The Fanconi anemia (FA) group C gene product (FANCC) functions to protect cells from cytotoxic and genotoxic effects of cross-linking agents. FANCC is also required for optimal activation of STAT1 in response to cytokine and growth factors and for suppressing cytokine-induced apoptosis by modulating the activity of double-stranded RNA-dependent protein kinase. Because not all FANCC mutations affect STAT1 activation, the hypothesis was considered that cross-linker resistance function of FANCC depends on structural elements that differ from those required for the cytokine signaling functions of FANCC. Structure-function studies were designed to test this notion. Six separate alanine-substituted mutations were generated in 3 highly conserved motifs of FANCC. All mutants complemented mitomycin C (MMC) hypersensitive phenotype of FA-C cells and corrected aberrant posttranslational activation of FANCD2 in FA-C mutant cells. However, 2 of the mutants, S249A and E251A, failed to correct defective STAT1 activation. FA-C lymphoblasts carrying these 2 mutants demonstrated a defect in recruitment of STAT1 to the interferon γ (IFN-γ) receptor and GST-fusion proteins bearing S249A and E251A mutations were less efficient binding partners for STAT1 in stimulated lymphoblasts. These same mutations failed to complement the characteristic hypersensitive apoptotic responses of FA-C cells to tumor necrosis factor-α (TNF-α) and IFN-γ. Cells bearing a naturally occurring FANCC mutation (322delG) that preserves this conserved region showed normal STAT1 activation but remained hypersensitive to MMC. The conclusion is that a central highly conserved domain of FANCC is required for functional interaction with STAT1 and that structural elements required for STAT1-related functions differ from those required for genotoxic responses to cross-linking agents. Preservation of signaling capacity of cells bearing the del322G mutation may account for the reduced severity and later onset of bone marrow failure associated with this mutation.
Introduction
Fanconi anemia (FA) is an autosomal recessive disease characterized by bone marrow failure, variable congenital anomalies, and a predisposition to leukemia.1-3 Cells from patients with FA exhibit hypersensitivity to alkylating agents such as mitomycin C (MMC) and diepoxybutane (DEB). Indeed, the hypersensitivity to cytotoxic effects of DNA cross-linking agents is currently used as the basis for the diagnostic tests for FA.4,5 It is known that FA is genetically heterogeneous, with at least 7 complementation groups (A-G) identified thus far.6,7 The genes encoding the groups A (FANCA),8 C (FANCC),9 D2 (FANCD2),10 G (FANCG),11 and F (FANCF)12 have been cloned. The group C locus,FANCC, encodes a 63-kd protein of unknown function.9 Expression of the normal FANCCcomplementary DNA (cDNA) in FA-C cells prevents MMC-induced chromosomal aberrations, prevents G2 arrest in cell cycle, and prevents growth inhibition and apoptosis.13,14 FANCC forms complexes with at least 3 other FA proteins, namely FANCA, FANCF, and FANCG.15-17 Such complexes are believed to function in DNA damage repair or toleration in part by facilitating posttranslational activation of FANCD2, which can then translocate to nuclear damage foci.18
Although the FANCC protein can be found in the nucleus,19 it is principally cytoplasmic.19-22 Moreover, evidence indicates that FANCC binds to a number of cytosolic proteins in vitro, including the mitotic cyclin-dependent kinase cdc2,23 the molecular chaperone GRP94,24 NADPH cytochrome P-450 reductase,25and the signal transducer and activator of transcription STAT1.26 Although the functional consequences of these molecular interactions are not clear, they suggest that FANCC may play roles in cell cycle control, protein transport, regulation of detoxification, and survival signal transduction.
In our view, the role of FANCC in suppressing apoptotic responses in hematopoietic cells may account for the nearly universal development of bone marrow failure in patients with inactivating mutations. This notion derives from reports that suppression of FANCC gene expression represses clonal growth of normal erythroid and granulocyte-macrophage progenitor cells and that disruption of theFANCC gene, in mice, rendered hematopoietic progenitor cells hypersensitive to the apoptotic effects of interferon γ (IFN-γ) and tumor necrosis factor-α (TNF-α).3,14,27,28 Conversely, overexpression of FANCC suppresses apoptosis in human hematopoietic factor-dependent progenitor cell lines,29 in CD34+ cells from FA-C patients,14,30 and in hematopoietic progenitor cells from fancc knock-out mice.31
One function of FANCC that may account for its importance in hematopoietic control is its role in governing proper activation of STAT molecules. Recently, we reported that lymphoblasts from FA-C patients were defective in STAT1 activation in response to stimulation with IFN-γ.3,26 We also noted that in FA-C cells STAT1 failed to dock at the IFN-γ receptor α-chain (IFN-γRα) and activated STAT1 dimers failed to translocate to the nucleus but that expression of a normal FANCC protein corrected these defects. A variety of cytokines and hematopoietic growth factors induce the association of STAT1 with FANCC but FANCC/STAT1 associations do not occur in FA cells bearing an inactivating point mutation (L554P).26 These findings suggest a distinct role for FANCC in cytokine (growth factor) signal transduction pathways. More recently we reported that FANCC also functions to modulate the activity of double-stranded RNA-dependent protein kinase (PKR).32 Loss of either of these functions would be expected to increase rates of apoptosis in progenitor cells from patients with FA-C, in the case of STAT1 function by creating a “factor-deprived” phenotype and in the case of PKR by failing to suppress a proapoptotic factor. Although the functions of FANCC vis-à-vis STAT1 and PKR could theoretically derive simply from the recently described nuclear function of FANCC,18 in our view this seemed less likely than a model of FANCC multifunctionality. We therefore sought to develop evidence of separate structural requirements for the nuclear damage function of FANCC and the antiapoptotic functions that may be largely cytoplasmic. In this report, we present a mutational analysis of the human FANCCgene product, the results of which provide the first concrete structural evidence that FANCC has multifunctional roles in somatic cells and that the structural elements required for FANCC to protect cells against genotoxic stress differ from those required for optimization of STAT1 signaling and for modulating responses to proapoptotic cytokines.
Materials and methods
Cell cultures and IFN-γ stimulation
Epstein-Barr virus (EBV)-transformed lymphoblasts were maintained in RPMI media 1640 (Life Technologies, Grand Island, NY) supplemented with 15% heat-inactivated fetal calf serum. The normal lymphoblast line JY and mutant FA-C lines PD4 and HSC536N have been described elsewhere.20 26 In all experiments, cells were stimulated with 1 ng/mL human IFN-γ (R & D Systems, Minneapolis, MN) for the indicated time periods. For detection of STAT1 phosphorylation, cells were serum starved for 2 hours followed by IFN-γ treatment.
Construction of site-directed FANCC mutants
All alanine (A; single-letter amino acid codes) substitutions in the FANCC protein were introduced by recombinant polymerase chain reaction (PCR) mutagenesis33 using Pfu enzyme (Stratagene, La Jolla, CA) with oligonucleotides encoding the following mutations. For F64A and T66A, the internal primers, encoding amino acid residues 61 to 69 of FANCC (GenBank sequence accession number X66893),9 were used to modify the protein at positions 64 and 66 to alanine. The 314-bp PCR-generated fragments encoding the F-to-A and T-to-A substitutions were used to replace the corresponding fragment in pLFACSN34 to create the mutants F64A and T66A, respectively. To mutagenize the central and carboxyl motifs of FANCC, the 1452-bp HindIII to stop codon-containing XbaI fragment of FANCC was subcloned into the corresponding sites of pUC18 to create pUCFAC-D1.34 For S249A and E251A substitutions, the internal primers encoding amino acid residues 245 to 254 of FANCC, having a new codon in place of the codon for serine (S) or glutamic acid (E), were used to mutate residues 249 or 251 to alanine. The 290-bp PCR products digested with Bsu36I and ApaI were then ligated into Bsu36I- and ApaI-digested pUCFAC-D1. Finally, the HindIII-XbaI fragment from the resulting plasmids was used to replace the corresponding fragment in pLFACSN to create mutants S249A and E251A. The same technique was used to create mutants F525A and Y531A. In this case, the internal primer encoded amino acid residues 522 to 534 and the PCR-generated 279-bp PstI-XbaI fragments were used to replace the corresponding ones in pUCFAC-D1. The naturally occurring FANCC truncated mutant, M55, was derived from the original expression vector pREP4-M55.35 This mutant cDNA encodes a FANCC polypeptide missing the amino-terminal 54 amino acids because of an abnormal initiation at methionine 55.29 All mutation-containing alleles were subjected to sequence analysis to confirm that the desired changes had occurred.
MMC sensitivity assay and cytogenetic analysis
The pLXSN36 plasmids containing the normalFANCC or its mutant versions (10 μg) were transduced into lymphoblasts by a previously described protocol.26 Sets of isogenic lines were selected for G418 (300 μg/mL) resistance, and cellular sensitivity to MMC was assayed by plating cells at a density of 2 × 105/mL in 24-well plates. After incubation with various concentrations of MMC for 5 days, viable cell counts by trypan blue exclusion were performed. Each sample was tested in duplicate or triplicate. Cytogenetic analysis for chromosomal aberrations was performed on the HSC536N cell lines with and without the addition of MMC and DEB. MMC and DEB were added at a final concentration of 40 ng/mL and 100 ng/mL, respectively, and the cultures, protected from light, were incubated at 37°C for 48 hours. After exposure to 0.25 μg/mL colcemid (Gibco-BRL, Grand Island, NY) for 1 hour, cells were treated with 0.075 M KCl for 10 minutes and then fixed with a 3:1 mixture of methanol/acetic acid. Slides of fixed cells were stained with Wright stain and about 50 metaphases from each sample were scored for chromosomal breaks and radial formation.
Subcellular fractionation, immunoprecipitation, and immunoblotting
Cells were lysed with lysis buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.6, 1% sodium deoxycholate, 1% Triton X-100, and 0.1% sodium dodecyl sulfate [SDS]) supplemented with the following protease inhibitors: 1% aprotinin, 1 μg/mL leupeptin, 1 mM phenylmethylsulfonylfluoride (PMSF), and 2 mM sodium orthovanadate. Cellular fractionation was performed as described elsewhere.19 For cell lysates used specifically to assess IFN receptor complex, cells were lysed in digitonin lysis buffer (1% digitonin, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 50 mM NaF, and 5 mM sodium pyrophosphate) containing the protease inhibitors mentioned above. Whole cell lysates ( WCLs; 1 mg total protein) were precleared with 50 μL 50% protein A-Sepharose suspension (Pharmacia Biotech, Piscataway, NJ) for 1 hour at 4°C. After separation of the protein A-Sepharose the lysate was incubated with anti–IFN-γRα (2 μg/mL) (Antigenix America Antibodies, New York, NY) for 3 to 5 hours at 4°C. Immunocomplexes were then bound to protein A-Sepharose beads (50 μL 50% slurry) during a 1- to 2-hour incubation at 4°C, recovered by centrifugation at 1000 rpm for 1 minute, and washed with 0.1% digitonin wash buffer. For immunoblotting, samples were heated at 94°C for 5 minutes in 2 × Laemmli SDS sample buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred onto nitrocellulose membranes. Immunoblots were subsequently incubated with anti–tyrosine phosphorylated STAT1 (P-STAT1) (New England Biolabs, Beverly, MA), anti-STAT1 α/β polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), or polyclonal antisera against FANCD218 or FANCC.19 37 After washing, the blots were incubated with appropriate secondary antibody for 30 minutes at room temperature, and developed by using an enhanced chemiluminescence (ECL) kit (Amersham, Buckinghamshire, United Kingdom).
GST-FANCC fusion protein purification and in vitro binding assays
The GST-FANCC fusion constructs were created by insertion of the entire open reading frame (ORF) of FANCC and its mutant versions into the BamHI and SmaI sites of a yeast overexpression vector pGST.38 Expression and purification of the GST-fusion proteins were performed as described elsewhere.26 For binding, about 10 μg GST-fusion proteins bound to glutathione-Sepharose beads was incubated with WCLs from untreated or IFN-γ–treated JY cells (1 mg total proteins) prepared in 1% digitonin buffer. The final concentration of digitonin in the binding mixture was adjusted to 0.5%. After shaking at 4°C for 40 minutes, the beads were recovered and washed 3 times with 0.1% digitonin buffer before boiling in Laemmli SDS sample buffer, and immunoblotted as described above.
Analysisof apoptosis
To quantify apoptotic cells, we used a polyclonal antibody to the active form of caspase 3 in a flow cytometric assay to detect cells in the early stages of apoptosis. The flow cytometric assays were performed by the procedure described elsewhere.32
Electromobilityshift assay
Nuclear extracts were prepared by the method of Dignam and coworkers.39 Oligonucleotides encoding the γ-IFN activation site (GAS) from the IRF1 gene40 and the p21 gene41 labeled with [γ-32P]-ATP to 2.5 × 104 cpm/ng using T4 polynucleotide kinase (Boehringer Mannheim, Indianapolis, IN). Binding reactions (20 μL) contained 5 μg nuclear extract and 0.2 ng labeled oligo were incubated at room temperature for 30 minutes, then resolved on a 4% nondenaturing polyacrylamide gel. In competition reactions, 25-fold molar excess of unlabeled oligonucleotides carrying the GAS were incubated with the binding mixtures. For supershift assays, binding reactions were incubated for a further 45 minutes in the presence of 5 μg STAT1 antibody before gel loading.
Results
Strategy for FANCC mutagenesis
The location of mutations in the FANCC gene found in FA-C patients reveal that both termini as well as the central region of the FANCC protein are critical for normal biologic function.1,2 We identified 3 motifs, located in the amino- or carboxyl-terminal and central regions of the FANCC protein (Figure 1), which are highly conserved among FANCC homologs of human, mouse, rat, and bovine origin.9,42,43 Sequence analysis of these conserved motifs by the nnpredict program44 predicted helical (motifs I and II) or β-strand (motif II) structures that have a propensity to interact with nucleic acids or proteins. Although such analyses are of a subjective nature, they served as a guide in our studies because analysis of FANCC primary sequence shows no homologies to known proteins or known functional motifs. We made alanine substitutions at amino acid positions F64 and T66 (motif I), S249 and E251 (motif II), and F529 and Y531 (motif III) in the human FANCC protein (Figure 1). The alanine mutagenesis strategy presumes that, although a functional group of the natural amino acid may be removed, alanine substitutions are less apt to perturb the overall structure or stability of the protein.45 This method thus allows for identification of amino acid residues within functional motifs of potential importance in the FANCC protein.
Motifs conserved among FANCC homologs.
Human (hFANCC), mouse (mFANCC), bovine (bFANCC), and rat (rFANCC) are compared. Bold letters correspond to identical amino acids. X indicates nonconserved residue. The number of residues between the different motifs is indicated in parentheses. The asterisks indicate the positions at which alanine substitutions were made.
Motifs conserved among FANCC homologs.
Human (hFANCC), mouse (mFANCC), bovine (bFANCC), and rat (rFANCC) are compared. Bold letters correspond to identical amino acids. X indicates nonconserved residue. The number of residues between the different motifs is indicated in parentheses. The asterisks indicate the positions at which alanine substitutions were made.
Complementation activity of the FANCCmutants
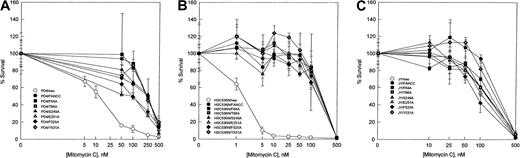
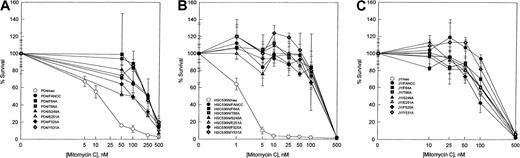
To examine the abilities of the FANCC mutants to complement the characteristic MMC hypersensitivity of FA cells, alanine mutantFANCC cDNAs, as well as the wild-type FANCC gene, were subcloned into the retroviral vector pLXSN36 and transduced into lymphoblasts derived from FA-C patients. The PD4 lymphoblast cell line is heterozygous for a frameshift mutation in one allele, predicting a truncated peptide of the amino-terminal 44 amino acids, and a nonsense mutation in the other allele, predicting a terminated peptide of 185 residues.20 The HSC536N cell line contains a T to C substitution in one allele, resulting in an L554P amino acid change; the other allele is not expressed because it is deleted.9,46 As shown in Figure2, PD4 cells and HSC536N cells transduced with the normal FANCC gene corrected the defect of the mutant FA-C cells in resistance to MMC exposure. Parental cells alone or transduced with the retroviral vector exhibited hypersensitivity to MMC with an effective drug concentration yielding 50% reduction in cell viability (EC50) of 5 nM MMC for PD4 cells (Figure 2A) and approximately 2 nM MMC for HSC536N cells (Figure 2B). All 6 alanine-substituted mutants conferred MMC resistance in that the survival curves in both PD4 and HSC536N cells were similar to those of the parental cell lines transduced with the normal FANCCgene. Although these alanine-substituted mutants were able to complement the MMC hypersensitivity in mutant PD4 and HSC536N cells, their ability to exhibit a dominant negative phenotype in vivo could not be excluded. For instance, mutations in the ATP- binding domain of DNA mismatch repair protein MSH2 do not affect mismatch recognition but cause a dominant negative effect when overexpressed in wild-type cells.47 Therefore, we introduced the 6 mutants along with the normal FANCC as well as the retroviral vector alone into the normal lymphoblast cell line JY. As shown in Figure 2C, no alanine-substituted mutation influenced survival of JY cells exposed to MMC. Cytogenetic analysis has shown that FA-C lymphoblasts had an increased frequency of chromosomal aberrations induced by DNA cross-linking agents. We analyzed radial chromosomes formed in retroviral-transduced HSC536N cells treated with MMC. As shown in Table1, whereas HSC536N and HSC536N/neo cells had a characteristic high frequency of radial formation (> 20%), all 6 alanine-substituted mutant-transduced cell lines as well as HSC536N/FANCC had significantly reduced numbers of chromosomal radials (< 5%). Similar results were obtained with analysis of chromosomal breaks induced by MMC and DEB in these retrovirally transduced cell lines (data not shown). These cytogenetic data are consistent with the MMC survival curves demonstrated in Figure 2. Taken together, these results indicate that the alanine substitutions in the 3 conserved motifs (Figure 1) of the FANCC protein are not defective in their capacity to protect FA-C cells from cross-linking agent-induced toxicity.
MMC hypersensitivity.
Effect of alanine-substituted FANCC mutants on MMC sensitivity is shown in FA-C lymphoblast cell lines PD4 (A) and HSC536N (B) and normal lymphoblast cell line JY (C). Cells were treated with varying concentrations of MMC for 5 days before stained with trypan blue dye to quantify viable cells. Cell viability is expressed as percentages of viable cells relative to the untreated control culture. Each data point represents the mean ± SD of at least triplicate determinations.
MMC hypersensitivity.
Effect of alanine-substituted FANCC mutants on MMC sensitivity is shown in FA-C lymphoblast cell lines PD4 (A) and HSC536N (B) and normal lymphoblast cell line JY (C). Cells were treated with varying concentrations of MMC for 5 days before stained with trypan blue dye to quantify viable cells. Cell viability is expressed as percentages of viable cells relative to the untreated control culture. Each data point represents the mean ± SD of at least triplicate determinations.
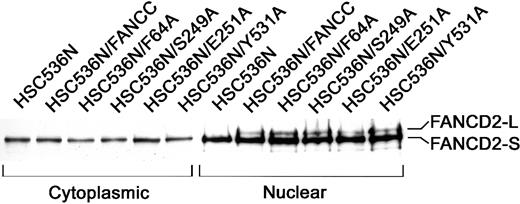
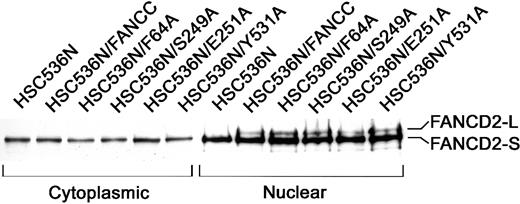
Because an intact FA protein complex is essential for cellular resistance to MMC, we further tested whether these alanine mutations affected the FA complex formation. Because the formation of a functional FA complex is required for the posttranslational activation of FANCD2,18 we chose to examine the activation state of FANCD2 in nuclear extracts of lymphoblasts bearing these alanine mutants as the most direct way to demonstrate the function of the FA complex. As shown in Figure 3, the corrected FA-C cells (HSC/FANCC) and all 6 alanine mutants contained 2 nuclear FANCD2 isoforms: the short form (FANCD2-S) which is the primary translation product,10 and posttranslationally modified (by monoubiquitination18) FANCD2-L form (lanes 8-12). FA-C lymphoblasts (HSC536N) expressed only nuclear FANCD2-S (lane 7). As expected, no activated FANCD2-L was observed in any cytoplasmic sample (lanes 1-6). This result suggests that these alanine-substituted FANCC proteins are normally assembled into the multisubunit nuclear complex that consequently promotes the activation of FANCD2 and functions to protect cells from MMC genotoxicity.
Effects of alanine-substituted FANCCmutations on activation of FANCD2.
The indicated lymphoblast lines were lysed and fractionated into cytoplasmic and nuclear fractions. Equal amounts from each fraction were analyzed by immunoblotting with antibody against FANCD2. The high-molecular-weight isoform, FANCD2-L, represents the activated FANCD2. All samples were analyzed for β-tubulin levels to ensure effective cellular fractionation.
Effects of alanine-substituted FANCCmutations on activation of FANCD2.
The indicated lymphoblast lines were lysed and fractionated into cytoplasmic and nuclear fractions. Equal amounts from each fraction were analyzed by immunoblotting with antibody against FANCD2. The high-molecular-weight isoform, FANCD2-L, represents the activated FANCD2. All samples were analyzed for β-tubulin levels to ensure effective cellular fractionation.
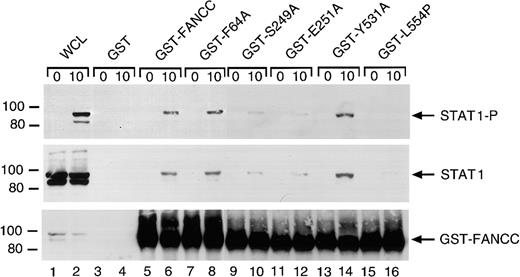
Effects of FANCC mutations on STAT1 phosphorylation in HSC536N cells
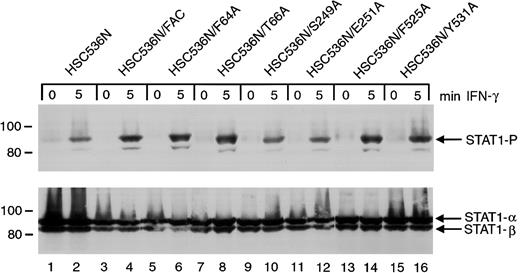
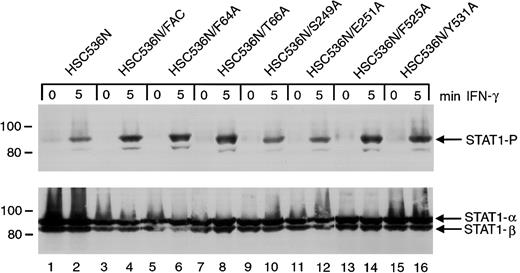
The ability of the 6 alanine-substituted mutants to complement the MMC hypersensitivity of FA-C cells is an indication that the mutants function normally in facilitating DNA damage responses or DNA repair. However, a different situation may exist for the signaling functions of FANCC. We have previously shown that STAT1 phosphorylation induced by IFN-γ is markedly reduced in HSC536N cells when compared to wild-type cells and the complemented HSC536N/FANCC cells, and FANCC acts to facilitate the docking of STAT1 to the activated IFN-γ receptor.3 26 We sought to elucidate the effects of the FANCC mutant proteins on STAT1 phosphorylation defects in FA-C cells. Retrovirally transduced HSC536N cells were serum-starved for 2 hours then treated with 1 ng/mL IFN-γ for 5 minutes, and STAT1 phosphorylation was determined by immunoblot analysis with an antibody specific for tyrosine-phosphorylated STAT1 (Figure4). Despite correction of the MMC hypersensitivity phenotype by all 6 alanine mutations, immunoblot analysis of cell lysates revealed that defective STAT1 phosphorylation was not corrected in HSC536N cells transduced with FANCCcDNAs containing the 2 mutations (S249A and E251A) located in the motif II of FANCC (Figure 4, lanes 10 and 12). In contrast, mutations located in the amino-terminal motif (F64A and T66A) and in the carboxyl-terminal motif (F525A and Y531A) complemented the STAT1 phosphorylation defect (lanes 6, 8 and 14, 16, respectively). Results were identical in 4 separate experiments. This suggests that the central region of the FANCC protein is required for STAT1 activation in response to IFN-γ stimulation. The 6 HSC536N cell lines transduced with alanine mutants of FANCC mutations were also tested for expression of FANCC protein. All lines expressed more FANCC protein than that detected in the parent line (data not shown).
Effects of alanine-substituted FANCCmutations on STAT1 phosphorylation in HSC536N cells.
Upper panel: The HSC536N FANCC mutant cells (lanes 5-16) were exposed to 1 ng/mL IFN-γ for times indicated and compared to HSC536N cells (lanes 1 and 2) as well as HSC536N/FANCC cells complemented by transduction with wild-type FANCC (lanes 3 and 4). Cell lysates were separated by SDS-PAGE in a 7.5% polyacrylamide gel and immunoblotted with an antibody specific for STAT1 protein phosphorylated on Y701. Lower panel: The blot described in the upper panel was stripped and reprobed for total STAT1 protein.
Effects of alanine-substituted FANCCmutations on STAT1 phosphorylation in HSC536N cells.
Upper panel: The HSC536N FANCC mutant cells (lanes 5-16) were exposed to 1 ng/mL IFN-γ for times indicated and compared to HSC536N cells (lanes 1 and 2) as well as HSC536N/FANCC cells complemented by transduction with wild-type FANCC (lanes 3 and 4). Cell lysates were separated by SDS-PAGE in a 7.5% polyacrylamide gel and immunoblotted with an antibody specific for STAT1 protein phosphorylated on Y701. Lower panel: The blot described in the upper panel was stripped and reprobed for total STAT1 protein.
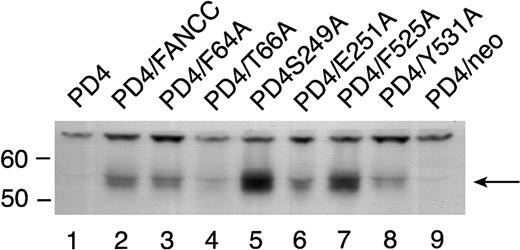
FANCC mutant protein expression in PD4 cells
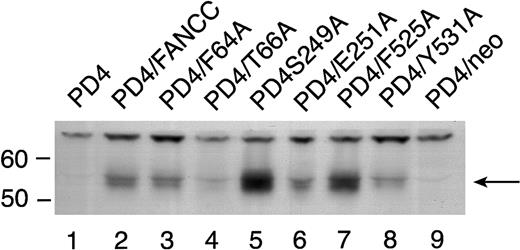
To rule out the possibility that the reduced amount of STAT1 phosphorylation in the 2 mutants located in the middle domain was simply due to reduced stability of these mutant proteins, we expressed the 6 mutant FANCC proteins in PD4 cells. PD4 cells do not express endogenous full-length FANCC protein because they contain 2 naturally occurring truncated FANCC mutations (delG322, and C to T at bp 808).20 Cell lysates from PD4 cells carrying the FANCC mutants were probed with antiserum generated against an N-terminal polypeptide to the FANCC protein revealed varying amounts of FANCC protein in some of the cell lines (Figure5). All effectively corrected the MMC hypersensitivity phenotype in PD4 cells (Figure 2A) despite the fact that some expressed low levels of mutant FANCC protein (Figure 5, T66A [lane 4] and Y531A [lane 8]). Variable amounts of protein expression did not correlate with reduced STAT1 phosphorylation seen in the HSC536N cells (Figure 4), because the quantity of FANCC protein in the S249A and E251A mutants was not reduced (Figure 5, lanes 5 and 6).
Expression of alanine-substituted FANCC mutant protein in PD4 cells.
PD4 cell lysates (100 μg each) were subjected to SDS-PAGE in a 10% polyacrylamide gel. PD4 alanine-substituted mutant cells (lanes 3-8) were compared to PD4 cells (lane 1), PD4/FANCC cells complemented with wild-type FANCC (lane 2), and PD4/neo cells transduced with vector alone (lane 9). The immunoblot was probed with rabbit polyclonal antiserum generated against the N-terminus of the FANCC protein.
Expression of alanine-substituted FANCC mutant protein in PD4 cells.
PD4 cell lysates (100 μg each) were subjected to SDS-PAGE in a 10% polyacrylamide gel. PD4 alanine-substituted mutant cells (lanes 3-8) were compared to PD4 cells (lane 1), PD4/FANCC cells complemented with wild-type FANCC (lane 2), and PD4/neo cells transduced with vector alone (lane 9). The immunoblot was probed with rabbit polyclonal antiserum generated against the N-terminus of the FANCC protein.
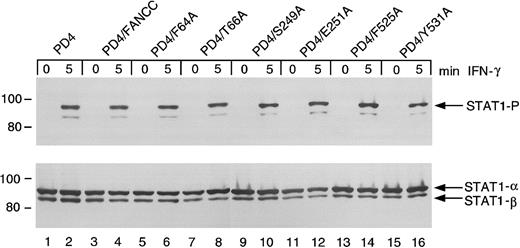
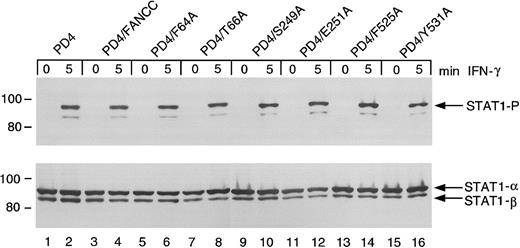
Effects of FANCC mutations on STAT1 phosphorylation in PD4 FA-C cells
The FANCC mutant-expressing PD4 cells above were further tested for STAT1 phosphorylation defects. Unlike HSC536N cells, PD4 cells do not exhibit suppressed STAT1 phosphorylation in response to IFN (Figure6, lanes 1 and 2). The expression of FANCC mutant proteins (lanes 5-16) and wild-type FANCC (lanes 3 and 4) neither suppresses nor augments induced STAT1 phosphorylation. The S249A and E251A constructs, which failed to complement STAT1 phosphorylation in the HSC536N cells, are not dominant negative mutants for STAT1 activation in PD4 cells (lanes 10 and 12). Based on results of studies described below, the gene product derived from the 322delG mutation by reinitiation at residue 55 (M55), explains the normal STAT1 phosphorylation phenotype detected in the PD4 cells. This reinitiation product loses only a small portion of the amino-terminus of the FANCC protein leaving the motif II domain and its surrounding peptide sequences intact (see below).
Effects of alanine-substituted FANCCmutations on STAT1 phosphorylation in PD4 cells.
Upper panel: PD4 alanine-substituted FANCC mutant cells (lanes 5-16) were exposed to 1 ng/mL IFN-γ for times indicated and compared to PD4 cells (lanes 1 and 2) as well as PD4/FANCC cells complemented by transduction with wild-type FANCC (lanes 3 and 4). Cell lysates were separated by SDS-PAGE in a 7.5% polyacrylamide gel and immunoblotted with an antibody specific for tyrosine phosphorylated STAT1 protein. Lower panel: The blot described in the upper panel was stripped and reprobed for total STAT1 protein.
Effects of alanine-substituted FANCCmutations on STAT1 phosphorylation in PD4 cells.
Upper panel: PD4 alanine-substituted FANCC mutant cells (lanes 5-16) were exposed to 1 ng/mL IFN-γ for times indicated and compared to PD4 cells (lanes 1 and 2) as well as PD4/FANCC cells complemented by transduction with wild-type FANCC (lanes 3 and 4). Cell lysates were separated by SDS-PAGE in a 7.5% polyacrylamide gel and immunoblotted with an antibody specific for tyrosine phosphorylated STAT1 protein. Lower panel: The blot described in the upper panel was stripped and reprobed for total STAT1 protein.
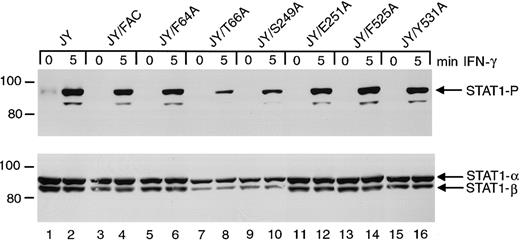
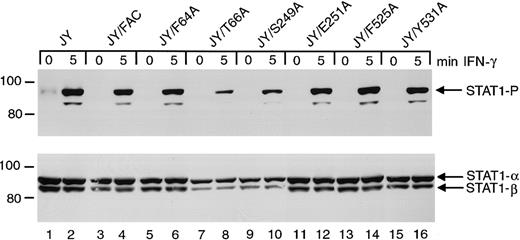
Effects of FANCC mutations on STAT1 phosphorylation in normal JY cells
To test whether these FANCC mutations disrupt normal cell functions, we introduced the mutant cDNAs into normal JY lymphoblasts. Immunoblot analysis of cell lysates indicated that STAT1 phosphorylation in JY cells is unaffected by the expression of the FANCC mutant proteins (Figure 7). The S249A and E251A mutations have no detectable effect on STAT1 phosphorylation in these normal cells and are therefore not negatively dominant for STAT1 activation. Reduced amounts of phosphorylated STAT1 in lanes 8 and 10 result from reduced total STAT1 protein in this particular experiment.
Effects of alanine-substituted FANCCmutations on STAT1 phosphorylation in normal JY cells.
Upper panel: JY alanine-substituted FANCC mutant cells (lanes 5-16) were exposed to 1 ng/mL IFN-γ for times indicated, and compared to JY cells (lanes 1 and 2) as well as JY/FANCC cells transduced with wild-type FANCC (lanes 3 and 4). Cell lysates were separated by SDS-PAGE in a 7.5% polyacrylamide gel and immunoblotted with an antibody to tyrosine phosphorylated STAT1. Lower panel: The blot described in the upper panel was stripped and reprobed with an antibody to total STAT1 protein. Reduced amounts of phosphorylated STAT1 in lanes 8 and 10 appear to be a result of reduced total STAT1 protein in this experiment. In all subsequent experiments this particular finding was not noted.
Effects of alanine-substituted FANCCmutations on STAT1 phosphorylation in normal JY cells.
Upper panel: JY alanine-substituted FANCC mutant cells (lanes 5-16) were exposed to 1 ng/mL IFN-γ for times indicated, and compared to JY cells (lanes 1 and 2) as well as JY/FANCC cells transduced with wild-type FANCC (lanes 3 and 4). Cell lysates were separated by SDS-PAGE in a 7.5% polyacrylamide gel and immunoblotted with an antibody to tyrosine phosphorylated STAT1. Lower panel: The blot described in the upper panel was stripped and reprobed with an antibody to total STAT1 protein. Reduced amounts of phosphorylated STAT1 in lanes 8 and 10 appear to be a result of reduced total STAT1 protein in this experiment. In all subsequent experiments this particular finding was not noted.
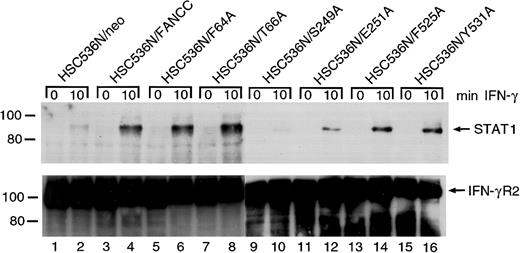
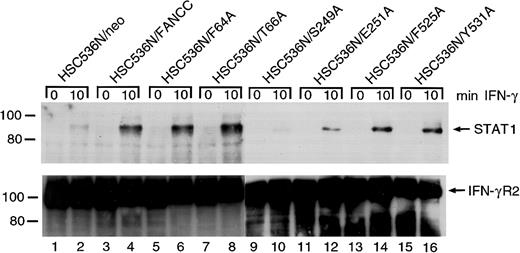
FANCC mutations are defective in STAT1 recruitment to the IFN-γ receptor
It has been reported that in IFN-γ–stimulated cells latent STAT1 is recruited to a docking site on IFN-γRα, where STAT1 is then activated by tyrosine phosphorylation.48,49Consistent with this, STAT1 has been found associated with tyrosine-phosphorylated IFN-γRα both in vitro and in vivo.50-52 We have shown that FANCC can facilitate STAT1 activation by recruiting STAT1 to the IFN-γ receptor.26The apparent failure of S249A and E251A mutants to correct the STAT1 defect is an indication that in these 2 mutants STAT1 may not be recruited to the receptor. To confirm this notion we stimulated the retrovirally transduced cells with IFN-γ, prepared WCLs in 1% digitonin lysis buffer, and performed coimmunoprecipitation using antibody to IFN γRα. The presence of STAT1 in anti–IFN-γRα immunoprecipitates was detected by probing with anti-STAT1 antibody. Whereas HSC536N cells transduced with the vector alone (HSC536N/neo) did not contain STAT1 in the anti–IFN-γRα precipitate (Figure8, top, lanes 1 and 2), IFN-γ induced coimmunoprecipitation of STAT1 with IFN-γRα in the normalFANCC-transduced cell line HSC536N/FANCC (lane 4). Whereas the 4 alanine-substituted mutations in both termini of FANCC showed normal STAT1 recruitment, cells transduced with the 2 alanine-substituted mutations located in the central region of FANCC, S249A and E251A, contained markedly less STAT1 in the receptor complex (lanes 6, 8, 14, and 16 versus 10 and 12). This result correlated with the observation in which S249A and E251A mutants only partially corrected the deficiency of STAT1 activation in HSC536N cells (Figure4). All lanes contained approximately equal amounts of immunoprecipitated IFN-γRα protein loaded onto the gels (Figure8, bottom).
Effect of alanine-substituted FANCCmutations on IFN-γ–induced formation of STAT1–IFN-γRα complex in B lymphocytes.
Cells were stimulated with IFN-γ for 10 minutes and solubilized in digitonin lysis buffer as described in “Materials and methods.” The WCLs were then incubated with anti–IFN-γRα antibody. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting. Upper panel: Cell lysates prepared from HSC536N isogenic lines were immunoprecipitated with anti–IFN-γRα and probed with anti-STAT1 monoclonal antibody. Lower panel: The blot described in the top panel was stripped and reprobed with anti–IFN-γRα.
Effect of alanine-substituted FANCCmutations on IFN-γ–induced formation of STAT1–IFN-γRα complex in B lymphocytes.
Cells were stimulated with IFN-γ for 10 minutes and solubilized in digitonin lysis buffer as described in “Materials and methods.” The WCLs were then incubated with anti–IFN-γRα antibody. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting. Upper panel: Cell lysates prepared from HSC536N isogenic lines were immunoprecipitated with anti–IFN-γRα and probed with anti-STAT1 monoclonal antibody. Lower panel: The blot described in the top panel was stripped and reprobed with anti–IFN-γRα.
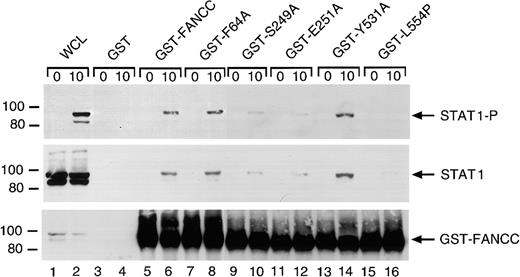
FANCC mutations suppress FANCC-STAT1 interaction
The FANCC protein has been shown to associate with STAT1 in WCLs from IFN-γ–stimulated cell lines.26 We therefore examined the effect of the alanine substitutions on FANCC-STAT1 interaction. We stimulated the normal lymphoblasts (JY) with IFN-γ and incubated WCLs with the normal or mutated FANCC proteins bound to glutathione-Sepharose beads through GST. FANCC-interacting cellular proteins were then isolated from WCLs by glutathione-Sepharose affinity precipitation. Western blotting with antibody to P-STAT1 showed that IFN-γ induced tyrosine phosphorylation of STAT1 in IFN-γ–stimulated WCLs (Figure 9, top panel, lane 2). GST-fusion protein affinity binding assays showed that IFN-γ induced the association of STAT1 with the normal FANCC and the alanine-substituted mutants F64A and Y531A, but significantly less STAT1 association occurred with the S249A and E251A mutants (lanes 6, 8 and 14, 16 versus 10 and 12). STAT1 showed an even weaker association with GST-fused L554P, a naturally occurring FANCC mutation found in FA-C patients46 (lane 16). STAT1 did not bind to GST alone (lanes 3 and 4). Reprobing the blot with antibody to total STAT1 indicated that IFN-γ did not have measurable effect on the expression of the STAT1 protein (Figure 9, middle panel, lanes 1 and 2), and confirmed that FANCC-STAT1 interaction was suppressed by S249A, E251A, and L554P mutations (lanes 10, 12, and 16). The bottom panel of Figure 8 demonstrates that input amounts of GST-fusion proteins were comparable in the binding reactions.
Two alanine-substituted FANCC mutations suppress IFN-γ–induced association of FANCC and STAT1 in B lymphocytes.
WCLs of JY cells stimulated with IFN-γ for 10 minutes (1 mg protein) were incubated with yeast-expressed GST-fusion proteins (10 μg) bound to glutathione-Sepharose beads. The glutathione-Sepharose affinity precipitates were analyzed by SDS-PAGE followed by immunoblotting. Upper panel: The Western blot was probed with antibodies to P-STAT1. Middle panel: The blot described in the top panel was stripped and reprobed with anti-STAT1 antibody. Lower panel: The same blot was reprobed with anti-FANCC to verify the input of the GST-fusion proteins. Lanes 1 and 2 were run as WCLs (60 μg of total proteins each) to show the effects of IFN-γ on expression and activation of STAT1.
Two alanine-substituted FANCC mutations suppress IFN-γ–induced association of FANCC and STAT1 in B lymphocytes.
WCLs of JY cells stimulated with IFN-γ for 10 minutes (1 mg protein) were incubated with yeast-expressed GST-fusion proteins (10 μg) bound to glutathione-Sepharose beads. The glutathione-Sepharose affinity precipitates were analyzed by SDS-PAGE followed by immunoblotting. Upper panel: The Western blot was probed with antibodies to P-STAT1. Middle panel: The blot described in the top panel was stripped and reprobed with anti-STAT1 antibody. Lower panel: The same blot was reprobed with anti-FANCC to verify the input of the GST-fusion proteins. Lanes 1 and 2 were run as WCLs (60 μg of total proteins each) to show the effects of IFN-γ on expression and activation of STAT1.
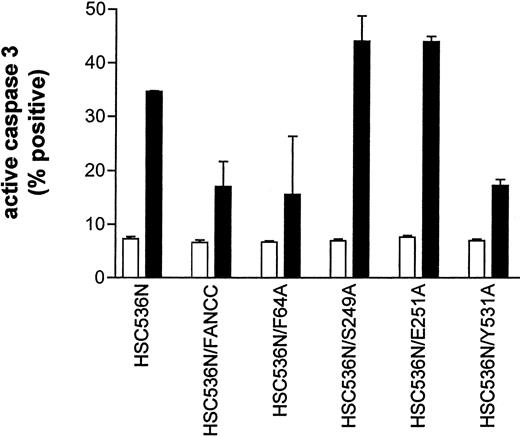
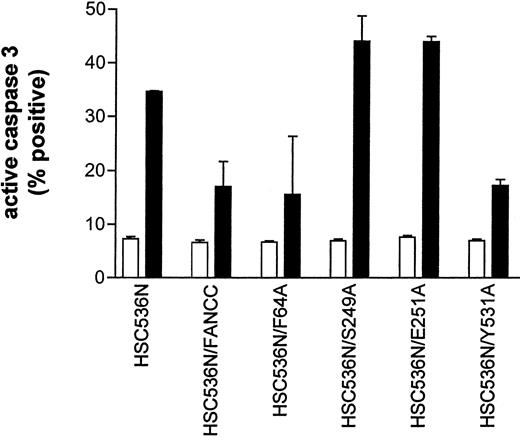
The results described above implicate a potential defect in survival signaling in S249A and E251A mutants. We thus sought to identify the functional significance of these alanine mutations. We expressed mutant FANCC proteins in HSC536N lymphoblasts, then tested the transductants for sensitivity to IFN-γ and TNF-α using a flow cytometric method for caspase 3 activation. Figure10 shows that the S249A and E251A mutants had 2-fold more apoptotic cells than the corrected cells after treatment with IFN-γ and TNF-α (compare columns S249A and E251A with FANCC) and were as hypersensitive to IFN-γ and TNF-α as the parent HSC536N cells (compare columns S248A and E251A with HSC536N). However, cells expressing alanine substitutions in the amino- and carboxy-terminal motifs, F64A and Y531A, were resistant to IFN-γ and TNF-α (columns F64A and Y531A). These data demonstrate that the central domain of FANCC is essential for protection of hematopoietic cells from apoptosis induced by IFN-γ and TNF-α.
Alanine mutants S249A and E251A sensitize hematopoietic cells to IFN-γ and TNF-α cytotoxicity.
HSC536N lymphoblasts expressing normal FANCC or mutatedFANCC were incubated with IFN-γ (10 ng/mL) for 15 hours before treatment with 10 ng/mL TNF-α for 48 hours and harvested for flow cytometric analysis (▪, treated; □, untreated). Treatments of HSC/VEC and HSC/FANCC with Camptothecin were used as positive controls, 50 mM for 9 hours) and induced positivity of 65.1% and 22.5%, respectively. Total events of 10 000 for each sample were analyzed.
Alanine mutants S249A and E251A sensitize hematopoietic cells to IFN-γ and TNF-α cytotoxicity.
HSC536N lymphoblasts expressing normal FANCC or mutatedFANCC were incubated with IFN-γ (10 ng/mL) for 15 hours before treatment with 10 ng/mL TNF-α for 48 hours and harvested for flow cytometric analysis (▪, treated; □, untreated). Treatments of HSC/VEC and HSC/FANCC with Camptothecin were used as positive controls, 50 mM for 9 hours) and induced positivity of 65.1% and 22.5%, respectively. Total events of 10 000 for each sample were analyzed.
M55 mutation restores STAT1 activation but fails to correct MMC hypersensitivity of FA-C cells
We next sought to determine whether MMC and STAT functions could be disconnected by naturally occurring FANCC mutants. We investigated a naturally occurring FANCC truncated mutant, M55, which lacks the amino-terminal 54 residues owing to an abnormal initiation at methionine 55.35 Note that the deletion point of M55 is close to motif I but leaves intact the central motif II (Figure11A), which is required for optimal STAT1 activation (Figures 4, 8, and 9) and antiapoptotic function. We determined the effect of the M55 truncated mutant on STAT1 activation and MMC sensitivity in FA-C lymphocytes. Electromobility shift assay (EMSA) using a GAS (a STAT1 binding site from the IRF1gene40) oligonucleotide probe, showed that STAT1 DNA-binding activity was restored when M55 was expressed in IFN-γ–stimulated HSC536N cells. Control HSC536N cells were completely defective in STAT1 DNA-binding (Figure 11B, compare lane 1 with lane 2). Nuclear extracts from PD4 cells also showed GAS-binding activity (lane 4), consistent with the result described in Figure 6. The complexes formed with the GAS in extracts from FANCC-corrected HSC536N and PD4 cells were disrupted by excess unlabeled GAS (Figure11B, lanes 5 and 6). The identity of STAT1-DNA complexes was confirmed by a STAT1-specific antibody that altered the migration of the STAT1-DNA band in the EMSA (lane 7). Furthermore, EMSA using the GAS sequences derived from the p21 gene promoter41 produced essentially the same results (Figure 11C) as demonstrated in Figure11B. FA-C mutant HSC536N cells transduced with the M55 cDNA did not correct MMC hypersensitivity of FA-C cells as compared to the parental mutant cells (Figure 11D). The slight increase in survival of M55-transduced HSC536N cells probably reflects a dose effect because transduction of HSC536N cells even with the noncomplementing L554P mutant cDNA also resulted in slight resistance to MMC exposure (Figure11D). Neither the M55 nor the L554P mutant corrected MMC-induced cytogenetic abnormalities in HSC536N cells. In addition, expression of M55 did not reduce survival of the normal JY lymphocytes exposed to MMC (data not shown). Although Youssoufian and colleagues53reported that overexpression of the L554P mutant rendered human 293 cells hypersensitive to MMC, we did not observed this phenomenon in L554P-expressing lymphoblasts (Figure 11D) and HeLa cells (data not shown). One possible explanation for the differences is the expression levels of the mutant protein. Youssoufian and coworkers used expression vector pED6, which resulted in 4- to 15-fold higher levels of expression than the endogenous FANCC gene. We used retroviral vector pLXSN that expressed only 1- to 2-fold higher FANCC mutant proteins relative to the endogenous FANCC proteins, which apparently were not sufficient to cause a dominant negative effect. Another possibility is that we used different cell lines.
M55 mutation restores STAT1 DNA-binding activity but fails to complement MMC hypersensitivity of FA-C cells.
(A) A schematic diagram of full-length FANCC and truncated mutant M55 that lacks amino-terminal 54 amino acids. The 3 motifs described in Figure 1 are indicated as black boxes. (B,C) EMSA analyses of STAT1 activation in M55-transduced FA-C lymphocytes. Normal FANCC- or M55-transduced HSC536N and PD4 cells were treated with IFN-γ (10 ng/mL) for 30 minutes and EMSA was performed using 5 μg of nuclear extracts and [32P]-labeled oligonucleotides carrying the GAS from the IRF1 gene (B) or from the p21 gene (C). Competitive binding was carried out by incubation with 25-fold excess molar unlabeled GAS (lanes 5 and 6) oligonuleotides. Lane 7 indicates that binding reactions were incubated with an STAT1 antibody (5 μg). Arrows denote STAT1-GAS complexes. (D) Effect of M55 mutation on MMC sensitivity in FA-C lymphoblast cell line HSC536N. Cells were treated with varying concentrations of MMC for 5 days before staining with trypan blue dye to quantify viable cells. Cell viability is expressed as percentages of viable cells relative to the untreated control culture. Each data point represents the mean ± SD of at least triplicate determinations.
M55 mutation restores STAT1 DNA-binding activity but fails to complement MMC hypersensitivity of FA-C cells.
(A) A schematic diagram of full-length FANCC and truncated mutant M55 that lacks amino-terminal 54 amino acids. The 3 motifs described in Figure 1 are indicated as black boxes. (B,C) EMSA analyses of STAT1 activation in M55-transduced FA-C lymphocytes. Normal FANCC- or M55-transduced HSC536N and PD4 cells were treated with IFN-γ (10 ng/mL) for 30 minutes and EMSA was performed using 5 μg of nuclear extracts and [32P]-labeled oligonucleotides carrying the GAS from the IRF1 gene (B) or from the p21 gene (C). Competitive binding was carried out by incubation with 25-fold excess molar unlabeled GAS (lanes 5 and 6) oligonuleotides. Lane 7 indicates that binding reactions were incubated with an STAT1 antibody (5 μg). Arrows denote STAT1-GAS complexes. (D) Effect of M55 mutation on MMC sensitivity in FA-C lymphoblast cell line HSC536N. Cells were treated with varying concentrations of MMC for 5 days before staining with trypan blue dye to quantify viable cells. Cell viability is expressed as percentages of viable cells relative to the untreated control culture. Each data point represents the mean ± SD of at least triplicate determinations.
Discussion
An increasingly persuasive body of evidence indicates that FANCC functions in distinct pathways controlling protective responses to alkylating agents, and survival responses to cytokines and human growth factors.1,3,5,26,32 The functional heterogeneity of the Fanconi proteins, known to exist as multimeric complexes with each other,54,55 resembles the heterogeneity of the ATM protein. The latter plays roles in signal transduction, cell cycle checkpoints, and DNA damage response.56,57 Through the use of alanine-scanning mutagenesis, we have identified herein a central motif of the FANCC protein that appears to be required to mediate (1) the recruitment of STAT1 to the IFN-γRα chain, (2) the interaction between FANCC and STAT1, (3) the activation and nuclear translocation of STAT1, and (4) the protection of hematopoietic cells against high-level cytotoxicity induced by IFN-γ and TNF-α (Figures 4, 8,9, 10, and 11), an effect we have recently shown to be due, in large part, to the capacity of FANCC to suppress PKR activation.32 However, alanine substitutions in this motif retain the ability to complement the hypersensitivity of FA-C lymphoblasts treated with DNA cross-linking agents MMC and DEB (Figure2 and Table 1). These data provided clear structural evidence that FANCC plays multifunctional roles in cellular damage responses and transduction of growth control signals.
We also determined that certain mutations in FA-C patients might result in the disconnection of one function of FANCC from another. Specifically, a naturally occurring mutant of FANCC, M55, was capable of restoring STAT1 activation in FA-C cells, but failed to correct MMC hypersensitivity of FA-C cells (Figure 11). Moreover, lymphoid cells from a child bearing this mutation exhibit normal STAT1 activation responses to IFN. Based on these findings, we argue that FANCC structural determinants for damage/repair responses may differ from those for STAT1 signaling.
Although alanine substitutions usually preserve the native conformation of the wild-type protein,45 such substitutions may remove the contacts that mediate interactions between the 2 interacting proteins or between protein and DNA. The loss of critical amino acid-amino acid contacts due to the alanine substitutions may explain our observation that the S249A and E251A substitutions in the central motif suppress the FANCC-STAT1 interaction (Figures 8 and 9). Although the substitutions themselves are not bulky, the mutations might, nonetheless, influence tertiary structure of FANCC. Specifically, the highly conserved S249 and E251 residues, which are polar and charged, respectively, exist within a predicted β-strand. These residues may stabilize the β-strand via hydrogen bonds to the peptide backbone. It is reasonable to presume that disruption of these hydrogen bonds by alanine substitutions causes aberrant folding of FANCC, which may lead to defects in the interaction with STAT1 and in the recruitment of STAT1 to IFN-γRα. This observation is congruent with the recent finding that the central region of FANCC is responsible for its association with the molecular chaperone GRP94.24 GRP94 has been long known to be involved in trafficking cytoplasmic proteins to the cell membrane and the nucleus.58 It is plausible that FANCC acts as a cochaperone and acts in concert with GRP94 to deliver STAT1 to the IFN-γRα complex.
High degrees of homology between FANCCs from various organisms is scattered in small blocks of several amino acid residues and spans the entire peptide sequence. When we tested the alanine-substituted mutants for responses to the cytotoxic effects of MMC and DEB, we were surprised to find that this function of FANCC is remarkably resistant to alanine substitutions (Figure 2 and Table 1). The ability of the FANCC protein to accommodate these substitutions without an impact on the MMC resistance function of the mutants may explain the recent finding that bovine FANCC is able to complement a FA-C patient-derived lymphoblast cell line HSC536N, even though the sequence between the 2 FANCCs are only 72% identical.42 Similarly, introduction of the normal human FANCC cDNA into a mouse hematopoietic progenitor cell line suppressed apoptosis induced by growth factor deprivation29 and into a mouse FA-C mutant fibroblast cell line rescued normal resistance to MMC (M. C. Heinrich and G.C.B., unpublished data, 1999), despite the fact that the human and mouse FANCCs are even more distant (62% identical).
Our results indicate that hypersensitivity to MMC or DEB in FA cells is not coupled to the cytokine signaling functions of FANCC, including the defect of STAT1 activation because the 2 alanine mutants in the central region of FANCC retain the ability to confer normal resistance to the cytotoxic effects of DNA cross-linking agents (Figure2 and Table 1) but render cells defective in the activation of STAT1 (Figures 4, 8, and 9). Furthermore, we confirmed that FANCC mutations with “disconnected” functions actually occur in nature. The data obtained with a naturally occurring FANCC mutant, presented in Figure11, demonstrate that the truncated M55 mutant preserves the ability to facilitate STAT1 activation but fails to correct MMC hypersensitivity of FA-C cells. Taken together, our results suggest that separate structural elements of FANCC permit the protein to specifically respond to different extracellular signals. It is likely that FANCC acts coordinately with additional, as yet unrecognized factors, and with other FA proteins, by using separate structural elements, to function in different cellular processes including the response or repair (or both) of specific DNA damage and the control of cell growth by facilitating the cellular response to growth and survival factors.
Growth factor signal transduction is defective in FA cells, a finding that may contribute to the bone marrow failure characteristic of this disease. Several lines of evidence indicate that bone marrow failure in FA results from increased apoptosis owing to hypersensitivity to inhibitory factors and diminished responses to survival cues.3,26,29,59 Our results, together with those of others, support a model in which one of the functions of FANCC is to ensure optimal cellular responses to growth factors, at least in part by facilitating STAT1 activation. It is known that depletion of growth factors or cytokines leads to progressive apoptosis in many cell types including fibroblast60 and hematopoietic cells,29,61 which can be rescued by the addition of defined factors. These factors include STAT1 activators such as IFN-γ,62 colony-stimulating factor,63platelet-derived growth factor,60 interleukin 6,64 and stem cell factor.59 Therefore, deficient STAT1 signaling in response to critical hematopoietic growth factors may play an important role in the bone marrow defect in FA. This model will be particularly difficult to formally test, however, because the FANCC domain required for the STAT1 cofactor function is the same as the domain required to suppress STAT1-independent apoptotic responses in IFN- and TNF-treated cells.
Regardless of the precise mechanism, domain II is unambiguously essential for the antiapoptotic function of FANCC. In fact, our results also provide a potential molecular basis for the clinical observation that bone marrow failure seems milder in children with the 322delG mutation in which all 3 conserved motifs, including motif II, remains intact. Finally, the current diagnostic tests for FA would necessarily fail to identify lesions (eg, point mutations in domain II) that disrupt signaling functions of Fanconi proteins without interdicting their capacity to protect cells from cross-linking agents. Although we have not yet identified such mutations in nature, evidence of such lesions should be now be sought in children whose phenotype is suggestive of FA but whose MMC or DEB tests are normal.
We thank Dr Manuel Buchwald for providing the lymphoblast cell line HSC536N from a type C Fanconi anemia patient, Dr A. D. Miller for the retroviral vector pLXSN, Dr David C. Hinkle for the pGST vector, Dr Brian Druker for anti–phosphotyrosine antibody 4G10 and for helpful discussions, and Tara Koretsky for valuable technical assistance. We thank Dr Alan D'Andrea for providing the M55FANCC cDNA and anti-FANCD2 antibody as well as for helpful discussions.
Supported by National Institutes of Health grant HL48546 and a Department of Veterans Affairs Merit Review Grant to G.C.B.
Q.P. and T.A.C. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Grover C. Bagby, MD, Oregon Cancer Center, Department of Medicine (Division of Hematology and Medical Oncology) and Department of Molecular and Medical Genetics, Oregon Health Sciences University, Portland, OR 97201; e-mail: grover@ohsu.edu.


![Fig. 11. M55 mutation restores STAT1 DNA-binding activity but fails to complement MMC hypersensitivity of FA-C cells. / (A) A schematic diagram of full-length FANCC and truncated mutant M55 that lacks amino-terminal 54 amino acids. The 3 motifs described in Figure 1 are indicated as black boxes. (B,C) EMSA analyses of STAT1 activation in M55-transduced FA-C lymphocytes. Normal FANCC- or M55-transduced HSC536N and PD4 cells were treated with IFN-γ (10 ng/mL) for 30 minutes and EMSA was performed using 5 μg of nuclear extracts and [32P]-labeled oligonucleotides carrying the GAS from the IRF1 gene (B) or from the p21 gene (C). Competitive binding was carried out by incubation with 25-fold excess molar unlabeled GAS (lanes 5 and 6) oligonuleotides. Lane 7 indicates that binding reactions were incubated with an STAT1 antibody (5 μg). Arrows denote STAT1-GAS complexes. (D) Effect of M55 mutation on MMC sensitivity in FA-C lymphoblast cell line HSC536N. Cells were treated with varying concentrations of MMC for 5 days before staining with trypan blue dye to quantify viable cells. Cell viability is expressed as percentages of viable cells relative to the untreated control culture. Each data point represents the mean ± SD of at least triplicate determinations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/5/10.1182_blood.v98.5.1392/5/m_h81711459011.jpeg?Expires=1765889094&Signature=Gpqr9zG5D09O4yXiVB8GrHcu4a8hz4NyNNNkIZdMeNXqUF9rQNNj9Xxaa6iIXAPpARxIAE838pWpiQjDQuqULZxPWyQe0xKsxcb3wUZrim4U4R-NE7EtayZb527v2lc~zPlFqkoJPxj-eLqgg1lVvxBUv1wcYsGo5Mi30FclpYGIHprXdVxk4yGS456MCssdtL0~Jcq~Xb8HZy2E-ic28s9AoMz4eU1dgz-CPMRebqTNwTxAYrpQunzJ4cB~orxv-D3fDzI~cYeA4rXMGE-ovAm2MpUB09ZcUwa4pIjjLTr0SKTqVonfptVo-q1~S1KhymZFNBLIKbjYPOiJqelbEw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Fig. 11. M55 mutation restores STAT1 DNA-binding activity but fails to complement MMC hypersensitivity of FA-C cells. / (A) A schematic diagram of full-length FANCC and truncated mutant M55 that lacks amino-terminal 54 amino acids. The 3 motifs described in Figure 1 are indicated as black boxes. (B,C) EMSA analyses of STAT1 activation in M55-transduced FA-C lymphocytes. Normal FANCC- or M55-transduced HSC536N and PD4 cells were treated with IFN-γ (10 ng/mL) for 30 minutes and EMSA was performed using 5 μg of nuclear extracts and [32P]-labeled oligonucleotides carrying the GAS from the IRF1 gene (B) or from the p21 gene (C). Competitive binding was carried out by incubation with 25-fold excess molar unlabeled GAS (lanes 5 and 6) oligonuleotides. Lane 7 indicates that binding reactions were incubated with an STAT1 antibody (5 μg). Arrows denote STAT1-GAS complexes. (D) Effect of M55 mutation on MMC sensitivity in FA-C lymphoblast cell line HSC536N. Cells were treated with varying concentrations of MMC for 5 days before staining with trypan blue dye to quantify viable cells. Cell viability is expressed as percentages of viable cells relative to the untreated control culture. Each data point represents the mean ± SD of at least triplicate determinations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/5/10.1182_blood.v98.5.1392/5/m_h81711459011.jpeg?Expires=1765969530&Signature=ZuTZmTvC1yPlhYTeYDJSO5adBVK~XuN6hw9XGmkgr0AbKG~rbiFwUzJy7p4rVr1HUFI6qE~xx0VUBP2BPTW5vKwSX~uKhhVjdHQQTYHvRJD75PQhEtz~nEBQs9AmyTtGLnfPJVKSJMEjOdif8c87-GDGQmkZlH~BEPMQeelFAqlJUw2ld-JnDUY1DnLebVTwsNiR~qtAWr32oHWI5sz2jI~MJUsydSFWRCrr5g4nIQiXNgqrZtHK8O3PqER9wFosRO-xya7ohwvL7kMAYMWtkzKIbbmSgP0XeVEg~OS6qXmnx7b8wJPFVb0Kngu-2WBP9i5csjDMFnXSmDXXAzuuvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)