Abstract
Variants of factor XI containing Gln226 to Arg (Q226 to R) and Ser248 to Asn (S248 to N) substitutions were first identified in an African American family with a history of excessive bleeding. The substitutions have recently been identified in unrelated individuals, suggesting they are relatively common. Both amino acids are located in the third apple domain of factor XI, an area implicated in binding interactions with factor IX and activated platelets. Recombinant factor XI–R226 and factor XI–N248 were compared with wild-type factor XI in assays for factor IX activation or platelet binding. Factor XI–R226 activates factor IX with a Michaelis-Menten constant (Km) about 5-fold greater than wild-type protein. The catalytic efficiency of factor IX activation is similar to wild-type protein, however, due to an increase in the turnover number (kcat) for the reaction. Iodinated factor XI–N248 binds to activated platelets with a dissociation constant (Kd) more than 5-fold higher than wild-type protein (55 nM and 10 nM, respectively). Activation of factor XI–N248 by thrombin in the presence of activated platelets is slower and does not progress to the same extent as activation of the wild-type protein under similar conditions. Factor XI–N248 activates factor IX normally in a purified protein system and has relatively normal activity in activated partial thromboplastin time (aPTT) assays. Factor XI–N248 is the first factor XI variant described with a clear functional difference compared with wild-type protein. Importantly, the defect in platelet binding would not be detected by routine clinical evaluation with an aPTT assay.
Introduction
Coagulation factor XI is the zymogen of a plasma protease that contributes to hemostasis by activating factor IX.1 In congenital factor XI deficiency in humans, low plasma activity is almost always associated with a comparable loss of antigen (cross-reactive material–negative [CRM−] deficiency).2,3 Few factor XI–deficient patients with low activity and disproportionately high circulating levels of factor XI antigen (CRM+ deficiency) have been reported,4-6 and the genetic abnormalities in these individuals have not been determined. This distinguishes factor XI deficiency from hemophilias A and B (deficiency of factor VIII or IX, respectively) and von Willebrand disease, where many circulating dysfunctional variants have been described.7 Another peculiarity of factor XI deficiency that distinguishes it from hemophilia is the poor correlation between plasma factor XI activity and bleeding symptoms.6,8 9 It is likely that several factors are responsible for this phenomenon. One possibility is that commonly used in vitro tests for factor XI activity do not accurately measure properties of the protein most important for in vivo hemostasis.
Previously, we reported 2 amino acid substitutions in factor XI (Q226R and S248N), identified in an African American with a history of excessive bleeding and a modest deficiency in plasma factor XI activity.10 This patient is a compound heterozygote, with the amino acid substitutions residing on separate alleles. The patient's mother, a heterozygote for the S248N substitution, also experienced excessive bleeding. Both amino acid substitutions may turn out to be relatively common, because they have subsequently been identified in DNA samples from individuals unlikely to be related to the index case.11,12 Amino acids 226 and 248 reside in the third apple (A3) domain of the factor XI heavy chain.13 A3 appears to be a key region for interactions between factor XI and other molecules involved in hemostasis. Putative binding sites for factor IX,14,15 platelet membranes,16,17 and heparin18,19 have been described within this domain. Our original investigation of recombinant factor XI containing either the Q226R or S248N substitution indicated comparable activity to wild-type factor XI in conventional activated partial thromboplastin time (aPTT) assays.10 In this report we describe studies examining interactions of the proteins with factor IX and activated platelets. The results demonstrate that the S248N substitution is associated with a defect in platelet binding that interferes with factor XI activation.
Materials and methods
Materials
293 human fetal kidney fibroblasts were from the American Type Culture Collection (CRL 1573; Rockville, MD). Cellgro Complete serum-free medium was from Mediatech (Herndon, VA). Gelcode Blue stain for sodium dodecyl sulfate (SDS)–polyacryamide gels was from Pierce (Rockford, IL). Chromogenic substrates S-2366 (L-pyroglutamyl-L-prolyl-L-arginine-p-nitroanaline) and S-2765 (N-α-benzyoxycarbonyl-D-arginyl-glycyl-L-arginine-p-nitroanaline) were from Diapharma (West Chester, OH). Recombinant human factor VIII (Recombinate) was from Baxter/Hyland (Glendale, CA). Purified thrombin and factors IX, X, IXa, Xa, and XIIa were from Enzyme Research Laboratories (South Bend, IN). High-molecular-weight kininogen was prepared by the method of Kerbiriou and Griffin.20 Rabbit brain cephalin was prepared from rabbit brain acetone powder (Sigma, St Louis, MO) by the method of Bell and Alton.21 A 1:10 dilution of this preparation is optimal for clotting assays and was used in all experiments. Thrombin receptor agonist SFLLRN-amide was synthesized at the Protein Chemistry Facility of the University of Pennsylvania. Factor XI was purified from human plasma by a previously published method.22
Preparation, activation, and radiolabeling of recombinant protein
The site-directed mutagenesis procedure used to produce expression constructs for factor XI–R226 and factor XI–N248 has been described.10 The 2 variant complementary DNAs (cDNAs) and wild-type factor XI cDNA were expressed in 293 fibroblasts according to published methods14,15 and purified from 500 mL serum-free conditioned media (Cellgro Complete) by antibody affinity chromatography.14,15 Proteins were checked for purity by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentrations were determined by dye binding assay (Bio-Rad, Hercules, CA). Proteins were activated by diluting to 200 to 400 μg/mL in 25 mM Tris-HCl (pH 7.4), 100 mM NaCl (TBS) containing 5 μg/mL human factor XIIa and incubating at 37°C. Conversion of the 80-kd zymogen to the 45-kd and 35-kd chains of factor XIa was followed by reducing SDS-PAGE. Purified proteins were radiolabeled by a modification of the Iodogen method.17 Briefly, 2 μL 125I NaOH (18.5-37 MBq/μL) was added to 100 μL protein (0.1-1.0 mg/mL) in a microfuge tube coated with Iodogen reagent (20 μL) for 20 minutes. The reaction was stopped by adding metabisulfate (50 μg/mL). The125I protein was purified by centrifugation at 200g for 20 minutes through a 1 mL Sephadex G-50 column equilibrated in 0.01 M Tris (pH 8.0). The specific activity of125I factor XI was 1.1 MBq/μg. Radiolabeled protein retained more than 98% of biologic activity in an aPTT assay.
Activities of activated recombinant proteins in chromogenic substrate assays
Cleavage of S-2366 by activated recombinant proteins.
Activated proteins were diluted to 3.0 nM in TBS containing 50 to 1000 μM S-2366, and change in absorbance at 405 nm was followed on a ThermoMax microtiter plate reader (Molecular Devices, Sunnyvale, CA). Michaelis-Menten constants (Km andVmax) were determined by standard methods using the average of 2 separate experiments. Values forVmax were converted to nanomoles of para-nitroanaline (pNA) generated per second using an extinction coefficient of 9800 optical density units (405 nm) per mole of pNA. Turnover number (kcat) was calculated from the ratio of Vmax to enzyme concentration.
Activation of factor IX by recombinant proteases.
Activation of factor IX by activated factor XI was evaluated as described.14 15 Briefly, enzyme (1 nM) was incubated at 37°C for 1 minute with factor IX (0.05-5.0 μM) in TBS containing 0.1% bovine serum albumin (TBSA) and 5 mM CaCl2. Activation was stopped by adding ethylenediaminetetraacetic acid to 25 mM. The reaction was diluted 1:100 in TBSA, and 10 μL was added to 30 μL factor X (450 nM) and to 50 μL of a mixture of factor VIII (8 units/mL), CaCl2 (10 mM), and rabbit brain cephalin. Incubations were at 37°C for 2.5 minutes, and then ethylenediaminetetraacetic acid was added as above. Fifty microliters of each reaction was mixed with 50 μL of 1.0 mM S-2765, and change in absorbance at 405 nm was followed on the microtiter plate reader. Results were compared with a control curve constructed with known amounts of purified factor IXa. Michaelis-Menten constants were determined using the average of 2 experiments. This complex assay is necessary because there is no commercially available chromogenic substrate for factor IXa with suitable sensitivity to carry out kinetic studies.
Platelet-binding experiments
Gel-filtered human platelets were prepared from fresh blood as previously described,23 and binding experiments were performed by a modification of the method of Greengard et al.24 Briefly, platelets (108/mL) were activated by 5 μM SFFLRN-amide for 5 minutes at 37°C and then supplemented with ZnCl2 (25 μM), CaCl2 (2 mM), and high-molecular-weight kininogen (50 nM).125I-labeled recombinant factor XI at various concentrations was added to the activated platelets, and the suspension was incubated for 30 minutes at 37°C. Aliquots (100 μL) of the platelet suspensions were layered on Dow Corning methyl silicon oil (3 parts 550-density oil/2 parts 200-density oil), and platelets were separated from unbound protein by centrifugation in a microfuge. The total 125I factor XI bound to the platelet pellet was measured with a Wallace 1470 Wizard gamma counter. Nonspecific binding was determined by measuring 125I factor XI bound to the platelet pellet in the presence of a 100-fold molar excess of unlabeled factor XI. Specific binding of factor XI to platelets is the difference between total binding and nonspecific binding.
Factor XI activation by thrombin on the platelet surface
Activation of recombinant wild-type and mutant factor XI was measured by a chromogenic substrate assay.25 26Gel-filtered platelets (108/mL) were activated by 5 μM SFFLRN-amide for 1 minute at 37°C and then supplemented with ZnCl2 (25 μM), CaCl2 (2 mM), and high-molecular- weight kininogen (50 nM). Recombinant factor XI (60 nM final concentration) and human α-thrombin (1 nM final concentration) were added, and incubation was continued at 37°C. Reactions were carried out in a 200 μL volume of TBSA. At various time points, reactions were diluted to 1 mL with 800 μL TBSA containing 600 μM S-2366. Change in absorbance at 405 nm was followed on a spectrophotometer, and the amount of factor XIa generated was determined by comparing results with a control curve constructed with a known standard of purified factor XIa.
Results
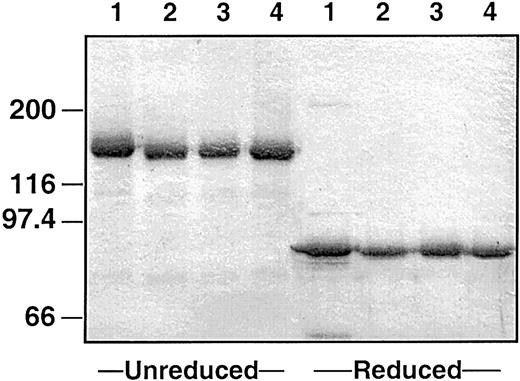
Recombinant proteins
Recombinant wild-type factor XI, factor XI–R226, and factor XI–N248 were expressed in 293 fibroblasts using a vector containing the cytomegalovirus promoter.14 For all constructs, stable clones were established that express protein at 1 to 2 μg/mL media (determined by enzyme-linked immunosorbent assay).15 An SDS–polyacrylamide gel of purified proteins is shown in Figure1. Factor XI derived from human plasma is about 160 000 d in size and is composed of 2 identical 80 000-d peptides connected by a disulfide bond (Figure 1, lane 1).27 28 All recombinant proteins run as single 160 000-d bands under nonreducing conditions and 80 000-d bands under reducing conditions, indicating that they are properly formed dimers. When incubated with factor XIIa, all proteins were converted to factor XIa in a similar manner (data not shown).
Plasma-derived and recombinant human factor XI.
Two-microgram samples of (lane 1) plasma-derived factor XI, (lane 2) wild-type recombinant factor XI, (lane 3) factor XI–R226, and (lane 4) factor XI–N248 were size-fractionated under reducing and nonreducing conditions on a 7.5% polyacrylamide-SDS gel. The gel was stained with Gelcode Blue. The positions of molecular mass standards are given in kilodaltons on the left.
Plasma-derived and recombinant human factor XI.
Two-microgram samples of (lane 1) plasma-derived factor XI, (lane 2) wild-type recombinant factor XI, (lane 3) factor XI–R226, and (lane 4) factor XI–N248 were size-fractionated under reducing and nonreducing conditions on a 7.5% polyacrylamide-SDS gel. The gel was stained with Gelcode Blue. The positions of molecular mass standards are given in kilodaltons on the left.
Activities of recombinant proteins in chromogenic substrate assays for factor IX activation
Initially all activated proteins were tested for their capacity to cleave the chromogenic substrate S-2366. This is a test for the integrity of the factor XIa catalytic domain. Data shown in Table1 are based on mean values of duplicate experiments (values for the 2 experiments differed by < 5%). Similar kinetic parameters were obtained for all proteins, indicating that the amino acid substitutions do not affect the conformation of the substrate-binding pocket in the catalytic domain. The activated proteins were then tested in an assay for their capacity to activate factor IX. Results, again, represent means of 2 separate experiments (values for the 2 experiments differed by about 15%, consistent with our prior experience with this assay). Wild-type factor XIa and factor XIa–N248 give similar results in this assay, indicating that the N248 substitution does not interfere with interactions with factor IX in solution (Table 1). The reaction involving factor XIa–R226 demonstrated an increased Km, (about 5-fold), indicating a moderate defect in factor IX binding. Interestingly, thekcat for the reaction is increased 5-fold compared with wild-type factor XIa (Table 1) so that the catalytic efficiency (kcat/Km) is similar to that of wild-type protein.
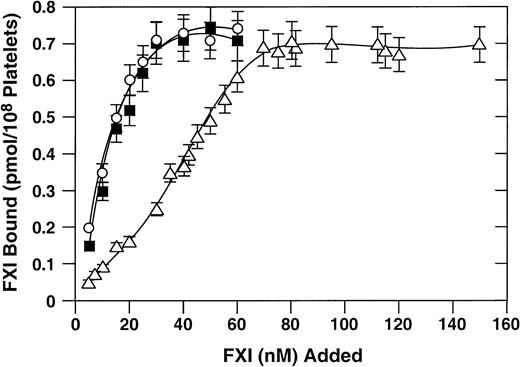
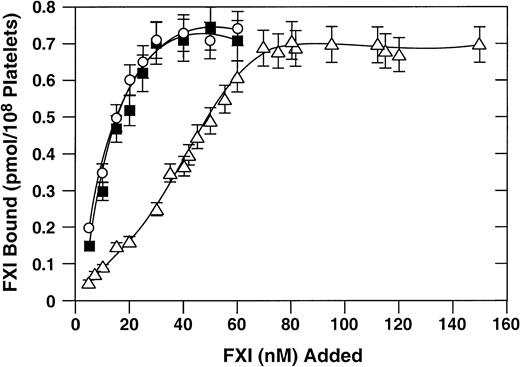
Binding of recombinant proteins to activated platelets
125I-labeled recombinant factor XI proteins were tested for their capacity to bind to activated platelets. Previous studies have demonstrated that binding of factor XI or factor XIa to platelets requires a protein cofactor (high-molecular-weight kininogen or prothrombin) and divalent cations (zinc or calcium).25,26 In the presence of high-molecular-weight kininogen and zinc, wild-type factor XI and factor XI–R226 bind to platelets with similar dissociations constants (Kd) (10 nM; Figure2). These results are in agreement with previously published results for binding of plasma-derived factor XI to activated platelets.17 The Kd for factor XI–N248 is 5- to 6-fold higher (about 55 nM), indicating a modest platelet-binding defect.
Binding of recombinant factor XI to activated platelets.
Direct binding studies were carried out as described in “Materials and methods.” 125I-labeled recombinant factor XI preparations at various concentrations were tested for binding to activated platelets in the presence of 50 nM high-molecular-weight kininogen and 25 μM ZnCl2. Wild-type recombinant factor XI (○), factor XI–R226 (•), and factor XI–N248 (▵) are shown. Data are mean values (± SEM) of total binding results for 3 separate experiments, each performed in triplicate.
Binding of recombinant factor XI to activated platelets.
Direct binding studies were carried out as described in “Materials and methods.” 125I-labeled recombinant factor XI preparations at various concentrations were tested for binding to activated platelets in the presence of 50 nM high-molecular-weight kininogen and 25 μM ZnCl2. Wild-type recombinant factor XI (○), factor XI–R226 (•), and factor XI–N248 (▵) are shown. Data are mean values (± SEM) of total binding results for 3 separate experiments, each performed in triplicate.
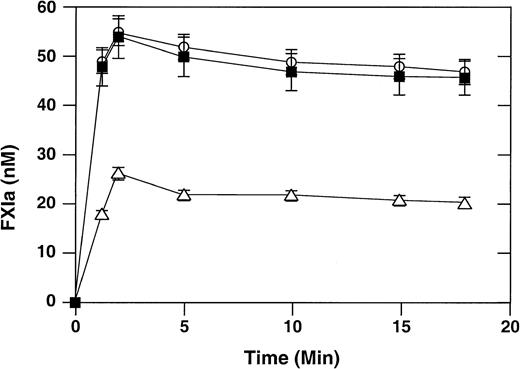
Activation of factor XI by thrombin on the surface of activated platelets
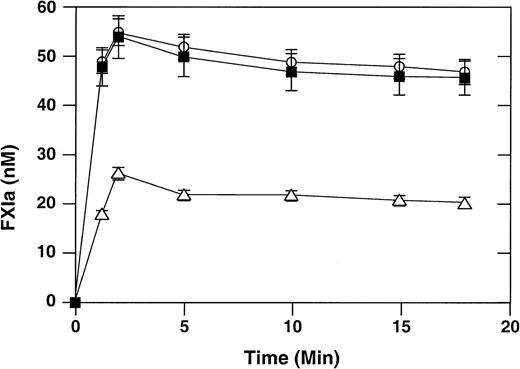
It has been shown that binding of factor XI to activated platelets facilitates factor XI activation by the proteases thrombin, factor XIIa, and factor XIa, with thrombin being the most potent activator.25,26 We compared thrombin-mediated activation of wild-type recombinant factor XI with activation of factor XI–R226 and factor XI–N248 in the presence of activated platelets using the chromogenic substrate S-2366 (Figure 3). The rate of activation of factor XI–R226 is comparable to that of the wild-type protein. The initial rate of activation of factor XI–N248 is reduced about 3-fold compared with wild-type factor XI. Interestingly, the extent of factor XI activation is also diminished, with only one half as much protein being converted to the active form, compared with wild-type factor XI. The reason for this pattern of activation is not clear, but we have observed it previously with other recombinant factor XI molecules defective in platelet binding.17
Activation of factor XI by thrombin in the presence of activated platelets.
Recombinant factor XI (60 nM) and α-thrombin (1 nM) were incubated at 37°C in the presence of activated platelets (0.5 × 108/mL), ZnCl2 (25 μM), CaCl2 (2 mM), and high-molecular-weight kininogen (50 nM), as described in “Materials and methods.” At various time points, aliquots were tested for factor XIa generation using the chromogenic substrate S-2366. Wild-type recombinant factor XI (○), factor XI–R226 (▪), and factor XI–N248 (▵) are shown. Data are mean values (± SEM) for 3 separate experiments, each performed in triplicate.
Activation of factor XI by thrombin in the presence of activated platelets.
Recombinant factor XI (60 nM) and α-thrombin (1 nM) were incubated at 37°C in the presence of activated platelets (0.5 × 108/mL), ZnCl2 (25 μM), CaCl2 (2 mM), and high-molecular-weight kininogen (50 nM), as described in “Materials and methods.” At various time points, aliquots were tested for factor XIa generation using the chromogenic substrate S-2366. Wild-type recombinant factor XI (○), factor XI–R226 (▪), and factor XI–N248 (▵) are shown. Data are mean values (± SEM) for 3 separate experiments, each performed in triplicate.
Discussion
Previously, we identified 2 novel polymorphisms (one on each allele) in the factor XI genes of a 9-year-old African American boy with a history of excessive bleeding and mild factor XI deficiency (activity 42%-55% of normal, antigen 70% of normal).10Both polymorphisms result in amino acid substitutions (Q226 to R and S248 to N). The patient's mother, a heterozygote for S248N, also had a history of excessive bleeding associated with levels of plasma factor XI activity and antigen in the low-normal range (activity 67%-75% of normal, antigen 80% of normal).10 To our knowledge, these are the first reported examples of amino acid substitutions in factor XI associated with significant levels of circulating plasma protein in patients and with normal expression in an in vitro tissue culture system.10 An additional family member heterozygous for N248 also has low-normal plasma factor XI activity (67%) (R.E.W. and D.G., unpublished data, November 1997). Subsequently, factor XI–R226 and factor XI–N248 have been detected in individuals unlikely to be related to the family reported in the initial study. The polymorphism in exon 7 (A774G [The numbering system for amino acids in the factor XI protein, and for nucleotide residues in the factor XI cDNA, follows the work of Fujikawa et al.29]) responsible for the Q226R substitution was identified by Cargill et al in a large screening study for single nucleotide polymorphisms.11S248N is caused by a G840A change in exon 8.10 We have identified this change in a person of East African (Ethiopian Jewish) ancestry.12 In fact, G840A is on the same haplotype background in family members and the East African individual, demonstrating that the alleles have a common origin. Factor XI–R226 and factor XI–N248 may therefore be relatively common in persons of African ancestry.
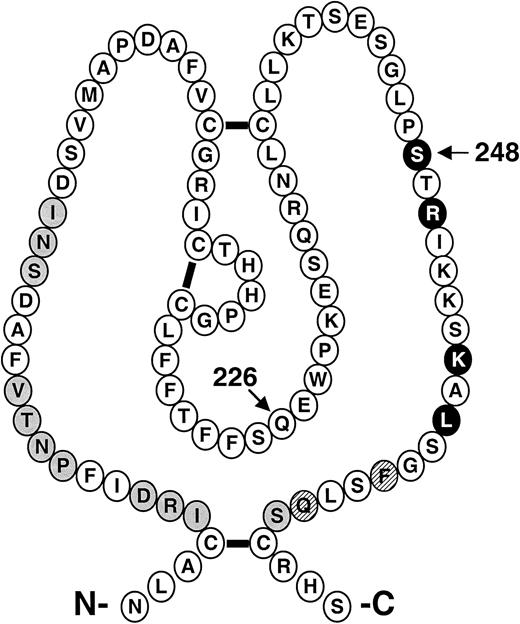
Recombinant factor XI–R226 and factor XI–N248 have specific activities (90% and 70% of normal, respectively) that are relatively comparable to recombinant wild-type factor XI and plasma factor XI (100% normal activity by definition) when tested in an aPTT assay.10 Factor XI, like other coagulation proteases, consists of an N-terminal noncatalytic region and a C-terminal trypsinlike catalytic domain.1,29,30 The noncatalytic portion of the molecule (the heavy chain) is composed of four 90-to-91 amino acid repeats called apple domains (designated A1 through A4 starting at the N-terminus).13,29 Amino acids 226 and 248 both reside in the A3 domain, an area implicated in several binding interactions with importance for hemostasis. The positions within A3 of putative binding sites for factor IX14,15 and platelets16 17 are shown in Figure4. We conducted specific assays involving factor IX activation or interactions with platelets to determine if the R226 or N248 substitutions had effects on these functions.
Schematic diagram of the factor XI A3 domain.
The primary amino acid sequence and disulfide bond structure of the A3 domain from human factor XI is shown.13 The positions of amino acids for putative binding sites for platelets (●) and for factor IX ( ) are highlighted. ○ indicate that amino acids are required for both platelet and factor IX binding. The arrows indicate the positions of the Q226R and S248N amino acid substitutions.
) are highlighted. ○ indicate that amino acids are required for both platelet and factor IX binding. The arrows indicate the positions of the Q226R and S248N amino acid substitutions.
Schematic diagram of the factor XI A3 domain.
The primary amino acid sequence and disulfide bond structure of the A3 domain from human factor XI is shown.13 The positions of amino acids for putative binding sites for platelets (●) and for factor IX ( ) are highlighted. ○ indicate that amino acids are required for both platelet and factor IX binding. The arrows indicate the positions of the Q226R and S248N amino acid substitutions.
) are highlighted. ○ indicate that amino acids are required for both platelet and factor IX binding. The arrows indicate the positions of the Q226R and S248N amino acid substitutions.
The activation of factor IX by factor XIa–R226 in a purified protein system differed considerably from activation by wild-type factor XIa. The Km for the reaction was higher with the R226 variant, suggesting a decrease in binding affinity. Interestingly, this appeared to be offset by a proportional increase inkcat so that the catalytic efficiency of the factor XIa-R226–mediated reaction is similar to wild-type factor XIa. This may explain our failure to detect a functional difference in an aPTT assay in our earlier study and suggests that the R226 substitution is not responsible for the bleeding phenotype in our patients.10 It is possible that substitution of an amino acid with a large positively charged side group for an uncharged polar residue causes a conformational change in the molecule sufficient to account for the results of the kinetic analysis. Alanine substitutions around amino acid 226 result in a protein that is expressed poorly in in vitro culture, consistent with this area being critical for proper protein conformation.15 It is conceivable that the R226 substitution is associated with reduced hepatic secretion, explaining why the propositus in our original report10 (a heterozygote for factor XI–R226) had slightly lower plasma factor XI antigen levels than his mother.
Recent work suggests that the platelet membrane is a physiologic site for factor XI activation17,25 and that amino acid 248 on factor XI is involved in the binding interaction with activated platelets.17 In studies with full-length recombinant factor XI proteins, alanine substitution at amino acid 248 decreases factor XI affinity for platelets about 10-fold (Figure4).17 Consistent with this finding, the N248 substitution is associated with a 5- to 6-fold reduction in affinity for platelets. Wild-type factor XI binds to activated platelets with aKd of about 10 nM.17 Because the normal plasma factor XI concentration is 30 nM, most factor XI binding sites on an activated platelet should be occupied. A 5- to 6-fold decrease in affinity for the platelet binding site could therefore have a significant effect on receptor occupancy. Because thrombin-mediated activation of factor XI on the platelet surface is directly related to occupancy of factor XI binding sites,26 the result could be a substantial reduction in factor XIa generation. Indeed, our examination of factor XI activation by thrombin in the presence of platelets demonstrated a decrease in rate and extent of activation for factor XI–N248. It has been proposed that factor IX is activated on the platelet surface by factor XIa.31 Technical limitations make it difficult to accurately measure factor IXa generation on the platelet surface, and the present study does not address the possibility that factor XIa-N248 has reduced capacity to activate factor IX in this setting. Previous work from our laboratories demonstrated that factor XI–N248 has relatively similar coagulant activity in an aPTT assay in the presence of purified phospholipid or activated platelets when kaolin is used to trigger clotting through contact activation.17 This indicates that factor IX activation by factor XIa on the platelet surface is not impaired significantly by the N248 substitution. The data suggest, therefore, that the functional abnormality in factor XI–N248 is a failure to activate properly rather than a defect in interactions with factor IX.
S248N is the first naturally occurring polymorphism in the human factor XI gene that has been shown to be associated with a functional abnormality. The significance of this finding in regard to the bleeding disorder seen in the family in which the polymorphism was originally described, however, is not clear. While the proteins we used for the in vitro studies are recombinant homodimers, the composition of factor XI in the patients' plasmas is almost certainly more complex. The propositus of our earlier study was a compound heterozygote for R226 and N248.10 Because factor XI is a homodimer, this patient may have had circulating homodimers of factor XI–R226 and factor XI–N248 as well as heterodimers with one chain of each. His mother, a heterozygote for the N248 substitution, would also have a mixture of different types of dimers. The consequences of such an arrangement on hemostasis are difficult to predict. Both patients share a history of excessive bleeding and factor XI–N248, suggesting that the substitution could be contributing to bleeding. Because the polymorphism appears to be common, further patients may be identified, which will help clarify the relationship, if any, between N248 and excessive bleeding.
It is well established that factor XI activity in plasma correlates poorly with the propensity to bleed in factor XI–deficient patients.6,8 This is undoubtedly due, in part, to the relatively mild nature of many bleeding episodes as well as varying definitions of what constitutes abnormal bleeding.32However, it is likely that the discrepancy in some instances is related to the nature of the mutation in the factor XI molecule. In aPTT assays, factor XI is an integral part of a pathway that drives the formation of the fibrin clot. However, in vivo, it is more likely that factor XI functions primarily in maintaining clot integrity over time rather than in initial fibrin generation.1 33-35 The aPTT, therefore, may not be an adequate test of all relevant factor XI activities. Furthermore, in the case of a mutation such as N248, because platelets are not incorporated into aPTT assays, the defect may be missed. Novel assays, perhaps containing platelets, may be necessary to adequately assess all important physiologic aspects of factor XI function.
The authors thank Jean McClure for artwork. D.G. is an Established Investigator of the American Heart Association.
Supported by grants HL58837 and HL02917 (D.G.) and HL46213, HL56153, and HL56914 (P.N.W.) from the National Heart, Lung, and Blood Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David Gailani, Hematology/Oncology Division, Vanderbilt University, 538 MRB II/2220 Pierce Ave, Nashville, TN 37232-6305; e-mail: dave.gailani@mcmail.vanderbilt.edu.




 ) are highlighted. ○ indicate that amino acids are required for both platelet and factor IX binding. The arrows indicate the positions of the Q226R and S248N amino acid substitutions.
) are highlighted. ○ indicate that amino acids are required for both platelet and factor IX binding. The arrows indicate the positions of the Q226R and S248N amino acid substitutions.