Abstract
Chronic exposure to benzene is associated with hematotoxicity and acute myelogenous leukemia. Inhibition of topoisomerase IIα (topo II) has been implicated in the development of benzene-induced cytogenetic aberrations. The purpose of this study was to determine the mechanism of topo II inhibition by benzene metabolites. In a DNA cleavage/relaxation assay, topo II was inhibited byp-benzoquinone and hydroquinone at 10 μM and 10 mM, respectively. On peroxidase activation, inhibition was seen with 4,4′-biphenol, hydroquinone, and catechol at 10 μM, 10 μM, and 30 μM, respectively. But, in no case was cleavable complex stabilization observed and the metabolites appeared to act at an earlier step of the enzyme cycle. In support of this conclusion, several metabolites antagonized etoposide-stabilized cleavable complex formation and inhibited topo II–DNA binding. It is therefore unlikely that benzene-induced acute myelogenous leukemia stems from events invoked for leukemogenic topo II cleavable complex-stabilizing antitumor agents.
Introduction
Human DNA topoisomerase II (topo II), a nuclear enzyme responsible for modulating the topologic state of DNA, is critical for DNA replication, chromosomal condensation/decondensation, and chromosomal segregation at mitosis.1 Several anticancer agents (eg, doxorubicin, etoposide) exert their cytotoxic effects through topo II.1 However, clinical utility of these agents is limited by the risk of secondary acute myelogenous leukemia (AML) developing due to DNA aberrations that form from the processing of this unusual form of DNA damage.2 The vast majority of these lesions involve the myeloid-lymphoid leukemia (MLL) locus at 11q23.
Benzene is a known human leukemogen and chronic exposure has been associated with many blood dyscrasias, including AML.3 The cytogenetic abnormalities associated with benzene-induced secondary AML most commonly involve the loss of part of or whole chromosomes 5 and 7. Less frequent aberrations occur in chromosomes 8, 17, and 21.4 These cytogenetic effects have been attributed not to benzene itself, but rather its metabolites.5 Primary metabolism of benzene occurs in the liver where it is biotransformed by cytochrome P-450 2E1 to 1,2,4-benzenetriol, catechol, and hydroquinone.6,7 Phenolic metabolites are further activated by myeloperoxidase in the bone marrow to quinone derivatives.8 Biomolecular analysis demonstrating that these latter benzene metabolites inhibit the DNA decatenation activity of topo II led to the hypothesis that the mechanism underlying benzene's clastogenic effects involves the inhibition of topo II.9 However, the mechanism of topo II inhibition remains to be elucidated.
The purpose of this study was to determine the precise mechanism of topo II inhibition by benzene metabolites in vitro. Preliminary work from our laboratory suggested that many of these metabolites were incapable of stabilizing topo II–DNA complexes.10 In this report, we demonstrate that native and peroxidase-activated benzene metabolites interfere with topo II–DNA binding activity and antagonize etoposide-mediated cleavable complex stabilization. Therefore, the hypothesis invoking topo II in benzene leukemogenesis is inconsistent with our mechanistic findings, and with current models of the role of cleavable complexes in topo II–directed xenobiotic-induced leukemias.
Materials and methods
Reagents
Purified human topo IIα was from Topogen (Columbus, OH). 1,2,4-benzenetriol (99%), 2,2′-biphenol (99%), and catechol (99%) were from Acros, Fisher Scientific (Pittsburgh, PA). Benzoquinone, 4,4′-biphenol (99%), etoposide, hydroquinone, H2O2, and horseradish peroxidase (HRP) type IV were from Sigma (St Louis, MO). Stock solutions and dilutions of benzene metabolites were prepared in deionized water except for 2,2′-biphenol and 4,4′-biphenol (in dimethyl sulfoxide), andp-benzoquinone (prepared as a stock solution in dimethyl sulfoxide with subsequent dilutions prepared in deionized water).
Topo II–mediated DNA cleavage
The effects of benzene metabolites on stabilization of topo II–DNA complexes were evaluated using the method of Gantchev and Hunting.11 Reactions were modified to use 300 ng pBR322 plasmid DNA (Promega, Madison, WI) and 5 μg bovine serum albumin.
Electrophoretic mobility shift assay
Experiments were performed as described previously.12
HRP activation
An HRP/H2O2 protocol that models the bone marrow myeloperoxidase metabolic system has been described previously.13 Benzene metabolites were activated for 60 minutes at 22°C in a 15-μL reaction volume containing 2.0 mM H2O2 and 0.0075 units HRP.
Results and discussion
In preliminary experiments (data not shown) using a plasmid DNA relaxation assay, hydroquinone and p-benzoquinone were found to be dose-dependent inhibitors of topo II. We therefore sought to identify the mechanism of inhibition using the modified cleavage assay of Gantchev and Hunting.11 This assay detects topo II–DNA complexes through the appearance of a linear DNA band following proteinase digestion. Performing experiments in both the presence and absence of a known cleavable complex stabilizer permits the evaluation of additivity/synergism or antagonism between the benzene metabolites and etoposide. This assay also allows catalytic inhibition of topo II to be distinguished from cleavable complex stabilization by (1) the absence of linear band formation and (2) a dose-dependent loss of etoposide-stabilized linear band intensity in coincubation reactions.
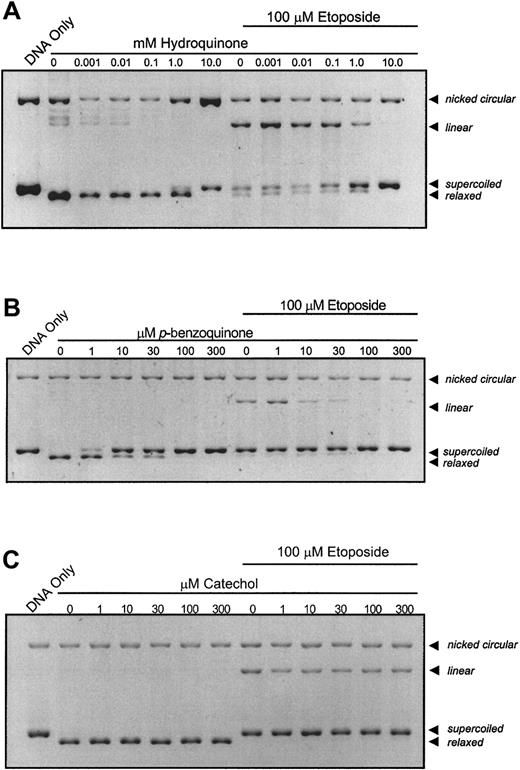
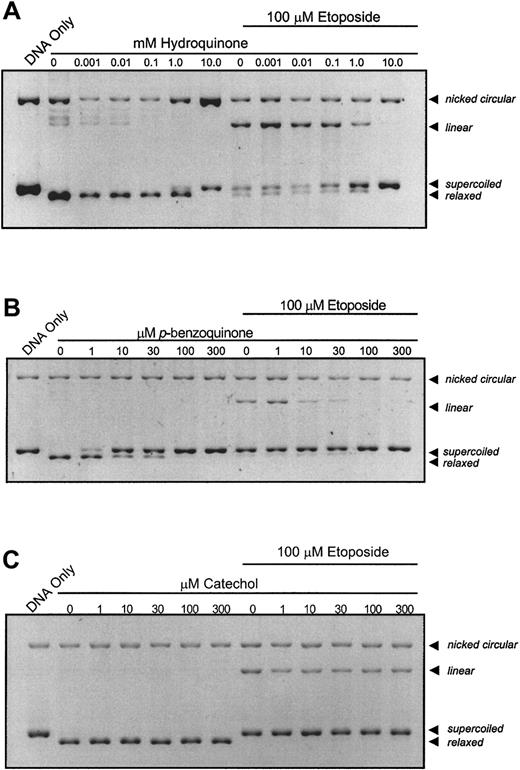
When topo II is incubated in the presence of plasmid DNA and increasing concentrations of hydroquinone (Figure1A) or p-benzoquinone (Figure1B), enzyme activity is inhibited at 10 mM and 10 μM, respectively, as indicated by the dose-dependent loss of relaxed DNA and maintenance of unreacted, supercoiled DNA. There is no detectable appearance of a linear DNA band with either compound, even at high metabolite concentrations. Hydroquinone consistently produces a dose-dependent increase in nicked circular DNA. This is not observed with other metabolites and may be attributed to hydroquinone reduction-oxidation cycling at high metabolite concentration. Of greatest relevance, dose-dependent loss of the linear DNA band occurs when reactants are coincubated with etoposide and increasing concentrations of benzene metabolite (Figure 1A,B). p-benzoquinone is more potent than hydroquinone and substantially antagonizes etoposide-stabilized cleavage complexes at one tenth the molar concentration of etoposide (Figure 1B). Further experiments have demonstrated that catalytic inhibition of topo II by p-benzoquinone is reversible (data not shown). In contrast, neither catechol nor the biphenolic metabolites, 4,4′-biphenol or 2,2′-biphenol, at concentrations up to 300 μM, produced consistent effects on either overall topo II DNA relaxation activity or etoposide-stabilized, cleavable complex formation (Figure 1C and data not shown).
Catalytic inhibition of topo II and abrogation of etoposide-stabilized, enzyme–DNA complexes by hydroquinone,
p-benzoquinone, and catechol. The pBR322 plasmid DNA (300 ng) was combined with topo II (6 U) following a 10-minute incubation with compound at indicated concentrations in the absence or presence of 100 μM etoposide. The dose-dependent loss of the relaxed band is indicative of inhibition of overall topo II catalytic activity. Although etoposide alone stabilizes enzyme-linked DNA complexes (indicated by the linear band), coincubation with increasing hydroquinone (A) and p-benzoquinone (B) concentrations antagonizes this effect in a dose-dependent fashion, whereas catechol (C) does not.
Catalytic inhibition of topo II and abrogation of etoposide-stabilized, enzyme–DNA complexes by hydroquinone,
p-benzoquinone, and catechol. The pBR322 plasmid DNA (300 ng) was combined with topo II (6 U) following a 10-minute incubation with compound at indicated concentrations in the absence or presence of 100 μM etoposide. The dose-dependent loss of the relaxed band is indicative of inhibition of overall topo II catalytic activity. Although etoposide alone stabilizes enzyme-linked DNA complexes (indicated by the linear band), coincubation with increasing hydroquinone (A) and p-benzoquinone (B) concentrations antagonizes this effect in a dose-dependent fashion, whereas catechol (C) does not.
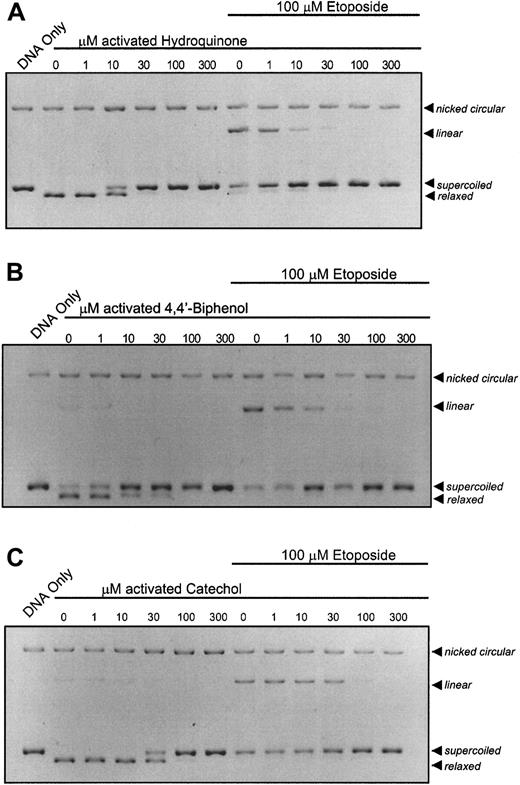
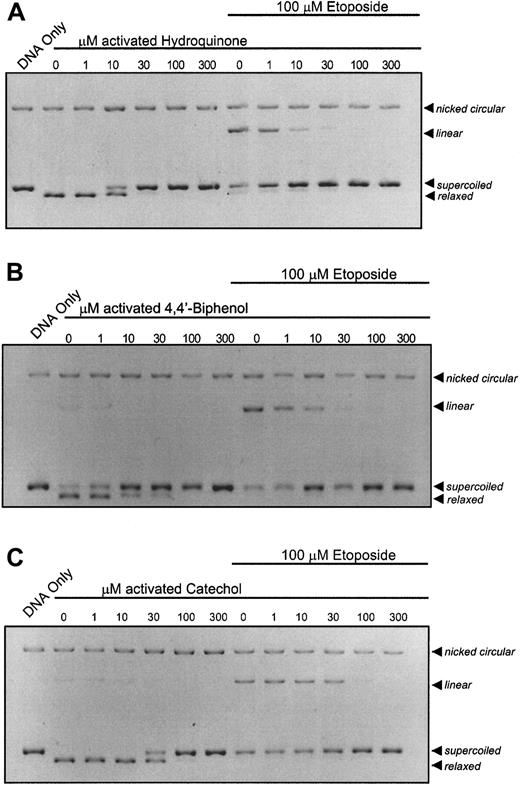
It is recognized that, in bone marrow, benzene metabolites can react with myeloperoxidase leading to the generation ofp-benzoquinone by peroxidation of hydroquinone. It was hypothesized that peroxidase activation of hydroquinone would increase its potency for topo II inhibition. HRP activation of hydroquinone increased its potency for topo II by 1000-fold, resulting in substantial inhibition of topo II activity at 10 μM (Figure2A). This increased potency compares favorably with inhibitory concentrations p-benzoquinone (Figure 1B), the terminal oxidation product of the hydroquinone oxidation.14 Activation of 4,4′-biphenol (Figure 2B) and catechol (Figure 2C) with HRP results in catalytic inhibition of topo II and antagonism of etoposide-stabilized cleavable complex formation at 10 μM and 30 μM, respectively.
Enhancement of the catalytic inhibition of topo II by hydroquinone; 4,4′-biphenol; and catechol following peroxidase activation.
The pBR322 plasmid DNA (300 ng) was combined with topo II (6 U) following a 10-minute incubation with activated compound (1, 10, 30, 100, 300 μM) in the absence or presence of 100 μM etoposide. (A) DNA cleavage assay performed with peroxidase-activated hydroquinone. (B) DNA cleavage assay performed with peroxidase-activated 4,4′-biphenol. (C) DNA cleavage assay performed with peroxidase-activated catechol. All samples, including untreated controls, contain identical concentrations of activating agents. Reactions were interpreted as described in Figure 1.
Enhancement of the catalytic inhibition of topo II by hydroquinone; 4,4′-biphenol; and catechol following peroxidase activation.
The pBR322 plasmid DNA (300 ng) was combined with topo II (6 U) following a 10-minute incubation with activated compound (1, 10, 30, 100, 300 μM) in the absence or presence of 100 μM etoposide. (A) DNA cleavage assay performed with peroxidase-activated hydroquinone. (B) DNA cleavage assay performed with peroxidase-activated 4,4′-biphenol. (C) DNA cleavage assay performed with peroxidase-activated catechol. All samples, including untreated controls, contain identical concentrations of activating agents. Reactions were interpreted as described in Figure 1.
To evaluate further the mechanism of topo II inhibition by these compounds, the effect of p-benzoquinone, hydroquinone, and activated hydroquinone on topo II–DNA binding was evaluated by electrophoretic mobility shift assay. Sequence specificity and involvement of topo II in this protein-DNA complex have been previously established.12p-benzoquinone completely inhibits topo-DNA interaction at 100 μM (Figure3A), whereas native hydroquinone has no effect (Figure 3B). HRP-activated hydroquinone inhibits topo II–DNA binding at all concentrations tested (Figure 3C). Although inclusion of HRP and H2O2 in control samples had no effect on protein-DNA complex formation, free radical production may have an additive effect with the activated compound.
Benzene metabolites inhibit topo II–DNA binding.
Oligonucleotides, containing a strong topo II binding site from positions 87 to 126 of pBR322, were annealed and end-labeled with [α-32P]dCTP. Nuclear extract (1 μg protein) from HeLa cells was assayed for binding activity to the 32P-labeled binding site in the presence of 0.1 μg poly(dI:dC) • (dI:dC). The DNA-protein complex (bound) was separated from free probe (free) by electrophoresis through a nondenaturing, 4% polyacrylamide gel in 0.25 × Tris borate–EDTA buffer. Nuclear extract was incubated with increasing concentrations (1, 10, 30, 100, and 300 μM) ofp-benzoquinone (A), hydroquinone (B), and peroxidase-activated hydroquinone (C) prior to initiating the reaction with labeled oligonucleotide.
Benzene metabolites inhibit topo II–DNA binding.
Oligonucleotides, containing a strong topo II binding site from positions 87 to 126 of pBR322, were annealed and end-labeled with [α-32P]dCTP. Nuclear extract (1 μg protein) from HeLa cells was assayed for binding activity to the 32P-labeled binding site in the presence of 0.1 μg poly(dI:dC) • (dI:dC). The DNA-protein complex (bound) was separated from free probe (free) by electrophoresis through a nondenaturing, 4% polyacrylamide gel in 0.25 × Tris borate–EDTA buffer. Nuclear extract was incubated with increasing concentrations (1, 10, 30, 100, and 300 μM) ofp-benzoquinone (A), hydroquinone (B), and peroxidase-activated hydroquinone (C) prior to initiating the reaction with labeled oligonucleotide.
The earlier demonstration of topo II inhibition by benzene metabolites9 raised an important and provocative question regarding the etiology of benzene leukemogenesis. Although it is generally termed a topo II inhibitor, etoposide is more accurately classified as a topo II poison that traps the enzyme in its catalytic DNA cleavage intermediate. This unusual type of DNA damage is central to the leukemogenic potential of this class of antitumor agents.15
A model has been proposed to distinguish between the cytotoxic and leukemogenic effects of topo II poisons. Invoking an earlier model of Liu,16 Strick and colleagues17 demonstrated that xenobiotic-induced topo II cleavable complexes were reversible by DNA religation or repair and suggested that this could, in rare cases, lead to illegitimate chromosomal translocations resulting in leukemia. However, persistence and accumulation of cleavable complexes is believed to lead to apoptosis. Hence, the initial processing of cleavable complexes (presumably by recombinational repair15 18) is critical to the leukemogenic potential of these agents.
In contrast to these topo II poisons, benzene metabolites and their peroxidase-activated congeners appear to inhibit topo II via a distinct and upstream mechanism. Therefore, metabolites should more appropriately be classified as catalytic inhibitors of topo II, similar to that described for the bisdioxopiperazines (ICRF-154, -187, and -193), aclarubicin, and merbarone. Although some topo II catalytic inhibitors can induce cellular DNA damage, no compound in this class has ever been linked directly to the therapy-related MLLrearrangements at 11q23 that are characteristic of de novo and etoposide-related translocations. To our knowledge, there is only one report19 in which a bisdioxopiperazine has been associated clinically with chromosome 11 damage [t(7;11)(p15;p15)], but this translocation is distinct from that seen with topo II poisons.
A diverse set of observations supports the conclusion that the clastogenicity of benzene and its metabolites plays a role in the pathogenesis of benzene-induced AML. Exposure of human cells to the polyhydroxy metabolites of benzene produces micronuclei and concentration-dependent hypoploidy involving chromosomes 5, 7, and possibly 8.20-24 These observations may be explained, at least in part, by the fact that hydroquinone and its principal oxidation product, p-benzoquinone, are potent spindle poisons that interfere with the equilibrium dynamics of microtubule assembly by inhibiting guanosine triphosphate (GTP) binding to tubulin.25-27 Additional biochemical processes are likely to be involved in triggering the underlying events. However, results presented herein indicate that topo II–dependent cleavable complex formation is not one of them.
The authors wish to recognize the excellent technical assistance of Brante P. Sampey in establishing and validating the Gantchev/Hunting topo II assay in the Kroll laboratory.
Supported in part by National Institutes of Health grants CA76201 (to D.J.K.) and ES06258 (to R.D.I.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David J. Kroll, Duke University Medical Center, Research Dr, 239 Jones Bldg, Box 3020, Microbiology, Durham, NC 27710; e-mail: kroll001@mc.duke.edu.

![Fig. 3. Benzene metabolites inhibit topo II–DNA binding. / Oligonucleotides, containing a strong topo II binding site from positions 87 to 126 of pBR322, were annealed and end-labeled with [α-32P]dCTP. Nuclear extract (1 μg protein) from HeLa cells was assayed for binding activity to the 32P-labeled binding site in the presence of 0.1 μg poly(dI:dC) • (dI:dC). The DNA-protein complex (bound) was separated from free probe (free) by electrophoresis through a nondenaturing, 4% polyacrylamide gel in 0.25 × Tris borate–EDTA buffer. Nuclear extract was incubated with increasing concentrations (1, 10, 30, 100, and 300 μM) ofp-benzoquinone (A), hydroquinone (B), and peroxidase-activated hydroquinone (C) prior to initiating the reaction with labeled oligonucleotide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/3/10.1182_blood.v98.3.830/5/m_h81511361003.jpeg?Expires=1769404119&Signature=zblls2-b4jThRiCQ6ugT4R0~JTlI7O6DrIng8IIDSQdCtdGB~p2OFM7q~sKQ27F5THo-qleLm8XPzeBHgNB0cAotQHc2FDKyg14ZdVwd~UCNSJvUR9Rz0YGusynnOIjez0dxxCrPZ3fHBrmtNt1jg5w~InLyuwpUeDvyMuj64ntkaadw~bBxwCvoeMOVPUHrMxJ0zN8kqPMr3-JafV-LI0ERnbRpOHJYNevZX6-04EQ5FjPRkzXYPaaVbuyIzftvWdJZ6JzFAKzGh8ltjmpnBVUrDif8522ObR7DYtOx2sr3durrXkheST1qRIW5gwkmajOYAWQh~V8ac19v9LbAAg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 3. Benzene metabolites inhibit topo II–DNA binding. / Oligonucleotides, containing a strong topo II binding site from positions 87 to 126 of pBR322, were annealed and end-labeled with [α-32P]dCTP. Nuclear extract (1 μg protein) from HeLa cells was assayed for binding activity to the 32P-labeled binding site in the presence of 0.1 μg poly(dI:dC) • (dI:dC). The DNA-protein complex (bound) was separated from free probe (free) by electrophoresis through a nondenaturing, 4% polyacrylamide gel in 0.25 × Tris borate–EDTA buffer. Nuclear extract was incubated with increasing concentrations (1, 10, 30, 100, and 300 μM) ofp-benzoquinone (A), hydroquinone (B), and peroxidase-activated hydroquinone (C) prior to initiating the reaction with labeled oligonucleotide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/3/10.1182_blood.v98.3.830/5/m_h81511361003.jpeg?Expires=1769404120&Signature=AAPLPVPgVrF1KT-~GLZIqFF5lfpOzNOANhHO0mNjMAZfHPEETf4z3b18E2cMTzCpdsXONl-ofvNFqYkyfkX8JSfA1zd3Z99Tlz73ScmodX~Oy2y2HdPj~IJgPnAiRfts9vz8Qf8BVldXllBV4BdEXeM5KKZIOZaqlqy2LuXE~IgcnNr7lVhQquI0qshrWcCqJDMVo6kvL9KUzQP1xsX11XAVeH6xtlyh86VbU-ddhoHgXkZ3G259OtOI5uTevEs60-~81gldanwUFv1eKOyow4YBxhO01tu3bMqUniurZizAdy-bnye4PuMyPdojBb9a7llzibP~Nao1FNVXcq~~Sw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)