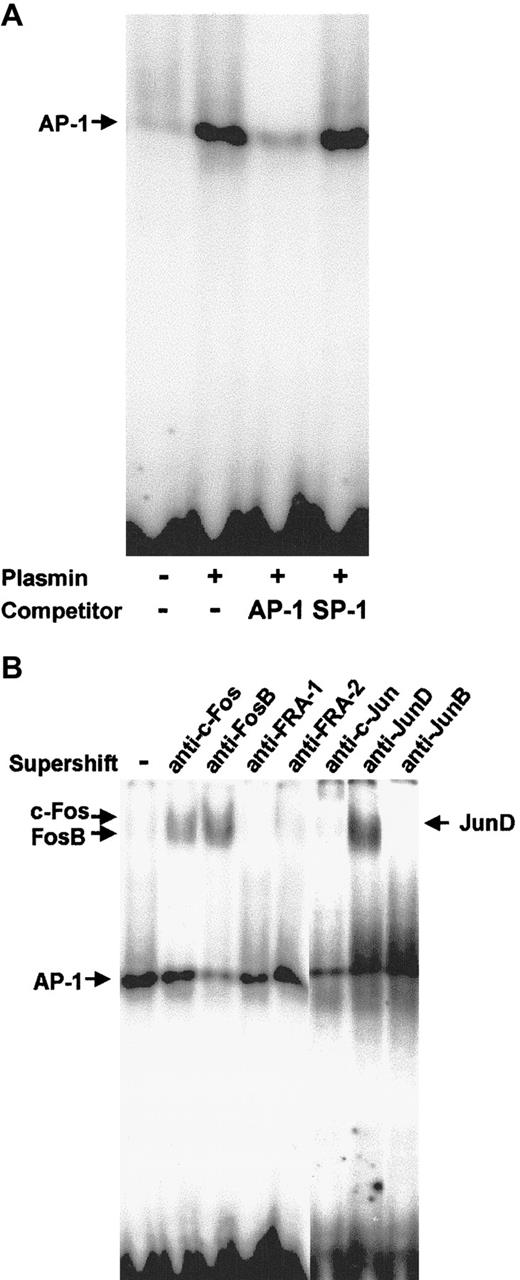
Nuclear proteins from monocytes stimulated for 2 hours with 0.43 CTA U/mL plasmin were subjected to EMSA with a DNA probe containing the AP-1 binding site. Plasmin induced strong binding to the AP-1 probe (Figure 10A). Binding specificity was confirmed using 100-fold molar excess of unlabeled AP-1 and nonspecific SP-1 probes. The identity of the Fos/Jun proteins involved in the plasmin-induced AP-1 DNA binding in human monocytes was characterized in supershift assays with specific antibodies (Figure 10B). Anti–c-Fos induced a slight supershift, whereas anti-FosB induced a stronger supershift. The AP-1 shift was also strongly retarded in the presence of anti-JunD, but not in the presence of JunB, FRA-1, or FRA-2 antibodies (Figure 10B).
AP-1 activation in plasmin-stimulated monocytes.
Binding to the AP-1 site was analyzed using EMSA. (A) Nuclear extracts (10 μg) from monocytes either unstimulated or stimulated for 2 hours with plasmin 0.43 CTA U/mL were incubated with the32P-labeled DNA probe containing the AP-1 binding site. In competition experiments the effects of specific (AP-1) and nonspecific (SP1) competitors at 100-fold molar excess were analyzed. (B) Composition of the AP-1 complexes was characterized by supershift analysis with antibodies against Jun and Fos proteins. Results of 1 of 3 experiments are shown.
AP-1 activation in plasmin-stimulated monocytes.
Binding to the AP-1 site was analyzed using EMSA. (A) Nuclear extracts (10 μg) from monocytes either unstimulated or stimulated for 2 hours with plasmin 0.43 CTA U/mL were incubated with the32P-labeled DNA probe containing the AP-1 binding site. In competition experiments the effects of specific (AP-1) and nonspecific (SP1) competitors at 100-fold molar excess were analyzed. (B) Composition of the AP-1 complexes was characterized by supershift analysis with antibodies against Jun and Fos proteins. Results of 1 of 3 experiments are shown.
Discussion
Monocytes play a central role in host defense, largely because of the release of effector molecules, such as cytokines and TF, that may in turn mediate systemic effects. Tight regulation of this process is indispensable for the prevention of undue tissue damage. Here we show that the proinflammatory activity of the serine protease plasmin extends to proinflammatory gene expression. Plasmin triggers monocytes to release TNF-α, IL-1α, and IL-1β and to express TF on the cell membrane. Because numerous immediate-early genes, including cytokines and TF, are transcriptionally regulated by NF-κB/Rel and AP-1 nuclear factors,13,16 38-40 we studied the role of distinct members of the NF-κB and AP-1 signaling pathways in the plasmin-induced monocyte activation.
Endotoxic LPS, one of the most potent stimuli of monocytes16,25,37 that signals through NF-κB,25,26,37-39 was used for comparison. Apart from LPS, signaling through other activators, such as TNF-α, IL-1β, or PMA, also converges in NF-κB activation in monocytes and monocytic cells, though the signaling cascades triggered by these stimuli appear to differ markedly.35-38 40
In monocytes, plasmin induces rapid expression of TNF-α, IL-1α, IL-1β, and TF mRNA, followed by corresponding protein biosynthesis. TNF-α was detected in the supernatant of plasmin-stimulated monocytes as early as 1 hour after stimulation. Although these data suggested a direct response to plasmin, indirect monocyte stimulation through autocrine loops were excluded in additional experiments. Thus, LTB4 released by plasmin-stimulated monocytes2,3 has been implicated in the augmentation of cytokine release from monocytes by some authors41 but not by others.42 Using 2 structurally different inhibitors of 5-lipoxygenase, zileuton, and MK-886, at concentrations that totally abolished plasmin-induced formation of LTB4, we could show that this lipid mediator did not contribute to TNF-α expression (Syrovets et al, unpublished results, July 1998). Another candidate would be IFN-γ that might be released by contaminating lymphocytes. Indeed, IFN-γ has been reported to exert a priming effect on LPS-induced production of TNF-α and IL-1β by human monocytes.43,44 However, for enhanced cytokine production, at least the costimulation of LPS with IFN-γ is required43; optimal TNF-α expression occurred only after pretreatment with IFN-γ.44 In any case, we could not detect any IFN-γ in the supernatants of plasmin-stimulated monocytes, excluding the possibility that IFN-γ contributed to the plasmin-induced cytokine expression.
LPS led to quicker and—specifically in the case of TNF-α—stronger mRNA expression than did stimulation with plasmin. However, except for IL-1β, monocytes stimulated with plasmin released similar or even higher amounts of cytokines and generated more TF. This discrepancy could be explained by the fact that cytokine production is regulated on multiple levels, such as mRNA stability, translation, processing and secretion.13,31,45 46
Regulation of mRNA stability is an important control mechanism of cytokine and TF gene expression.31,45 Although changes in the half-lives might differ only slightly, they can affect mRNA abundance by orders of magnitude over a short time period. LPS up-regulates mRNA of many cytokines and TF.16,18 This up-regulation is partially caused by mRNA stabilization and seems to play a role in the LPS induction of IL-1β.31 45 Because expression of cytokine and TF mRNA in unstimulated monocytes was barely detectable, we compared the stability of the cytokine and TF mRNA in plasmin and LPS-stimulated cells. Compared with LPS, the stability of all plasmin-induced mRNAs investigated was decreased, most profoundly in IL-1β. This indicates that in plasmin-treated monocytes, the stabilization of mRNA contributed less to gene expression than it did in LPS-stimulated cells.
Cytokine production is also regulated on the level of translation. Stimulation of monocytic cells with C5a, hypoxia, or clotting blood induces the synthesis of large amounts of IL-1β mRNA without significant translation into IL-1β protein.31 This dissociation between transcription and translation is characteristic of IL-1β but also of TNF-α. Although the above stimuli might not suffice as translational signals despite their being robust signals for transcription, LPS effectively triggers both processes.31In this context plasmin is obviously a less potent inducer of IL-1β than LPS, but not so for TNF-α. Further, the degree of mRNA polyadenylation might influence the rate of translation. Thus, LPS stimulation of RAW 264.7 macrophages leads to a 200-nucleotide increase in the 3′ poly(A) tail, which promotes the association of mRNA with polysomes and the initiation of translation.32 Because mRNA degradation is coupled to ongoing translation of mRNA,45 the reduced stability of the mRNA might reflect the increased translation of the corresponding cytokine or TF in plasmin-stimulated monocytes compared with LPS-treated cells. Comparing the amounts of mRNA with the corresponding protein expression, such a mechanism cannot be excluded for IL-1α, TNF-α, and TF. However, compared with LPS-stimulated controls, the 8-fold decreased stability of IL-1β mRNA in plasmin-treated cells was paralleled by a profoundly reduced protein expression, indicating that the accelerated mRNA decay was apparently not caused by an enhanced translation rate.
Processing and secretion processes may be particularly relevant in the case of IL-1α, IL-1β, and TNF-α, which are generated in pro-forms. Proteolytic processing of the precursors of IL-1α and IL-1β by calpains and caspase-1, respectively, results in generation of the mature forms.31 The precursor of TNF-α is anchored to the cytoplasmic membrane and subsequently cleaved into mature, soluble TNF-α.46 This processing may proceed through matrix metalloproteinases and also through serine proteases.46 The reported ability of the serine protease plasmin to promote secretion of biologically active IL-147might also contribute to the release of cytokines by plasmin-stimulated monocytes.
The NF-κB/Rel superfamily of nuclear factors provides a molecular basis for cellular responses to various stimuli. Binding sites for NF-κB/Rel were found in promoter regions of different proinflammatory genes, including IL-1α, IL-1β, TNF-α, and TF.12,18,48,49 Stimulus-specific regulation of proinflammatory genes is assured through different patterns of NF-κB binding activity.14 LPS induction of TNF-α in monocytic cells leads to nuclear translocation of p50/p65 heterodimers.16 Activation of the TF gene by LPS, however, requires binding of c-Rel/p65 heterodimers to the κB sites.18 In line with these findings, plasmin triggers the nuclear translocation of p50, p65, and c-Rel proteins crucial for the expression of cytokine and TF genes.
In unstimulated cells, NF-κB dimers are kept in the cytoplasm through interaction with inhibitory IκB proteins.12 IκBβ degradation is believed to be responsible for slow, sustained gene activation, whereas degradation of IκBα leads to rapid and acute NF-κB activation.12 In our experiments, plasmin led to rapid degradation of IκBα and simultaneous activation of NF-κB. The observed accumulation of the IκBα protein 2 hours after the onset of plasmin stimulation coincided with the decrease of the NF-κB binding, suggesting that the induction of IκBα gene transcription is responsible for the down-regulation of the NF-κB activation.12
Factor p105 is believed to have dual functions: the retention of NF-κB/Rel proteins in the cytoplasm and the generation of p50 through processing.14,50 Coordination of these functions during cell activation is poorly understood. However, it was suggested that the inducible proteolysis of p105 contributes to activation of NF-κB.35,36 Transgenic mice lacking p105, but still expressing p50, exhibit chronic inflammation, stressing the importance of p105 for immune homeostasis.14 IKKs physically interact with p105 and inducibly phosphorylate 3 C-terminal serines. After phosphorylation, p105 is degraded.12,36 We demonstrate that within 2 minutes, plasmin initiated rapid degradation of p105 that peaked at 10 minutes. However, there was no increase in p50, indicating complete degradation instead of processing. Interestingly, similar results were reported for TNF-α–stimulated HeLa cells in which TNF-α stimulation reduced the p105 half-life from 90 minutes to less than 20 minutes.36 Moreover, p105 proteolysis is also not accompanied by p50 accumulation.36 In our experiments, LPS induced only moderate and delayed proteolysis of p105 without any enhanced generation of p50 by processing. Taken together, these results suggest that p105 and IκBα are differentially sensitive targets and that, in plasmin-stimulated monocytes, proteolysis of p105 does not lead to the processing of p50. These observations are supported by the differential processing of ΙκΒ proteins in human monocytes stimulated either with LPS or TNF-α35 and by recent findings demonstrating that p105 is not a precursor of p50.51
Degradation of the NF-κB inhibitors requires their phosphorylation by specific kinases, IKKα and IKKβ.12 It has been shown that IKKβ, but not IKKα, activation is essential for the induction of κB genes by LPS.12,38 In addition, IKKβ phosphorylates IκBα with a 50 to 60 times higher activity than IKKα.52 However, IKKα is rapidly activated in TNF-α–stimulated monocytes, indicating that it might contribute to early effects during TNF-α signaling.37 Preparation of cell extracts in the presence of 1% Triton allowed us to separate both kinases and to analyze them differentially. At variance with TNF-α,37 there was no detectable IKKα activation after plasmin stimulation. As with LPS,37-39 plasmin preferentially induced activation of IKKβ in monocytes. In light of the critical role of IKKβ in the response to proinflammatory stimuli,12 this finding further supports the notion that plasmin acts on monocytes as a proinflammatory activator. The maximum activation of IKKβ occurred after 60 minutes and coincided with the degradation of IκBα and the activation of NF-κB. By contrast, LPS-stimulated cells reacted more rapidly, and maximum IKKβ activity and IκBα degradation was already observed after 30 minutes. Similar observations were made in LPS-stimulated THP-1 cells.39Thus, plasmin and LPS both act through the activation of IKKβ, leading to the proteolytic degradation of IκBα and p105, though the timing and extent of these processes are different, suggesting differential regulation.
In addition to the κB sites, the TF and IL-1β promoters contain 2 AP-1 sites required for optimal gene induction.15,18,53 In LPS-stimulated THP-1 cells, c-Jun–containing complexes interact with NF-κB proteins and synergistically enhance the induction of the TNF-α promoter.17 Stimulation of monocytic cells with LPS leads to AP-1 activation.16,18,25,26 The identity of the proteins that interact with the AP-1 site in the resting state and after LPS stimulation remains controversial. Some authors25 report constitutive binding of JunD and Fos proteins, which is supplemented by c-Jun upon LPS stimulation of THP-1 cells. Others26 find constitutive binding of JunD/FRA-2 heterodimers in THP-1 cells, followed by phosphorylation of JunD on LPS exposure. The functional consequences of the different subunit compositions of AP-1 are poorly understood, though it is acknowledged that different Fos and Jun proteins are distinct in their ability to affect transcription.19 Our findings suggest that, in contrast to THP-1 cells, fresh monocytes cultured under nonadherent conditions have only minimal constitutive AP-1 activity. However, stimulation with plasmin triggers a rapid increase in DNA binding, with most AP-1 complexes composed of JunD, c-Fos, or FosB dimers.
Despite the efforts from many laboratories, the molecular entity of the putative functional plasmin receptor remains elusive. Confirming previous results,3,4 our data show that activation requires the binding of plasmin through its lysine binding site and an intact catalytic center, implying proteolytic activation. Moreover, similar to protease-activated receptors (PARs), plasmin-induced signaling, at least in monocytes and endothelial cells, apparently proceeds through a pertussis toxin–sensitive G protein.3,4,8 Despite suspicious similarity to PARs, the latter are unlikely to mediate plasmin-induced signaling. Monocytes express PAR-1, PAR-2, and PAR-3.54,55 However, neutrophils that express PAR-256 do not migrate toward plasmin,4 suggesting that PAR-2 is not involved. In contrast to thrombin, which activates PAR-1 and PAR-3 and triggers an increase in Ca in monocytes and U937 cells,57,58 plasmin does not do so (Simmet et al, unpublished data, February 1999), nor does it induce any increase of inositol 1,4,5-trisphosphate.3 Moreover, in contrast to plasmin, thrombin is hardly able to stimulate cytokine expression in monocytes.59 Transcleavage assays revealed that low concentrations of plasmin induce the truncation of PAR-1, including removal of the SLLR epitope yielding an inactivated receptor.60 Others using recombinant extracellular segments of PARs, as well as analysis by MALDI mass spectrometry and surface plasmon resonance, similarly conclude that plasmin would disable PAR-1 and PAR-2.61 Because the functional plasmin receptor has not been identified, its signaling mechanism to NF-κB is presently unknown.
Plasminogen is ubiquitously distributed throughout the human body.1,62 Generation of plasmin during wound healing,62 thrombus organization, and inflammatory and allergic reactions9-11,63 suggests that plasmin-induced monocyte chemotaxis,4 cytokine release, and TF expression might contribute to monocyte recruitment and to the regulation of these processes. Taking into account previous findings on plasmin-induced lipid mediator release,2 3 we propose that plasmin acts as a pathophysiologically relevant proinflammatory stimulus for human monocytes. Our data provide strong evidence for a role of the plasminogen/plasmin system that may extend far beyond fibrinolytic activities. Indeed plasmin may participate in the orchestration of the cytokine network and the amplification of inflammatory processes.
Supported by the Deutsche Forschungsgemeinschaft SFB 451.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Thomas Simmet, Dept of Pharmacology of Natural Products and Clinical Pharmacology, University of Ulm, Helmholtzstrasse 20, D-89081 Ulm, Germany; e-mail: thomas.simmet@medizin.uni-ulm.de.