Abstract
Inherited deficiency of adenosine deaminase (ADA) results in one of the autosomal recessive forms of severe combined immunodeficiency. This report discusses 2 patients with ADA deficiency from different families, in whom a possible reverse mutation had occurred. The novel mutations were identified in the ADAgene from the patients, and both their parents were revealed to be carriers. Unexpectedly, established patient T-cell lines, not B-cell lines, showed half-normal levels of ADA enzyme activity. Reevaluation of the mutations in these T-cell lines indicated that one of the inherited ADA gene mutations was reverted in both patients. At least one of the patients seemed to possess the revertant cells in vivo; however, the mutant cells might have overcome the revertant after receiving ADA enzyme replacement therapy. These findings may have significant implications regarding the prospects for stem cell gene therapy for ADA deficiency.
Introduction
Adenosine deaminase (ADA) is an enzyme of the purine salvage pathway that catalyses the deamination of adenosine and 2′-deoxyadenosine to inosine and 2′-deoxyinosine, respectively. Inherited deficiency of ADA was serendipitously found to result in one of the autosomal recessive forms of severe combined immunodeficiency (SCID).1 Evaluation of a genotype-phenotype relationship revealed a deficiency of ADA results in varied impairment of immune status as well as clinical course.2 Residual ADA enzyme activity of each mutant correlated closely with biochemical and clinical phenotypes of patients.3
In 1990, the first clinical trial of gene therapy was performed in a patient with ADA deficiency.4 Since then, more than 10 patients with ADA deficiency have been treated by gene therapy. Although beneficial effects of the gene therapy were reported in some cases,4,5 complete cure of the disease by stem cell gene therapy has not yet been realized.6-8 Such results may suggest that inevitable ADA enzyme replacement interferes with growth advantage of gene-introduced cells compared with recent successful gene therapy of X-SCID patients.9
Here we report data on 2 patients with ADA deficiency from different families, in whom possible reverse mutation had occurred as reported previously.10 We detected the reversion in the patients' T-cell lines, and at least one of them seemed to possess the revertant cells in vivo; however, mutant cells might overcome the revertant after receiving ADA enzyme replacement therapy.
Study design
Patients
Patient 1, now a 13-month-old girl, is the first child of a healthy unrelated couple. She had an episode of respiratory infection at age 15 days, and after 5 days, she was admitted to a hospital with progressive respiratory distress and failure to thrive. Because the number of her peripheral blood lymphocytes was very low (200/μL), she was suspected to have SCID, and screening of ADA enzyme activity revealed a trace level of ADA. The patient is now being treated with ADA polyethylene glycol-modified bovine ADA (PEG-ADA).
Patient 2, now an 11-month-old girl, is the second child of a healthy couple without consanguinity. When she was 1 month old, she had a bacterial skin infection on her thigh. The episode was temporally controlled; however, she was hospitalized after a few days because the infection recurred. Laboratory examination revealed a very low number of white blood cells (700-1700/μL) and hypogammaglobulinemia (IgG, 162 mg/dL; IgA, <6.0 mg/dL; IgM, 13.5 mg/dL). Her red blood cells (RBC) were examined by ADA screening assay, and the results showed a trace level of the enzyme activity. During this study, she received bone marrow transplantation from her healthy brother when she was 5 months old.
Cell lines
The T-cell lines from the patients were established using herpesvirus saimiri as described elsewhere.11 B-cell lines were also established with Epstein-Barr virus. Because of the low number of peripheral lymphocytes, it took 2 to 3 months to establish both the patients' lines. We performed clonality analysis of T-cell lines by FACS (FACSCalibur; Becton Dickinson, San Jose, CA) using murine monoclonal antibodies to human T-cell receptor variable regions (Endogen, Cambridge, MA).
Mutation analysis of the patients
Genomic DNA was purified from leukocytes using Sepa Gene (Sanko Junyaku, Tokyo, Japan). Primers and polymerase chain reaction (PCR) conditions used were described previously.2,12,13 Each PCR-amplified fragment was purified and then directly sequenced as described.14 To verify the mutations detected, the fragments including the mutation were digested with restriction enzymesBfaI and BglI (New England Biolabs, Beverly, MA) and PvuII (Promega, Madison, WI). Approval was obtained from the institutional review board for these studies, and informed consent from all familial members in this article was provided according to the Declaration of Helsinki.
ADA enzyme assay and measurement of adenine nucleotides (AXP)
The ADA enzyme activity was assayed by radiochemical thin-layer chromatography methods as previously reported.2,5,15 The results were expressed as nanomoles of inosine and hypoxanthine produced per minute by 108 cells (U; nmol/min/108 cells). The levels of AXP and deoxyadenosine nucleotides (dAXP) in erythrocytes were measured as previously described.16
HLA typing studies
The DNA typing studies for identification of HLA class I, DR, and DRB1 alleles were performed by PCR sequence-specific primer methods and PCR-microtiter plate hybridization methods,17respectively.
Results and discussion
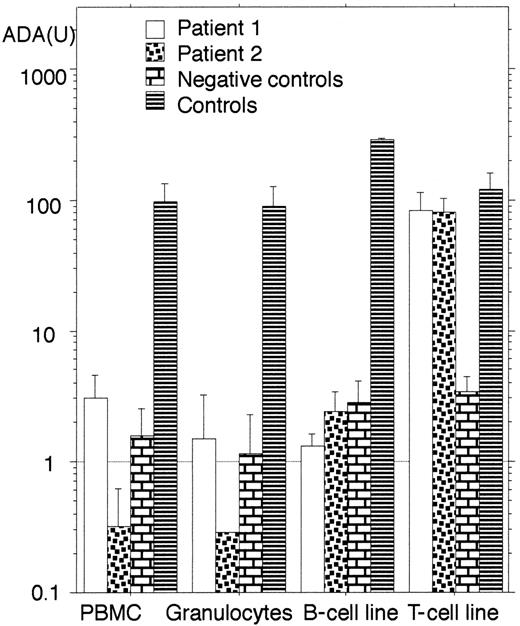
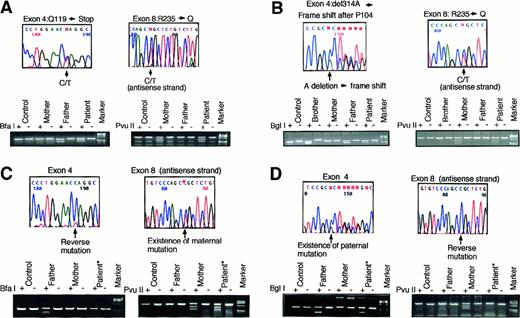
Both patients were diagnosed to be SCID with ADA deficiency. The results of ADA enzyme activity of their peripheral blood mononuclear cells (PBMC) and granulocytes are shown in Figure1, and dAXP levels in their RBC were: patient 1, 0.079 μmol/mL RBC; patient 2, 0.153 μmol/mL RBC; control, < 0.002 μmol/mL RBC. We also identified their mutations in the ADA gene. The mutations in patient 1 were nonsense mutation in exon 4 (355C>T, Q119X) (single-letter amino acid codes) and missense mutation in exon 8 (704G>A, R235Q). Molecular studies for her parents revealed they were carriers (Figure2A). The mutations in patient 2 were one base deletion in exon 4 (del314A) and missense mutation in exon 8 (704G>A, R235Q). Her parents also were carriers (Figure 2B). These results were verified by digestion of the PCR fragments with the restriction enzymes. These were novel mutations. As far as we could determine, these families were not related; however, their mothers shared the same mutant ADA allele (R235Q).
ADA enzyme activity in various cells from the patients.
The columns indicate the geometric mean of ADA activity (U; nmol/min/108 cells) and the positive error bars indicate the SDs. Negative controls except for T-cell line are obtained from another ADA-deficient patient before the gene therapy.5TJF2 cell line15 was used as negative control for T-cell line. Note that only T-cell lines from the patients show elevated enzyme activity.
ADA enzyme activity in various cells from the patients.
The columns indicate the geometric mean of ADA activity (U; nmol/min/108 cells) and the positive error bars indicate the SDs. Negative controls except for T-cell line are obtained from another ADA-deficient patient before the gene therapy.5TJF2 cell line15 was used as negative control for T-cell line. Note that only T-cell lines from the patients show elevated enzyme activity.
Mutation analysis.
Mutation analysis of the patient 1 (A) and patient 2 (B) with ADA deficiency and the reversion in T-cell lines from patient 1 (C) and patient 2 (D). Arrows indicate mutation sites. Each mutation was confirmed by digestion of the PCR fragment with a restriction enzyme indicated at the left end of each lower column. Each enzyme cuts only the fragment including the mutation, and both patterns with (+) and without (−) the enzyme are shown. Paternally inherited mutation of patient 1 and maternally inherited mutation of patient 2 were reversed. Digestion studies verified the reversions. An asterisk indicates the sample obtained from the T-cell line. Molecular marker used is φX 174HaeIII digested.
Mutation analysis.
Mutation analysis of the patient 1 (A) and patient 2 (B) with ADA deficiency and the reversion in T-cell lines from patient 1 (C) and patient 2 (D). Arrows indicate mutation sites. Each mutation was confirmed by digestion of the PCR fragment with a restriction enzyme indicated at the left end of each lower column. Each enzyme cuts only the fragment including the mutation, and both patterns with (+) and without (−) the enzyme are shown. Paternally inherited mutation of patient 1 and maternally inherited mutation of patient 2 were reversed. Digestion studies verified the reversions. An asterisk indicates the sample obtained from the T-cell line. Molecular marker used is φX 174HaeIII digested.
Unexpectedly, T-cell lines, not B-cell lines, from both patients showed half-normal levels of ADA activity (Figure 1). The results were consistent with Western blot analysis for ADA protein in these cell lines (data not shown). These unexpected findings forced us to study the mechanism of restoration of ADA enzyme activity in their T-cell lines.
The levels of ADA activities from both patients' T lines were half-normal. Therefore, we postulated that the restoration of ADA activity in the lines resulted from the reversion of one of the inherited mutations in the ADA gene, and this proved to be the case (Figure 2C,D). We also confirmed the reversions in RNA derived from their T-cell lines (data not shown). Both lines seemed to consist of absolute revertant cells from the sequencing results (Figure 2C,D).
The parents of both patients were carriers; therefore, the possibility of somatic mosaicism due to de novo mutation during embryogenesis was excluded. The reversed allele of patient 1 was derived from her father and that of patient 2 was from her mother.
The possibilities of mixture with their parents' cells in vivo or in vitro were excluded because no HLA class I, HLA DR, or DRB1 allele other than the patient's own type was detected in both the T-cell lines (data not shown).
The line of patient 1 showed polyclonal phenotype from the results of T-cell receptor panel analysis (Table 1). These results indicate the reversion of patient 1 had taken place in vivo in a T-cell precursor before the T-cell receptor genes were rearranged. In contrast, reverse mutation during in vitro culture could not be excluded in patient 2 because the established T-cell line showed monoclonal characteristics (Table 1). B-cell lines from both patients showed a trace level of ADA activity. Thus, even in the case of patient 1, cells belonging to B-cell lineage seemed not to be involved.
An ADA-deficient patient with reversion of inherited mutation was reported previously.10 In that case, reversion was confirmed in vivo and in vitro. The refusal of any treatment (including PEG-ADA replacement) for religious reasons seemed to make it easier for revertant cells to expand the population in vivo.
In the present patients, we had not realized the reverse mutation in the ADA gene until characterization of the established T-cell lines. However, there was supporting evidence for the presence of revertant cells in vivo. First, dAXP levels in their RBC were markedly lower than those found in SCID with ADA deficiency.3 Indeed, the expressed ADA activity from their common mutant allele of R235Q in Escherichia coli strain S∅3834 belongs to a group detected in SCID type3(> 0.01% of wild type18). Therefore, at least in patient 1, her ADA activity of PBMC seemed higher than the expected value from heteroallelic with R235Q and Q119X (Figure 1). Moreover, we noticed the effects of PEG-ADA replacement on patient 1. Before and 6 weeks after the replacement, the number of lymphocytes in patient 1 increased from 200 to 1200/μL, whereas ADA activity in PBMC decreased from 7.8 to 2.8 U, respectively, which could have resulted from PEG-ADA that would abolish the selective advantage of the revertant cells in vivo. These results were consistent with repetitive failures to reestablish a revertant T-cell line after the replacement. The same phenomenon might be implicated as to why stem cell gene therapy for ADA deficiency in combination with PEG-ADA replacement did not work well,6-8 whereas it succeeded for X-SCID.9
The occurrence of reverse mutation has been considered rare; however, increased numbers of examples of reverse mutations in patients with other diseases19-23 were reported, and we also very recently reported data on a patient with Wiskott-Aldrich syndrome.24 We must be more aware and pay more attention to detect the reverse mutation event in some genetic disorders because it would have important implications for somatic gene therapy.25
We thank all physicians who took care of the patients.
Supported by a Health Science Research grant from the Ministry of Health and Welfare of Japan; grant no. genome 029. M.S. Hershfield had support from NIH grant DK20902 and a grant from Enzon, Inc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tadashi Ariga, Dept of Gene Therapy, Hokkaido University School of Medicine, N15, W-7, Kita-ku, Sapporo, 060-8638, Japan; e-mail: tada-ari@med.hokudai.ac.jp.