Abstract
A comprehensive study of changes in messenger RNA (mRNA) levels in human neutrophils following exposure to bacteria is described. Within 2 hours there are dramatic changes in the levels of several hundred mRNAs including those for a variety of cytokines, receptors, apoptosis-regulating products, and membrane trafficking regulators. In addition, there are a large number of up-regulated mRNAs that appear to represent a common core of activation response genes that have been identified as early-response products to a variety of stimuli in a number of other cell types. The activation response of neutrophils to nonpathogenic bacteria is greatly altered by exposure to Yersinia pestis, which may be a major factor contributing to the virulence and rapid progression of plague. Several gene clusters were created based on the patterns of gene induction caused by different bacteria. These clusters were consistent with those found by a principal components analysis. A number of the changes could be interpreted in terms of neutrophil physiology and the known functions of the genes. These findings indicate that active regulation of gene expression plays a major role in the neutrophil contribution to the cellular inflammatory response. Interruption of these changes by pathogens, such as Y pestis, could be responsible, at least in part, for the failure to contain infections by highly virulent organisms.
Introduction
Neutrophils are the first cells to be recruited from the blood stream to sites of inflammation1,2 and are critically important for determining the outcome of some acute infections.3 They are postmitotic cells that synthesize lower levels of protein and RNA than most dividing cells, and they can interact and/or modulate inflammation. Nevertheless, on exposure to bacteria or other activating agents, neutrophils are known to synthesize and secrete a number of cytokines4,5 including interleukin-1 (IL-1),6 IL-8,7,8 oncostatin M,9 and small inducible cytokine A3/macrophage inflammatory protein 1α/(SCYA3/MIP1A).10-12
Neutrophils are readily isolated from human peripheral blood. The isolated cells are more than 99% pure, with the principal contaminant being eosinophils, which themselves have relatively low levels of macromolecular synthetic activity. The cells can be synchronously exposed to “natural” stimuli such as opsonized bacteria and offer an attractive system for the study of gene expression in terminally differentiated cells. Although the cell biology of neutrophil activation has been studied in some detail, studies of responses at the messenger RNA (mRNA) level have been circumscribed, focusing principally on one or a few cytokine mRNA species.
Approaches for simultaneously detecting changes in levels of many of the polyadenylated RNA in a cell population fall into 3 categories: hybridization to arrays of targets complementary to specific mRNAs, sequencing of many randomly chosen complementary DNA (cDNA) fragments, or display of specific cDNA fragments on gels. A method for display of 3′-end restriction fragments of each species of RNA13 has the advantages that the position of fragments corresponding to known genes is predictable and that no prior knowledge of the sequence is needed to detect previously “unknown” genes.
We have applied cDNA display to study changes in mRNA levels in neutrophils activated by exposure to various bacteria. Sufficient analyses were performed to detect, on a statistical basis, more than 90% of all changes in transcripts. We used time-course studies to get insight into the mechanisms underlying these changes. There is a dramatic and complex change in the gene expression profiles of activated neutrophils, indicating an important role for neutrophil gene regulation in the propagation and early evolution of the inflammatory response.
Materials and methods
Bacterial strains and culture
Yersinia pestis strains KIM5 and KIM614were derived from strain KIM (Kurdistan Iran man).15,16KIM6, a derivative of KIM5, lacks the 70-kb plasmid pCD1. This plasmid carries 60 genes, 47 of which have been implicated in a system that enables the bacteria to inject specific proteins directly into the cytoplasm of mammalian cells.17-20 The injection machinery, its substrate proteins, and its regulatory apparatus are encoded by this plasmid. Y pestis strains lacking pCD1 are completely avirulent. Escherichia coli K12 strain R594 (F−lac-3350 galK2 galT22 λ−rpsL179IN(rrnD-rrnE)1) was chosen to serve as a generic avirulent enterobacterial isolate.
Overnight cultures of Y pestis grown in modified Tryptose Blood Agar Base without the agar comprising 10 g tryptose, 3 g beef extract, and 5 g sodium chloride (NaCl) per liter supplemented with 2.5 mM calcium dichloride (CaCl2) were diluted to a density of 3 × 107bacteria per mL and incubated for 3 hours at 26°C in a water bath. At this point the temperature was shifted to 37°C, and the incubation was continued for an additional 2 hours. The bacteria were collected by centrifugation, washed with Hanks balanced salt solution (HBSS; without Ca++ or Mg++), resuspended in HBSS to a final density of 1.75 × 109 bacteria per mL and opsonized by the addition of 1.5 volumes of normal human serum incubated at 37°C for 20 minutes, washed twice with RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum (FCS), and resuspended to a final density of 7 × 108 bacteria per mL.
Overnight cultures of E coli K12 grown in LB were diluted 1:100 and incubated for 2 hours at 37°C. They were then washed and opsonized as described above, except that C7-deficient human serum (Sigma Chemical, St Louis, MO) was used. This precaution was not necessary with Y pestis, which is completely resistant to complement-mediated lysis.
Cell separation and activation by bacteria
We isolated neutrophils by using dextran sedimentation, centrifugation through Ficoll-Hypaque Plus, (Amersham Pharmacia Biotech, Uppsala, Sweden) and very brief hypotonic lysis of erythrocytes.21 All reagents, serum, buffers, and media were free of LPS (less than 0.01 ng/mL by limulus amoebocyte lysate assay [Sigma]). Monocytes were enumerated in neutrophil preparations by flow microfluorometry. A neutrophil suspension was incubated with fluorescein isothiocyanate (FITC)–conjugated anti-CD45 and phycoerythrin (PE)–conjugated anti-CD14 (Becton Dickinson, Mountain View, CA). The cells were then fixed with fluorescence-activated cell sorter (FACS) lysis buffer (Becton Dickinson) and analyzed with a FACScan flow cytometer (Becton Dickinson). Monocytes were identified on the basis of their forward and side light scattering properties and expression of CD45 and CD14. At least 105 events were analyzed for each sample.
Freshly isolated neutrophils and opsonized bacteria, suspended in RPMI plus 10% heat-inactivated FCS, were mixed to final densities of 2 × 106 cells per mL and 4 × 107 bacteria per mL, respectively. These cultures or control neutrophils were then incubated for 2 hours, or other indicated times, at 37°C with gentle agitation.
Monocytes were isolated from the peripheral blood mononuclear cells by a spontaneous aggregation method at 4°C.22 To activate monocytes, they were exposed for 2 hours to opsonized E coliK12 at a ratio of 20 bacteria per cell, which is the same procedure that was used for activation of neutrophils. Time-course experiments were analyzed with neutrophils incubated for at least 3 time points including 0 minutes (negative control), 10-30 minutes (early), and 120 minutes (late) with E coli K12 .
Northern blot analysis and in situ hybridization
Northern blot analysis of total cell RNA, extracted from neutrophils by the guanidine hydrochloride (HCl) method,21,23 was performed as described.24-26Levels of hybridization were measured quantitatively by the Phosphorimager System (Molecular Dynamics, Sunnyvale, CA) and normalized to the 18S ribosomal RNA signals. In situ hybridization was performed by a previously described method,27 using Cy3 and FluorX (Amersham Pharmacia Biotech, Piscataway, NJ) labeled oligonucleotide probes.
Gel display of 3′-end restriction fragments
cDNA displays of cells activated by bacteria were prepared as previously described in detail.21 28 Bands were displayed on sequencing gels run to display products of at least 100 bases in length. Bands were excised, amplified by polymerase chain reaction (PCR), and sequenced. The enzymes used to digest cDNA for comparison of the effects of Y pestis with those of E coli were BamHI, BclI, BglII,BsrGI, ClaI, EagI, EcoRI,HindIII, NcoI, PstI, andXbaI. Enzymes used for time-course studies wereApaI, BglII, HindIII, KpnI,SacI, SpeI, SphI, andXbaI.
For most experiments, every band that differed in relative intensity between the control pattern and any of the experimental patterns was sequenced. In different experiments using the same restriction enzymes, many bands could be confidently recognized as corresponding to previously sequenced bands on the basis of both band pattern and sequence.
Informatics
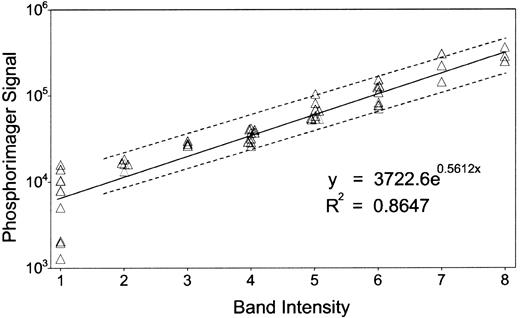
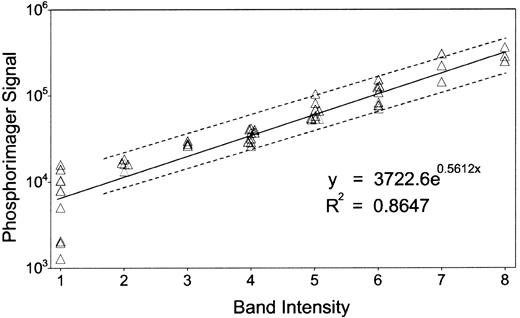
Most of the band intensities were quantified by eye, confirmed by a second investigator, and expressed as a single-digit numeric. A part of the bands was quantified by using the Phosphorimager. The integrated density of each band above background was calculated as a “volume” by the Phosphorimager, as described in Figure1. A least-square linear regression model of the logarithm of the “volume” (Ŷ) in terms of the visually quantified band intensity (X) givesŶ = 0.56X + 8.22, with Pearson correlation r = 0.93, an acceptable reliability estimate. The standard error of a given Ŷ0, estimated for a new observation X0, is computed as:
where n is the number of observations, is X averaged, and Ŷirepresents the predicted values corresponding to the observationsXi and
Thus for any given X0, a 1 − α confidence interval (CI) for Ŷ0 is the set of values of Y such that
where Tvalue (n − 2,α) is the 2-tailed t-value of a t-distribution with n − 2 degrees of freedom.
Correlation between band intensities and Phosphorimager quantification.
We confirmed the accuracy of our band quantification method by comparing our results with the Phosphorimager, whose sensitivity and reproducibility is comparable to scintillation counting.64About 50 bands from randomly chosen lanes were selected, and each of their intensities was quantified by eye, confirmed by a second investigator, and expressed as a single-digit numeric. The same gel was digitized by the Phosphorimager with 16-bit precision to form the image and analyzed by Imagequant software (Molecular Dynamics). We used the software program S-PLUS (Mathsoft, Cambridge, MA) for statistical analysis as described in “Materials and methods.” Dashed lines display the 95% CI.
Correlation between band intensities and Phosphorimager quantification.
We confirmed the accuracy of our band quantification method by comparing our results with the Phosphorimager, whose sensitivity and reproducibility is comparable to scintillation counting.64About 50 bands from randomly chosen lanes were selected, and each of their intensities was quantified by eye, confirmed by a second investigator, and expressed as a single-digit numeric. The same gel was digitized by the Phosphorimager with 16-bit precision to form the image and analyzed by Imagequant software (Molecular Dynamics). We used the software program S-PLUS (Mathsoft, Cambridge, MA) for statistical analysis as described in “Materials and methods.” Dashed lines display the 95% CI.
The accuracy of such confidence limits depends on the validity of the assumption of linearity and equally normal distributions ofY values across all values of X. However, measurements by the Phosphorimager at very low intensities are much less reliable. Therefore we fit the data to a linear regression model based on measurements at X > 1, yieldingŶ = 0.49X + 8.64 and a correlation ofr = 0.96. For all X0, Δ(X0) · Tvalue(38,0.05) ≈ 0.5.
Quantitative measurement of Northern blots of several mRNAs confirms that genes identified by gel display to be up- or down-regulated do indeed show increases or decreases of mRNA levels. These changes range from a 10-fold decrease to a 71-fold increase (Table1), and the logarithm of the values correlate to estimates from the gel display method (Pearson correlationr = 0.92).
Each sequence was searched against nr and dbest databases of the National Center for Biotechnology Information (NCBI) by the Basic Local Alignment Search Tool (BLAST) program (http://www.ncbi.nlm.nih.gov/BLAST/blast_overview.html).29Matches to known genes were confirmed to come from the 3′-untranslated regions of mRNA except where otherwise noted. The length of sequence obtained was compared with the size of bands on the display gel as a quality check.
A database was created by Shigeru Yamaga, jointly with Wen Ming Xiao (Gene Logic, Gaithersburg, MD), using Microsoft Access (Microsoft, Redmond, WA) as a database engine. An individual record was created for every differentially expressed band, and related information was entered as hypertext links to sequence files, search results of GenBank and TIGR databases, bands of overlapping sequence, references to relevant literature, keys for various classifications of bands, presence of polyA signals, quality of sequence, and scanned gel images, etc.
We used LocusLink ID,65 when available, as a unique key to known genes, and we used the terms listed as gene symbol (the HGMW-approved symbol, where applicable) and gene name. For ESTs, we used UniGene cluster numbers as a unique key.66Subsequently all sequences were clustered by a modified PHRAP approach.30 Public gene database search was completed on November 9, 2000.
Results
Quality of cell and RNA preparation
Morphologically, our neutrophil preparations were more than 99% pure except for the presence of 1% to 3% eosinophils; band forms accounted for less than 3% of the cells. No cells with the typical morphology of monocytes could be identified by light microscopy, nor did flow cytometry reveal any monocytes. Occasional preparations with more than 0.5% monocytes were discarded. The yield of total RNA from the neutrophil preparations averaged 13 μg/108 cells (range, 7-17 g/108 cells). We examined the distribution of IL-8 transcripts by in situ hybridization using a combination of 2 CY-3 (red)–labeled oligonucleotides complementary to different regions of the mRNA. IL-8 transcripts were detectable in virtually all neutrophils after incubation for 2 hours with E coli, although the intensity of RNA staining was somewhat variable from cell to cell. Neutrophils incubated in the absence of bacteria showed considerably less intense staining (data not shown).
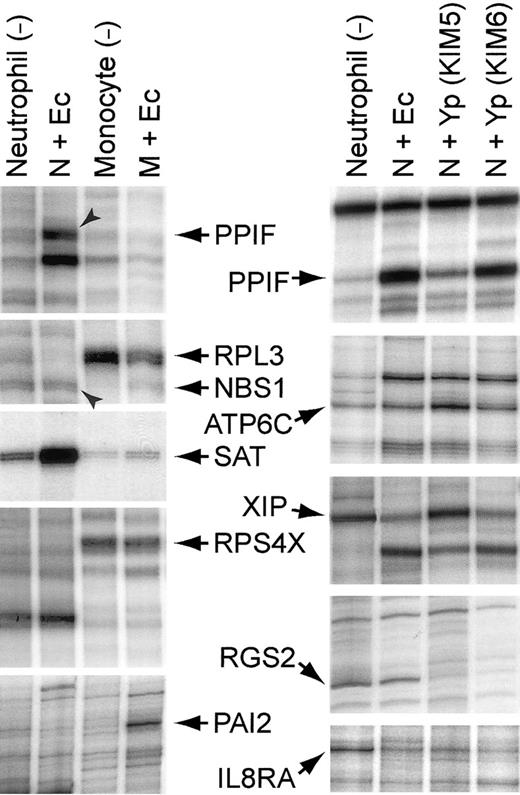
We prepared monocytes and neutrophils from the same blood sample. Both types of cells were exposed to E coli K12 for 2 hours and then harvested for cDNA display (Table2 and Figure2, left). In some cases RNA species that were among the most strongly induced in neutrophils were actually down-regulated in monocytes, excluding the possibility that monocytes activated by the bacteria were contributing to the observed pattern for these species. Northern blots also showed that RNA extracted from the neutrophils did not contain detectable transcripts forc-fms,31 the receptor for macrophage colony-stimulating factor (MCSF) (data not shown).
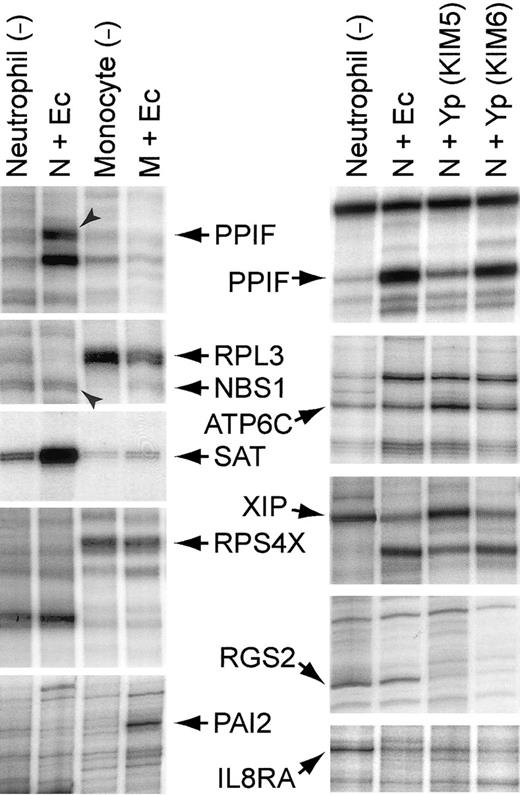
Representative segments of display gels of cDNA fragments: left, neutrophils and monocytes exposed to
E coli for 2 hours; right, neutrophils exposed to various bacteria for 2 hours. After incubating the leucocytes with or without bacteria at 37°C for 2 hours, total RNA was extracted, and double-stranded oligo-dT primed cDNA was synthesized by standard methods. The cDNA was digested with a restriction enzyme and ligated with a Y-shaped adapter. Ligated cDNA was amplified by PCR with a phosphorous 32 (32P)–labeled primer complementary to one arm of the Y-shaped adapter and a second primer complementary to the oligo-dT primer and containing one of the 12 possible dinucleotide extensions on its 3′-end, as previously described.1 2 Ec indicates E coli K12; Yp (KIM5), Y pestis KIM5 (pCD1+); Yp (KIM6), Y pestis KIM6 (pCD1−); N, neutrophil; and M, monocyte. Gene symbols are the same as those described in Tables 4-6, except for the following:PAI2, plasminogen activator inhibitor, type II (arginine-serpin) (GenBank Accession no. Y00630); RPL3, ribosomal protein L3 (X73460); and RPS4X, ribosomal protein S4, X-linked (M58458).
Representative segments of display gels of cDNA fragments: left, neutrophils and monocytes exposed to
E coli for 2 hours; right, neutrophils exposed to various bacteria for 2 hours. After incubating the leucocytes with or without bacteria at 37°C for 2 hours, total RNA was extracted, and double-stranded oligo-dT primed cDNA was synthesized by standard methods. The cDNA was digested with a restriction enzyme and ligated with a Y-shaped adapter. Ligated cDNA was amplified by PCR with a phosphorous 32 (32P)–labeled primer complementary to one arm of the Y-shaped adapter and a second primer complementary to the oligo-dT primer and containing one of the 12 possible dinucleotide extensions on its 3′-end, as previously described.1 2 Ec indicates E coli K12; Yp (KIM5), Y pestis KIM5 (pCD1+); Yp (KIM6), Y pestis KIM6 (pCD1−); N, neutrophil; and M, monocyte. Gene symbols are the same as those described in Tables 4-6, except for the following:PAI2, plasminogen activator inhibitor, type II (arginine-serpin) (GenBank Accession no. Y00630); RPL3, ribosomal protein L3 (X73460); and RPS4X, ribosomal protein S4, X-linked (M58458).
Changes in gene expression profile in neutrophils exposed to bacteria
We undertook an extensive comparison of the cDNAs generated from control neutrophils and neutrophils treated for 2 hours with one of 3 bacteria: E coli K12, Y pestis strain KIM5, orY pestis strain KIM6. A total of 17 different restriction enzymes were used for these displays, and fragments from each enzyme digest were displayed with each of the 12 possible 3′-terminal dinucleotides on the oligo-dT primer. On average, approximately 100 bands per lane could be evaluated visually. In most cases we analyzed the sequences of all bands whose inserts were in the size range of 75-600 base pairs (bp) and whose intensities differed by more than 2-fold between control and treated samples. Bands up to greater than 1 kb in length were analyzed if they were prominent and showed clear differences between samples. Based on the number of bands observed and on the frequency of randomly distributed restriction sites, we should have achieved an average of 1.5 representations of mRNA of intermediate abundance, with a higher frequency for the abundant mRNAs.
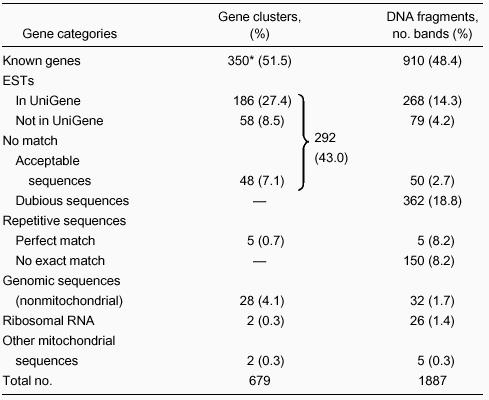
Striking differences were evident in patterns of cDNA display between control neutrophils and neutrophils exposed to bacteria (Figure 2, right). A total of 1887 bands were sequenced (Table3). Of these, approximately 19% did not give good sequence. A portion of these sequences still gave high probability matches to known sequences so that the bands could be identified. Any single prominent band is unlikely to represent more than a few percent of total mRNAs. This implies that bands corresponding to one mRNA molecule per cell are visible, except where obscured by darker bands. Redundancy in analyses occasionally occurred, particularly for some of the most prominent RNAs. Multiple bands representing the same transcript could arise by buckling out of nucleotides during oligo-dT priming, but often resulted from alternate sites of polyA addition in mRNAs.
In total, 350 known genes and 292 EST or anonymous sequences were found to change substantially in expression level by 2 hours after activation with bacteria. The anonymous cDNA could be derived from unrecognized alternate polyadenylation sites in genes represented elsewhere in the database, cDNA primed from A-rich internal sequences in mRNA or heterogeneous nuclear RNA, or genes not yet represented in the EST databases or GenBank. Five of the bands represented perfect copies of EST sequences derived from repetitive sequences. The perfect match in these cases suggests that the genomic template has been identified.
We obtained 48 nonrepetitive sequences that had no match in the gene databases. About half of these had a perfect polyA signal or a hexanucleotide that differed by a single base from the consensus AAUAAA sequence.32 Such deviation is commonly seen in mRNAs for known genes, so it is likely a large fraction of these represent polyadenylated RNA. Approximately 8% of the sequences corresponded to repetitive sequence, and most of them did not precisely match anything in the database. These frequently lacked even an approximate polyA addition signal. However, 4 different specific repetitive sequences were strongly induced in the neutrophils by exposure to bacteria. Increased transcription of repetitive sequences has been noted in stimulated cells and may have a physiologic role.33
Clustering neutrophil gene expression patterns
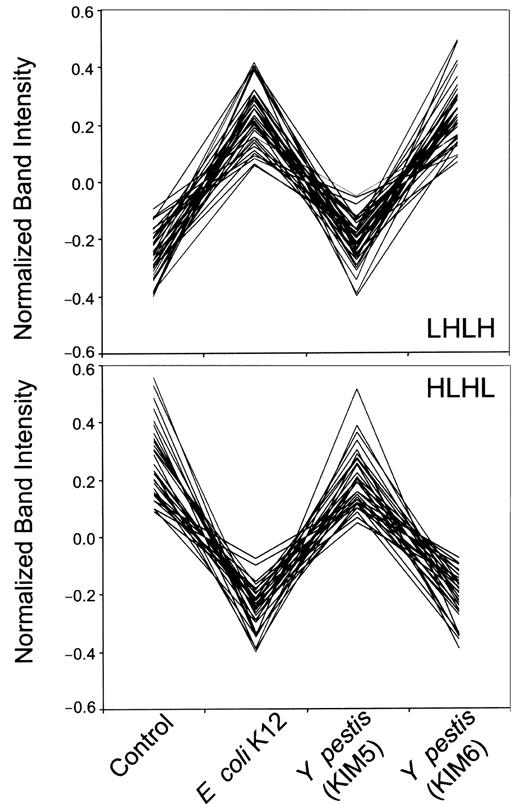
We grouped the neutrophil genes according to their expression profiles under 4 conditions in the following order: control, incubation with E coli K12 for 2 hours, and incubation with KIM5 (pCD1+) and KIM6 (pCD1−) strains of Y pestis. We clustered the genes according to their similarity to idealized expression patterns. For instance, the expression pattern of an ideal gene that is overexpressed (high) for the virulent KIM5 condition and under-expressed (low) for the control, E coli K12, and avirulent KIM6 conditions would be low-low-high-low (described as “LLHL”). Overall we have 24 − 2 idealized patterns excluding HHHH and LLLL. The Pearson correlation was used as the measure of similarity of each gene expression pattern,x = (x1,x2,x3,x4) to each of the 14 idealized patternsy = (y1,y2,y3,y4). The order of the entries for each gene expression vector xor y is control, E coli, KIM5, and then KIM6. The query gene is assigned to a cluster designated by the idealized pattern that has the maximal correlation with that gene. Figure3 shows 2 representative normalized gene expression patterns of neutrophils, LHLH (upper) and HLHL (lower).
Comparison of the expression patterns of 2 clusters of genes from neutrophils stimulated with various bacteria.
Patterns LHLH (upper, 48 genes) or HLHL (lower, 44 genes) correspond to those described in Table 4. Each set of 4 bands (control, E coli K12 and KIM5 and KIM6 strains of Y pestis; Figure2, right panel) in adjacent lanes with the same electrophoretic mobility in a differential display gel was quantified by its intensity and normalized so that the average of 4 bands equals zero, and the variance of 4 bands equals one. Each line on these graphs corresponds to one dot in Figure 4 and represents one gene in Table 4.
Comparison of the expression patterns of 2 clusters of genes from neutrophils stimulated with various bacteria.
Patterns LHLH (upper, 48 genes) or HLHL (lower, 44 genes) correspond to those described in Table 4. Each set of 4 bands (control, E coli K12 and KIM5 and KIM6 strains of Y pestis; Figure2, right panel) in adjacent lanes with the same electrophoretic mobility in a differential display gel was quantified by its intensity and normalized so that the average of 4 bands equals zero, and the variance of 4 bands equals one. Each line on these graphs corresponds to one dot in Figure 4 and represents one gene in Table 4.
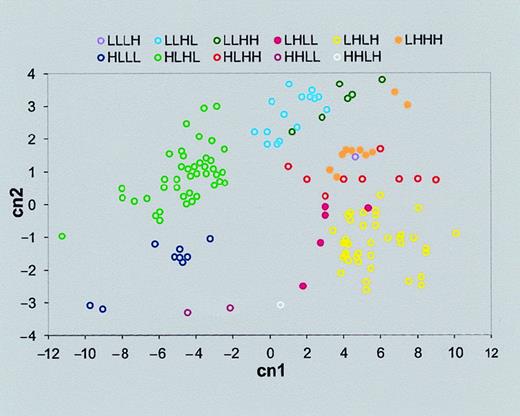
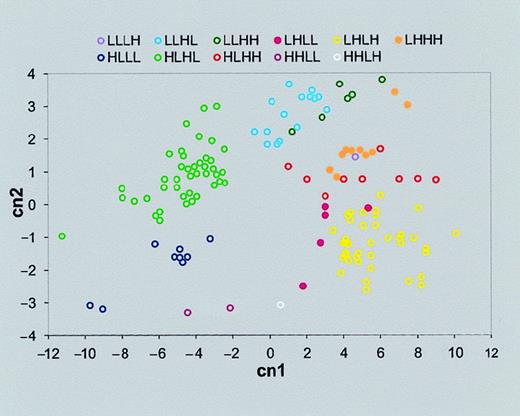
To show the affinity between genes classified to the same cluster, principal components analysis (PCA) was performed. Genes tend to coalesce in homogeneous clusters determined by their similarity to an ideal expression pattern (Figure 4). Thus, our criterion for classifying genes according to their similarity to predetermined idealized expression patterns allows us to recognize well-separated clusters. We note that this is equivalent to the first iteration of the standard k-means clustering technique.34There are 2 differences from k-means: (1) our clustering method does not require reassignment of new centers for all clusters, as is done at each k-means iteration step, and (2) the centers are predetermined by the idealized expression profiles as opposed to a random centers' initialization, which is the first step of the k-means algorithm.
Gene clusters separated by PCA.
PCA allows us to present the multidimensional data (in this case, 4-dimensional data of each gene expression pattern) in a simple 2-dimensional graph. First we derived the 4 principle components, which are a linear combination of the original variables (certain gene expression intensities of neutrophils of control or stimulated with one of 3 bacteria: E coli K12 and KIM5 and KIM6 strains ofY pestis). Then we found that the first 2 principal components capture most of the variation of the data (95.2% in our case). Therefore the data can be displayed (with a minor loss of information) in a 2-dimensional graph, with these 2 largest key principal components as the x- and y-axes. The axes titles “cn1” and “cn2” represent the first 2 principal components. The label of each cluster is the same as those in the “Expression pattern” row of Tables 4 and 5. As can be seen, a large fraction of the total differences in expression patterns of the genes can be visualized in this 2-dimensional graph.
Gene clusters separated by PCA.
PCA allows us to present the multidimensional data (in this case, 4-dimensional data of each gene expression pattern) in a simple 2-dimensional graph. First we derived the 4 principle components, which are a linear combination of the original variables (certain gene expression intensities of neutrophils of control or stimulated with one of 3 bacteria: E coli K12 and KIM5 and KIM6 strains ofY pestis). Then we found that the first 2 principal components capture most of the variation of the data (95.2% in our case). Therefore the data can be displayed (with a minor loss of information) in a 2-dimensional graph, with these 2 largest key principal components as the x- and y-axes. The axes titles “cn1” and “cn2” represent the first 2 principal components. The label of each cluster is the same as those in the “Expression pattern” row of Tables 4 and 5. As can be seen, a large fraction of the total differences in expression patterns of the genes can be visualized in this 2-dimensional graph.
Genes differently expressed in neutrophils exposed toY pestis
We also compared the effects of 2 strains of Y pestis, the causative agent of plague, on neutrophils. The high virulence of this pathogen is in part due to its ability to prevent the accumulation of neutrophils at foci of infection early in the course of disease.15,35,36 An important contribution of the type III secretion system to suppressing neutrophil accumulation is the inhibition of cytokine production.37 38
The most common pattern of mRNA change was a substantial increase in response to E coli or KIM6, but there was no change in response to KIM5 (Table 4, LHLH). Most of the cytokines we identified showed this pattern. A second common pattern is that mRNAs present in the control and KIM5-treated cells were depressed in the cells treated with E coli and KIM6 (Table 4, HLHL). This pattern also confirms that most of the cells received a stimulus as a result of exposure to the bacteria. A smaller number of mRNAs were induced or substantially up-regulated only by KIM5 (Table 4, LLHL). Overall the effects of nonpathogens on genes listed in Table 4 were quite parallel, presumably because the bacteria present common stimuli.
The expression of a smaller number of mRNAs appeared to be influenced by Y pestis, regardless of pCD1 but not by E coli(Table 5, LLHH and HHLL). Some genes were affected only byE coli (LHLL and HLHH), and a number were regulated alike by all 3 bacteria (LHHH and HLLL). Many of the changes in the levels of mRNA could be interpreted in terms of the known behavior of neutrophils. Activation of neutrophils by the nonpathogenic gram-negative bacteria induced expression of a variety of cytokines and receptors. Several known cytokines have not been previously associated with neutrophils or were first described in this context after the present analyses were completed. These include SCYA20 (LARC/MIP3A), oncostatin M, GRO1, and GRO2.
Putative membrane trafficking regulators were up-regulated in a functionally coordinate fashion. Thus mRNAs encoding the 3 small guanosine 5′-triphosphatases (GTPases), RAB1, RAB5A, and RAB7, were all up-regulated by E coli and KIM6. Interestingly, KIM5 slightly up-regulated RAB1 and RAB5A, but strongly up-regulated RAB7, a small GTPase implicated in transport from late endosomes to lysosomes. ARHGDIB, a guanine nucleotide dissociation inhibitor that would presumably delay reconversion of the GDP-bound form to the active GTP-bound form of these proteins, was down-regulated by nonpathogens, while KIM5 did not change its expression (Table4, HLHL).
Apoptosis of neutrophils in vitro is delayed by various activating stimuli. Examination of the RNAs up-regulated by the nonpathogens offers several potential mechanisms for this effect. BCL2A1 was strongly induced, as previously reported for activated monocytes.39 BCL2A1 has antiapoptotic properties, but unlike BCL2, it does not inhibit the accumulation of differentiated myeloid cells from the 32D cell line.40MCL1 is another strongly up-regulated product implicated as an antiapoptotic protein. IER3 is a p53 responsive gene that protects cells from FAS- or TNF-induced apoptosis.41 One of the most strongly up-regulated mRNAs was that for PPIF (cyclophilin F). This protein is a mitochondrial peptidyl-prolyl isomerase implicated in mitochondrial pore structure and perhaps permeability transitions.42 This is intriguing because cytochrome c release from mitochondria is a component of a caspase-activating system central to many forms of apoptosis. The mRNA encoding certain subunits of the vacuolar adenosine 5′-triphosphate (ATP)-dependent H+ pump, another potential downstream antiapoptotic factor, were also up-regulated. KIM5 had little effect on most of the above genes.
Changes in neutrophil gene expression were asynchronous
The changes in mRNA expression patterns at short time intervals following the addition of E coli K12 were also analyzed. Many of the striking increases in mRNA levels seen at 2 hours after exposure to bacteria were not reflected by changes in levels of the corresponding mRNA within the first 60 minutes, although the levels of some mRNAs progressively increased beginning within 30 minutes (Table 6, LHL and LHH). Display ofBglII-cut cDNAs prepared 3 and 4 hours after exposure toE coli showed a pattern that was for the most part similar to the 2-hour pattern (data not shown).
Some genes were transiently up-regulated, peaking at 30-60 minutes, but returning to baseline levels by 2 hours after treatment. Among the earliest response mRNAs for known genes was that for the dual-specificity protein kinase DYRK1A.43,44 This is the human homolog of Drosophila minibrain and potentially a homolog of the S cerevisiae gene YAK1, a possible negative regulator of growth and cell cycle progression.45By 60 minutes after activation, the pattern changed with down-regulation of some mRNAs and strong up-regulation of others, among which was the mRNA for ETR101,46 a proline-rich cytoplasmic protein known as a sometimes unstable early activation protein in other systems.
Discussion
The current study demonstrates that neutrophils are capable of extensive, rapid, and complex changes in gene expression. The changes in mRNA levels include both genes that are expressed and regulated in many cell types and genes that are expressed in a limited range of cells. Few of the regulated genes were strictly neutrophil-specific.
Activation of neutrophils by bacteria is a complex process that delivers multiple types of exogenous and endogenous signals to the cell. The bacterial lipopolysaccharide itself interacts with a specific receptor on the cell surface, and bacterially derived formyl peptides interact with the FMLP receptor (FPR1). Immunoglobulins and complement components associated with the bacteria stimulate an array of receptors present on neutrophils. An early consequence of neutrophil activation is the production of reactive oxygen species, and these in turn elicit a stress response from the cells. Neutrophil production of IL-1 or granulocyte-macrophage colony-stimulating factor (GM-CSF) presumably activates the corresponding receptors on the cell surface. The relative kinetics of induction of IL-8 and down-regulation of its receptors offer another potential for feedback effects on neutrophil activation.
In our study many known genes were induced on neutrophil activation including G0S2, ZFP36 (TTP/G0S24), PBEF (G0S9),ETR101, COPEB, FOSB (G0S3),FOS (G0S7), and the urokinase receptor(PLAUR). These corresponded to mRNA appearing in many other cell types during the transition from G0- to S-phase of the cell cycle or after other modes of activation. Other genes for widely used stress-response proteins, such as the heat shock products (HSPA10, HSPCA, HSPCB, and HSPF1) and the protein kinase MAP2K3, were also activated.
Two groups recently reported array analysis of changes in gene expression in fibroblasts in response to PDGF receptor47 or serum48 stimulation. For cells induced by receptor stimulation, more than 40% of the genes induced within 4 hours of stimulation were induced in neutrophils by 2 hours. This is an impressive overlap, as the fibroblasts were a transformed murine cell line held in 0.5% serum, whereas the neutrophils are postmitotic normal human cells maintained in high serum. This overlap emphasizes a commonality of very early response transcripts in mammalian cells, but suggests that quantitative considerations of the time and level of mRNA production may be central to understanding the differences in behavior of cell types.
None of the signal transduction or cell cycle genes induced in fibroblasts47 was regulated in neutrophils. However, more than 40% of immediate-early transcription factors were also up-regulated in neutrophils, and 7 of 8 genes classified as inflammation-related were also up-regulated in neutrophils. Itoh et al49 analyzed the 3′-end sequences of 1142 cDNA clones from neutrophils that were not intentionally activated and obtained sequences for 748 independent species. They listed 46 named genes for which they recovered 3 or more clones. In the present study we found that 90% of these genes were up-regulated on neutrophil activation.
Our data indicate that activated neutrophils are a source of physiologically significant trans-cellular signaling molecules. Measurements of IL-8 protein accumulation have shown that neutrophils produce IL-8 at about 1 ng per million cells per hour after exposure toE coli (J.D.G. and Y.V.B.K.S., unpublished results, February 1994). This corresponds to approximately 105molecules of IL-8 per cell per hour. In vitro, the cellular activating effects of IL-8 reach half-saturation levels at a concentration range of 0.5-1.0 nM. In vivo, human neutrophil counts commonly rise above 10 million cells per mL blood, enough to raise the concentration of IL-8 to physiologically effective levels within 1-2 hours. At sites of infection, tissue neutrophils are considerably more concentrated. Therefore, the levels of IL-8 production by neutrophils are physiologically very significant.
The levels of induced mRNA for a number of intracellular proteins are comparable to those for the more abundant cytokine mRNAs. This strongly suggests that the intracellular molecules are produced at levels that are physiologically significant, although the possibility of concomitant negative control of translation rate of specific mRNAs has not been investigated. More caution is necessary in interpretation of down-regulation of mRNA. The down-regulation will only correspond to changes in protein level if the protein normally has a short half-life or is specifically degraded following activation of the neutrophils. Some of the down-regulation is undoubtedly due to stopping transcription of relatively short-lived mRNAs. This change would not produce synchronous effects on all mRNA both because they have differing half-lives and because transcription may not be down-regulated simultaneously on different genes. Some mRNAs that are stable in cells treated with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) disappear rapidly after exposure to bacteria (data not shown). Studies with actinomycin D indicate that the mRNA for certain chemokine receptors is destabilized on LPS activation of cells,50and this destabilization is blocked by simultaneous, but not by delayed, addition of the transcription inhibitor. In any case, the events leading to destabilization are heterogeneous.
CC-chemokines, such as SCYA3, SCYA4, and SCYA20, were up-regulated. CXC chemokines, such as IL-8, GRO1, and GRO2, were also up-regulated. Although GRO1 and GRO2 share 90% identity at the deduced amino acid level, and both have melanoma growth-stimulating activity, their expression patterns were different. GRO1 was induced by KIM5 more strongly than by nonpathogenic bacteria, but the induction of GRO2 seen with KIM6 did not occur with KIM5.
Although both CXCR1 (IL-8RA, a receptor that is relatively specific for IL-8) and CXCR251 (IL-8RB, a receptor activated by other CXC chemokines including GRO1) were down-regulated, KIM5 fully inhibited gene expression of CXCR1, but not CXCR2. CXCR1 and CXCR2 are regulated in different modes by CXC chemokines and play diverse roles in mediating the inflammatory process.52 The putative G protein–coupled receptors CCRL2 (HCR) and HM74 were prominently up-regulated. HCR was previously identified in public databases as CCR6 (a receptor for SCYA20/LARC/MIP3A), but recently it has been described as a distinctive receptor, CCRL2. The sequence of CCRL2 in GenBank Accession no. U95626 is identified as CCR6, but differs from the sequence of U68030 CCR6 mRNA, so it remains uncertain whether CCRL2 is the receptor of SCYA20/LARC/MIP3A. The presence of both SCYA20/LARC/MIP3A and its receptor on the same cells would imply an autocrine loop. The strong induction of HM74 in human neutrophils suggests its utility as a clinical parameter and/or a drug target in inflammatory disorders. Overall, the responses to some stimuli were down-regulated, and new response pathways could be established. Whether these maintain or modulate the active state or have other functions remain to be determined, but they probably play important roles in the early evolution of the inflammatory process. A suggestion to explain the virulence of KIM5 is that the loss of production of the primary activating and chemo-attractant cytokine IL-8 would decrease the possibility that neutrophils which have ingested bacteria would attract additional neutrophils to sites of inflammation. The net effects of up-regulation of IL-1 and its receptor antagonist IL-1RN are uncertain, but they could provide an additional measure of feedback.
The balance between apoptotic and necrotic cell death in neutrophils plays an important role in the control of inflammation. Neutrophils accumulate in large numbers at sites of inflammation, forming tissue infiltrates and pus. Necrotic death of these cells releases toxic granule contents, such as elastase and collagenase, whereas removal of apoptotic neutrophils by macrophages protects surrounding tissues from such damage.53,54 However, inhibition of neutrophil apoptosis may augment host defense against infection by prolongation of functional longevity of the cells.55 When cultured in vitro, neutrophils rapidly undergo apoptosis, which is preceded by intracellular acidification.56 G-CSF and a variety of inflammatory mediators delay programmed cell death, in part by up-regulation of expression of Bcl-X1, but not other Bcl-2 family members.53 56-58 The current data suggest that other proteins, such as BCL2A1, MCL1, PPIF, TNFAIP3, and perhaps spermidine/spermine N1-acetyltransferase (SAT), may be important for the regulation of neutrophil apoptosis in response to infection.
Increases in mRNA for genes regulating transcription or translation were observed 2 hours after activation. These include theCOPEB gene, which is reported to stimulate expression of genes lacking a TATA box. In cells exposed to KIM5, 12 of 14 transcription-modifying genes examined were present at levels more similar to those of control neutrophils than of the neutrophils stimulated by the other bacteria.
In time-course studies we found that NFKBIA (IκBα) was induced byE coli K12 in 30 minutes, but NFKB1 (NF-κB) induction was observed after 60 minutes. In contrast to NFKBIA, NFKBIE (IκBε) was activated rather later. IκB is known as a negative regulator of NF-κB by formation of stable IκB/NF-κB complexes. This retains NF-κB in the cytoplasm until the NF-κB activation signal is received.59 This asynchronous activation of reciprocal transcription factors presumably reflects a transient activation of NFκB-dependent genes.
Activation by nonpathogens, but not by the pathogenic KIM5, caused down-regulation of mRNAs for some antibacterial products, including the phagocyte oxidase (PHOX) system (NCF1, NCF2, and NCF4) generating reactive oxygens,60 and the calgranulins (S100A8 and S100A9). In contrast, the free radical scavenging enzyme SOD2 was up-regulated by the nonpathogens. The reactive oxygen system is regulated by external stimuli61 and is auto-cytotoxic for neutrophils. Its down-regulation may contribute to the prolongation of life span of activated cells in inflammation.
Decay-accelerating factor (DAF) was up-regulated by nonpathogen, but MCP,62 a cofactor of serine protease factor I for inactivation of complements C3b and C4b, was down-regulated. Although both play a protective role in host cells against homologous complement, MCP is also the receptor for various viruses and bacterial pathogens. CD97,63 the receptor for DAF, is regulated oppositely to DAF, so that the cells may become desensitized to DAF.
Overall, the patterns of induction or disappearance of mRNAs for genes of known function can largely be rationalized in terms of the biologic role of neutrophils. Several different antiapoptotic mechanisms are set in play in an asynchronous fashion. This response would allow neutrophils that ingested nonpathogenic material to survive longer, potentially migrating to restricted tissue areas and also degrading ingested material. Additional defensive changes in the neutrophils include production of DAF. The cells change their own cytokine responsiveness and begin producing a range of new cytokines. These would not only transmit inflammatory signals and recruit unstimulated neutrophils, but they would also further stimulate activated neutrophils, contributing to the congregation of activated neutrophils and hence to abscess formation. Changes in the levels of intracellular signaling molecules might well change the responses to stimulation of pre-existing receptors. Membrane trafficking is accelerated perhaps related to ingestion of bacteria and discharge of preformed granules. There is also a previously unappreciated transition from early to delayed responses at the level of mRNA production.
In summary, nonpathogenic gram-negative bacteria induce a marked change in the patterns of gene expression in neutrophils, indicating massive changes in cytokine output and prolongation of cell survival. These changes imply that neutrophils are important effectors of the progression of the cellular inflammatory response. Interruption of these changes by pathogens, such as Y pestis KIM5, could be, at least in part, responsible for the failure to contain the infectious process.
Supplementary information is available on our Web site.67
We thank Andrea M. Neuman, Carolyn Padden, Angela Plette, Anne-Marie Quinn, and Connie Whitney for technical assistance and Dov Greenbaum for Web site set-up.
GENE SYMBOLS FOR TABLES 4, 5, AND 6
The gene symbols given in Tables 4, 5, and 6 are listed in alphabetical order. ACTB indicates actin, beta; ACTG1, actin, gamma 1; ADAM8, a disintegrin and metalloprotease domain 8 (CD156); ADORA2A, adenosine A2a receptor; AMPD2, adenosine monophosphate deaminase 2 (isoform L); ANPEP, alanyl (membrane) aminopeptidase (CD13);AQP9, aquaporin 9; ARHGDIB, rho GDP dissociation inhibitor (GDI) beta; ARPC1B, actin-related protein 2/3 complex, subunit 1A (41 kd); ATF4, activating transcription factor 4 (tax-responsive enhancer element B67); ATP2A3, ATPase, Ca++ transporting, ubiquitous; ATP2B1, ATPase, Ca++ transporting, plasma membrane 1;ATP5E, ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit; ATP6F, ATPase, H+ transporting, lysosomal (vacuolar proton pump) 21 kd;ATP6J, ATPase, H+ transporting, lysosomal (vacuolar proton pump), member J; ATP6L, ATPase, H+ transporting, lysosomal (vacuolar proton pump) 16 kd;ATP6S1, ATPase, H+ transporting, lysosomal (vacuolar proton pump), subunit 1; B2M, beta-2-microglobulin; BB1, malignant cell expression–enhanced gene/tumor progression–enhanced gene (human, UM-UC-9 bladder carcinoma cell line, mRNA, 1897 nt); BCL2A1, BCL2-related protein A1; BRF2, butyrate response factor 2 (EGF response factor 2) (TIS11D); BRI3, brain protein I3;BTF3, basic transcription factor 3a; BTG2, BTG family, member 2 (TIS21); CAMKK2, Ca/calmodulin-dependent protein kinase kinase 2, beta; CCRL2, chemokine (C-C motif) receptor-like 2; CD44, CD44 antigen (homing function and Indian blood group system); CD48, CD48 antigen (B-cell membrane protein); CD97, CD97 antigen; CDKN1A, cyclin-dependent kinase inhibitor 1A (p21, Cip1); CEBPB, CCAAT/enhancer binding protein (C/EBP), beta; CFLAR, CASP8 and FADD-like apoptosis regulator (I-FLICE); CLIC1, chloride intracellular channel 1; CLK1, CDC-like kinase 1;CLN2, ceroid-lipofuscinosis, neuronal 2, late infantile (Jansky-Bielschowsky disease); COPEB, core promoter element binding protein (CPBP); CPD, carboxypeptidase D;CROC4, transcriptional activator of the c-fos promoter;CTSD, cathepsin D (lysosomal aspartyl protease);CYBA, cytochrome b-245, alpha polypeptide (p22-PHOX);DAF, decay accelerating factor for complement (CD55, Cromer blood group system); DDIT3, DNA-damage–inducible transcript 3 (CHOP10); DIFF48, KIAA0386 gene product;KIAA0415, KIAA0415 gene product; DNM2, dynamin 2;DSIPI, delta sleep-inducing peptide, immunoreactor;DUSP6, dual-specificity phosphatase 6; DYRK1A, dual-specificity tyrosine-(Y)-phosphorylation–regulated kinase 1A (minibrain homolog); EDG6, endothelial differentiation, G-protein–coupled receptor 6; EGR1, early growth response 1 (TIS8, G0S30); EHD1, EH domain–containing 1 (HPAST);EIF4A1, eukaryotic translation initiation factor 4A, isoform 1; EMD, Emerin (Emery-Dreifuss muscular dystrophy);ERV1, endogenous retroviral sequence 1; ETR101, immediate-early protein; ETS2, v-ets avian erythroblastosis virus E26 oncogene homolog 2; EVI2B, ectropic viral integration site 2B; FACL1, fatty-acid-coenzyme A ligase, long-chain 1; FCAR, Fc fragment of IgA, receptor for (CD89);FCER1G, Fc fragment of IgE, high-affinity I, receptor for gamma polypeptide; FCGR3A, Fc fragment of IgG, low-affinity IIIa, receptor for (CD16); FLOT1, flotillin 1;FOS, v-fos FBJ murine osteosarcoma viral oncogene homolog (G0S7, c-FOS); FOSB, FBJ murine osteosarcoma viral oncogene homolog B (G0S3); FPR1, formyl peptide receptor 1;FTH1, ferritin, heavy polypeptide 1; FTL, ferritin, light polypeptide; G0S2, putative lymphocyte G0/G1 switch gene;GADD34, growth arrest and DNA-damage–inducible 34 (MyD116 homolog); GADD45B, growth arrest and DNA-damage–inducible, beta (MyD118 homolog); GC20, translation factor sui1 homolog; GCL, grancalcin; GCP2, gamma-tubulin complex protein 2; GCSH, glycine cleavage system protein H (aminomethyl carrier); GPR44, G protein–coupled receptor 44 (CRTH2); GPRK6, G protein–coupled receptor kinase 6;GRN, granulin; GRO1, GRO1 oncogene (melanoma growth stimulating activity, alpha); GRO2,GRO2 oncogene (MIP2A); GSTTLp28, glutathione-S-transferase–like (glutathione transferase omega);H1F2, H1 histone family, member 2; H3F3A, H3 histone, family 3A; HCLS1, hematopoietic cell–specific Lyn substrate 1; HDAC3, histone deacetylase 3; HEF1, enhancer of filamentation 1 (cas-like docking, Crk-associated substrate-related); HEM1, hematopoietic protein 1;HIF1A, hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor); HLA-A, MHC class I, A; HLA-C, MHC class I, C; HLA-E, MHC class I, E;HM74, putative chemokine receptor, GTP-binding protein;HMG17, high-mobility group (nonhistone chromosomal) protein 17; HMGIY, high-mobility group (nonhistone chromosomal) protein isoforms I and Y; HMOX1, heme oxygenase (decycling) 1; HNRPC, heterogeneous nuclear ribonucleoprotein C (C1/C2);HPCAL1, hippocalcin-like 1; HS1–2, putative transmembrane protein; HSPA10, heat shock 70-kd protein 10 (HSC71); HSPCA, heat shock 90-kd protein 1, alpha;HSPCB, heat shock 90-kd protein 1, beta; HSPF1, heat shock 40-kd protein 1; ICAM1, intercellular adhesion molecule 1 (CD54); ICAM3, intercellular adhesion molecule 3 (CD50); ICB-1, basement membrane–induced gene;IER3, immediate early response 3; IFITM1, interferon-induced trans-membrane protein 1 (9-27); IFNGR1, interferon-gamma receptor 1 (CDw119); IFNGR2, interferon-gamma receptor beta chain; IL1A, interleukin-1, alpha; IL1B, interleukin-1, beta; IL1RN, interleukin-1 receptor antagonist; IL8, interleukin-8;IL8RA, interleukin-8 receptor, alpha (CDw128a CXCR1);IL8RB, interleukin-8 receptor, beta (CXCR2);IRAK1, interleukin-1 receptor-associated kinase 1;IRF1, interferon regulatory factor 1; ITGA5, integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (CD49e);ITGB2, integrin, beta 2 (CD18, LFA-1); KIAA0370, KIAA0370 protein; KIAA0446, KIAA0446 gene product;KIAA1100, KIAA1100 protein; LCP2, lymphocyte cytosolic protein 2 (SH2 domain–containing leukocyte protein of 76 kd); LILRA2, leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 (LIR-7); LIMK2, LIM domain kinase 2; LOC51312, mitochondrial solute carrier;LOC51669, HSPC035 protein; LSP1, lymphocyte-specific protein 1; LTB, lymphotoxin beta (TNF superfamily, member 3); MACS, myristoylated alanine-rich protein kinase C substrate (MARCKS, 80K-L); MAP2K3, mitogen-activated protein kinase kinase 3; MAP3K8, mitogen-activated protein kinase kinase kinase 8 (COT);MCL1, myeloid cell leukemia sequence 1 (BCL2-related);MCP, membrane cofactor protein (CD46, trophoblast-lymphocyte cross-reactive antigen); MKNK1, MAP kinase–interacting serine/threonine kinase 1; MME, membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase, CALLA, CD10); MNDA, myeloid cell nuclear differentiation antigen;MYO1E, myosin IE; NACA, nascent-polypeptide–associated complex alpha polypeptide;NAF1, Nef-associated factor 1; NAPA,N-ethylmaleimide–sensitive factor attachment protein, alpha (a-SNAP); NBS1, Nijmegen breakage syndrome 1 protein (nibrin); NCF1, neutrophil cytosolic factor 1 (47 kd, chronic granulomatous disease, autosomal 1) (p47-PHOX);NCF2, neutrophil cytosolic factor 2 (65 kd, chronic granulomatous disease, autosomal 2) (p67-PHOX); NCF4, neutrophil cytosolic factor 4 (40 kd) (p40-PHOX); NDUFV2, NADH dehydrogenase (ubiquinone) flavoprotein 2 (24 kd);NFE2, nuclear factor (erythroid-derived 2), 45 kd;NFE2L2, nuclear factor (erythroid-derived 2)–like 2;NFKB1, nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105); NFKBIA, nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (I-kappa-B alpha); NFKBIE, nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon (I-kappa-B epsilon); NPM1, nucleophosmin (nucleolar phosphoprotein B23, numatrin); NS1-BP, NS1-binding protein; NUMB, numb (Drosophila) homolog; OLR1, oxidized low-density lipoprotein (lectin-like) receptor 1; OSM, oncostatin M;PBEF, pre–B-cell colony-enhancing factor (G0S9);PCOLN3, procollagen (type III) N-endopeptidase;PFN1, profilin 1; PIG7, LPS-induced TNF-alpha factor (LITAF); PLAUR, plasminogen activator, urokinase receptor (CD87); PLEK, Pleckstrin; PPGB, protective protein for beta-galactosidase; PPIA, peptidyl-prolyl isomerase A (cyclophilin A); PPIF, peptidyl-prolyl isomerase F (cyclophilin F);PRG1, proteoglycan 1, secretory granule; PSMC4, proteasome (prosome, macropain) 26S subunit, ATPase, 4 (TBP7);PSCDBP, Pleckstrin homology, Sec7 and coiled/coil domains, binding protein; PTGS2, prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclo-oxygenase) (COX2);PTPN6, protein tyrosine phosphatase, nonreceptor type 6;PTPRC, protein tyrosine phosphatase, receptor type, C (CD45); RAB1, RAB1, member RAS oncogene family;RAB5A, RAB5A, member RAS oncogene family; RAB7, RAB7, member RAS oncogene family; RAC1, Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1); RALGDS, Ral guanine nucleotide dissociation stimulator; RGS2, regulator of G-protein signaling 2, 24 kd (G0S8); RGS14, regulator of G-protein signaling 14;RPL18A, ribosomal protein L18a; RPN2, ribophorin II; RPS6KA1, ribosomal protein S6 kinase, 90 kd, polypeptide 1; RTN4, reticulon 4; S100A8, S100 calcium-binding protein A8 (calgranulin A); S100A9, S100 calcium-binding protein A9 (calgranulin B); S100A11, S100 calcium-binding protein A11 (calgizzarin); S100P, S100 calcium-binding protein P; SAT, spermidine/spermine N1-acetyltransferase; SCYA3, small inducible cytokine A3 (G0S19–1, LD78, MIP1A); SCYA4, small inducible cytokine A4 (LAG1, MIP1B); SCYA20, small inducible cytokine subfamily A (C-C), member 20 (LARC, MIP3A); SECTM1, secreted and trans-membrane 1; SELL, selectin L (lymphocyte adhesion molecule 1) (CD62L); SGK, serum/glucocorticoid-regulated kinase; SH3BP5, SH3-domain binding protein 5 (BTK-associated); SLC7A5, solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 (CD98);SLC11A2, solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 (NRAMP2); SLC16A3, solute carrier family 16 (monocarboxylic acid transporters), member 3 (MCT3);SOD2, superoxide dismutase 2, mitochondrial;SSR2, signal sequence receptor, beta (translocon-associated protein beta) (TRAPB); SUI1, putative translation initiation factor (EIF1–A121); TALDO1, transaldolase 1;TIMP1, tissue inhibitor of metalloproteinase 1 (EPA);TMSB4X, thymosin, beta 4, X chromosome; TNFAIP2, TNF-alpha–induced protein 2; TNFAIP3, TNF-alpha–induced protein 3; TNFAIP6, TNF-alpha–induced protein 6;TNFRSF1A, TNF receptor superfamily, member 1A (CD120A);TNFRSF10B, TNF receptor superfamily, member 10b (DR5);TOM1, target of myb1 (chicken) homolog; TPD52L2, tumor protein D52-like 2; TPM3, tropomyosin 3 (nonmuscle);TPT1, tumor protein, translationally controlled 1 (IgE-dependent histamine-releasing factor); TRIP8, thyroid hormone receptor interactor 8; TYROBP, TYRO protein tyrosine kinase binding protein; UBE2B, ubiquitin-conjugating enzyme E2B (RAD6 homolog); ULK1, unc-51 (C elegans)–like kinase 1;VATD, vacuolar proton pump delta polypeptide;VDUP1, up-regulated by 1,25-dihydroxyvitamin D-3 (HHCPA78);VPS35, vacuolar sorting protein 35 (yeast homolog);WBP2, WW domain binding protein 2; WBSCR1, Williams-Beuren syndrome chromosome region 1 (EIF4H); XIP, hepatitis B virus x-interacting protein (9.6 kd); ZFP36, zinc finger protein homologous to Zfp-36 in mouse (G0S24, TIS11, TTP);ZNF148, zinc finger protein 148 (pHZ-52); andZNF220, zinc finger protein 220 (MOZ).
Y.V.B.K.S. and S.Y. contributed equally to this work.
Submitted August 1, 2000; accepted November 30, 2000.
Supported by grants AI22176 (J.D.G.), CA42556 (S.M.W.), and DK54369 (P.E.N.) from the National Institutes of Health, Bethesda, MD, and by a research grant (P.E.N.) from the Arthritis Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sherman M. Weissman, Department of Genetics, Boyer Center for Molecular Medicine, Yale University School of Medicine, 295 Congress Ave, New Haven, CT 06536-0812; e-mail:sherman.weissman@yale.edu.