Lipopolysaccharide (LPS) induces human monocytes to express many proinflammatory mediators, including the procoagulant molecule tissue factor (TF) and the cytokine tumor necrosis factor alpha (TNF-α). The TF and TNF-α genes are regulated by various transcription factors, including nuclear factor (NF)-κB/Rel proteins and Egr-1. In this study, the role of the MEK-ERK1/2 mitogen-activated protein kinase (MAPK) pathway in LPS induction of TF and TNF-α gene expression in human monocytic cells was investigated. The MAPK kinase (MEK)1 inhibitor PD98059 reduced LPS induction of TF and TNF-α expression in a dose-dependent manner. PD98059 did not affect LPS-induced nuclear translocation of NF-κB/Rel proteins and minimally affected LPS induction of κB-dependent transcription. In contrast, PD98059 and dominant-negative mutants of the Ras-Raf1-MEK-ERK (extacellular signal–regulated kinase) pathway strongly inhibited LPS induction of Egr-1 expression. In kinetic experiments LPS induction of Egr-1 expression preceded induction of TF expression. In addition, mutation of the Egr-1 sites in the TF and TNF-α promoters reduced expression of these proinflammatory genes. It was demonstrated that LPS induction of the Egr-1 promoter was mediated by 3 SRE sites, which bound an LPS-inducible complex containing serum response factor and Elk-1. LPS stimulation transiently induced phosphorylation of Elk-1 and increased the functional activity of a GAL4–Elk-1TA chimeric protein via the MEK-ERK1/2 pathway. The data indicate that LPS induction of Egr-1 gene expression is required for maximal induction of the TNF-α and TF genes in human monocytic cells.
Introduction
The major outer membrane component of gram-negative bacteria, lipopolysaccharide (LPS) or endotoxin, is a potent activator of monocyte or macrophage function leading to responses that are both protective and injurious to the host.1 LPS induces the expression of the procoagulant molecular tissue factor (TF) and inflammatory cytokines such as tumor necrosis factor alpha (TNF-α).2 LPS binds to LPS-binding protein (LBP) in serum, followed by the binding of the LBP/LPS complex to CD14, which activates signal transduction pathways and transcription factors and induces gene expression.1 However, the requirement for CD14 can be bypassed by treating cells with high doses of LPS,3 suggesting that CD14 serves to present the LBP/LPS complex to a less abundant receptor. Recently, it was shown that theLps gene responsible for LPS hyporesponsiveness in C3H/HeJ mice is a mutant form of the Toll-like receptor (Tlr)4 gene.4 These studies and others demonstrated that TLR4 is the long-sought LPS signaling receptor.5
LPS has been shown to activate members of the mitogen-activated protein kinase (MAPK) family, including extacellular signal–regulated kinases (ERK)1/2,6,7 c-Jun amino terminal kinases (JNKs),8 and p38.9 Inhibition of MAPK kinase (MEK) in monocytes by a specific inhibitor U0126 reduced LPS induction of several inflammatory cytokines, including interleukin-1, interleukin-8, and TNF-α as well as prostaglandin E2, indicating a role for the ERK1/2 pathway in LPS signaling independent of the JNK and p38 pathways.10
LPS stimulation of monocytes activates many transcription factors, including the nuclear factor NF-κB/Rel family, which induce genes encoding various inflammatory mediators.2 In unstimulated monocytes the NF-κB/Rel proteins are retained in the cytoplasm by their interaction with the inhibitors IκBs.11 LPS stimulation of monocytes leads to the phosphorylation of IκBs by IκB kinases (IKKs), leading to the rapid translocation of NF-κB/Rel proteins to the nucleus.12 We and others have demonstrated that LPS activation of IKKβ is required for κB-dependent transcription and TNF-α expression in human monocytes and THP-1 cells.13-15
The transcription factor Egr-1 is an 80-kd nuclear phosphoprotein containing 3 zinc-finger DNA-binding domains.16 It is rapidly activated in various cell types in response to a variety of stimuli.17-19 For instance, LPS stimulation of murine macrophages induced a rapid induction of Egr-1 transcription, messenger RNA (mRNA), and protein.20 The Egr-1 promoter contains several serum response elements (SREs) and Ets binding sites that mediate induction.21-23 Serum response factor (SRF) and ternary complex factors (TCFs) form a ternary complex at these SRE and Ets sites. The TCF proteins are members of the Ets family, which include Elk-1, Sap1a, and Fli1.23 These proteins are phosphorylated and activated by upstream MAPKs in a cell type– and stimulus-specific manner.24,25 Phosphorylation of Elk-1 in its C-terminal domain induces conformational change that promotes binding of Elk-1 to Ets sites via its N-terminal Ets domain and binding to SRF dimers via its B box domain.26 Growth hormone stimulation of cells leads to phosphorylation of Elk-1 and formation of an Elk-1/SRF complex that mediates induction of Egr-1 gene expression.19
We have shown that Egr-1 plays a role in LPS induction of TNF-α gene expression in monocytic cells.27 We and others have shown that induction of TF gene expression in various cell types in response to a variety of stimuli, including phorbol esters, serum, vascular endothelial cell growth factor, shear stress, oxidized low density lipoprotein, and hypoxia, is mediated by Egr-1.28-32 In addition, preliminary studies suggest that Egr-1 plays a role in the induction of TF gene expression in LPS-stimulated monocytic cells.33-35 However, there have been no definitive studies showing that Egr-1 positively regulates TF gene expression in monocytic cells.
In this study, we investigated the role of the MEK-ERK1/2 signaling pathway in LPS induction of TF and TNF-α expression in human monocytic cells. The MEK1/2 inhibitor PD98059 reduced LPS induction of TF and TNF-α by inhibiting Egr-1 expression. LPS induction of the Egr-1 promoter was mediated by 3 SRE sites, which bound SRF and TCF Elk-1. TCFs, such as Elk-1, Sap-1, and Fli-1, are targets of the MAPK pathway.36 37 Our data indicated that phosphorylation of Elk-1 by the MEK-ERK1/2 pathway induces Egr-1 expression in LPS-stimulated monocytes and subsequent TF gene expression.
Materials and methods
LPS (Escherichia coli serotype 0111:B4) and the MEK inhibitor PD98059 were obtained from Calbiochem (Carlsbad, CA), and PMA was obtained from Sigma (St Louis, MO).
Cell culture
Human monocyte leukemia THP-1 cells were obtained from American Type Culture Collection (Manassas, VA). THP-1 cells were cultured in RPMI 1640 (Gibco BRL Life Technologies, Gaithersburg, MD) with 8% fetal calf serum (Omega Scientific, Tarzana, CA), L-glutamine (Irvine Scientific, Santa Ana, CA), and 2-mercaptoethanol. Human peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood from healthy volunteers by buoyant density gradient centrifugation on low endotoxin Ficoll-Hypaque.38 Monocytes were isolated by adhering to tissue culture plates for 1 hour at 37°C. Nonadherent cells were removed by washing.
TNF-α ELISA
THP-1 cells or PBMCs (1 × 106) were preincubated with various doses of PD98059 (0-25 μM) for 30 minutes at 37°C before addition of LPS for 5 hours at 37°C. Culture supernatants were stored at −80°C before performing assays. TNF-α protein levels were measured using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Biosource International, Burlingame, CA).
TF activity
THP-1 or PBMC pellets (1 × 106) were solubilized at 37°C for 15 minutes using 15 mM octyl-D-glucopyranoside. TF activity in cell lysates was assayed using a one-stage clotting assay as described.39 Clotting times were converted to milliunits of TF activity by comparison with a standard curve established with purified human brain TF. For reference, a clotting time of 50 seconds corresponds to 1000 mU of TF activity.
Western blotting
Cytosolic and nuclear extracts were prepared from THP-1 cells or PBMCs (5 × 106) as described.40 For analysis of TF protein expression, cells were lysed and lysates extracted with 1% Triton X-100 and proteins precipitated with acetone. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Hybond-enhanced chemiluminescence membrane (Amersham Pharmacia Biotech, Alameda, CA). TF was visualized using a 1:1000 dilution of an anti-TF antibody (No. 4501) (American Diagnostica, Greenwich, CT). Egr-1 was visualized using a 1:1000 dilution of an anti–Egr-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Phosphorylated ERK1/2 and total ERK were visualized using a 1:1000 dilution of antibodies against either phosphorylated ERK1/2 or nonphosphorylated ERK1/2, respectively (New England Biolabs, MA). Phosphorylated Elk-1 and nonphosphorylated Elk-1 were visualized using a 1:1000 dilution of antibodies against either phosphorylated Elk-1 (Santa Cruz Biotechnology) or nonphosphorylated Elk-1 (Cell Signaling Technology, Beverly, MA).
Northern blotting
Total cellular RNA was isolated from THP-1 cells (1 × 107) stimulated with LPS (10 μg/mL) using Trizol Reagent (Gibco). In some experiments, cells were preincubated with PD98059 (50 μM) for 30 minutes at 37°C prior to addition of LPS. RNA (10 μg) was analyzed by Northern blotting as described.41 A 641 human TF complementary DNA (cDNA) fragment, an 800 base pair (bp) human TNF-α cDNA fragment, or a 2500 bp human Egr-1 cDNA fragment29 was used as probes. The cDNA fragments were labeled with [α32P], deoxycytidine triphosphate (ICN, Costa Mesa, CA) using a Prime-It Kit (Stratagene Cloning Systems, San Diego, CA). Blots were rehybridized with the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (G3PDH; Clontech Laboratories, Palo Alto, CA). Bands were visualized by autoradiography.
Electrophoretic mobility shift assays
Nuclear extracts were prepared from THP-1 cells (5 × 106) as described previously.40Nuclear extracts were incubated with radiolabeled double-stranded oligonucleotide probes (Operon Technologies, Alameda, CA) containing an Egr-1 binding site (underlined), 5′-CCCGGCGCGGGGGCGATTTCGAGTCA-3′; a Sp1 site (underlined), 5′-ATTCGATCGGGGCGGGGCGAGC-3′; the murine immunoglobulin IgκB site (underlined), 5′-CAGAGGGGACTTTCCGAGA-3′; the TFκB site (underlined), 5′-GTCCCGGAGTTTCCTACCGGG-3′; an AP-1 site (underlined), 5′-CTGGGGTGAGTCATCCCTT-3′; the Egr-1 SRE3 site (underlined), 5′-AGCACCTTATTTGGAGTGGCCGGATATGGCCCGGCGCTTCC-3′; the Egr-1 SRE4 site (underlined), 5′-AGGATCCCCCGCCGGAACAACCCTTATTTGGGCAG-3′; the Egr-1 SRE5 site (underlined), 5′-TGCGACCCGGAAATGCCATATAAGGAGCAGGAAGGATCCCCT-3′; or the c-fos SRE site (underlined), 5′-GATCCAGGATGTCCATATTAGGACATCTA-3′. Protein-DNA complexes were separated from free DNA probe by electrophoresis through 6% nondenaturing acrylamide gels (Invitrogen, Carlsbad, CA) in 0.5 × Tris borate ethylenediaminetetraacetic acid buffer.40Supershift experiments employed 2 μL each of anti–Egr-1, anti-SRF, anti–Elk-1, and anti-p65 antibodies (Santa Cruz Biotechnology). Recombinant Egr-1 was provided by Dr E. Adamson (Burnham Institute, La Jolla, CA). Gels were dried, and protein complexes were visualized by autoradiography.
Plasmids
The p(κB)4-LUC contains 4 copies of an NF-κB site, and p(AP-1)4-LUC contains 4 copies of an AP-1 site. These sites were cloned upstream of the minimal simian virus 40 (SV40) promoter expressing the firefly luciferase (LUC) reporter gene in pGL2-Promoter (Promega, Madison, WI).40,42 The pTF-LUC contains 2106 bp of the human TF promoter.33 The wild-type human TF promoter (−278 to +14) and a version containing mutations in each of the 3 Egr-1 sites were cloned into pGL2-Basic (Promega, Madison, WI). The mutations of the Egr-1 sites have been described.29 The wild-type pTNF-α–LUC and the Egr-1 mutant contain 615 bp of the human TNF-α promoter.27 The pEgr-1–LUC contains 1200 bp of the murine Egr-1 promoter. The 5′ deletion series of the Egr-1 promoter has been described previously.21 The Egr-1 promoter mutants SRE3 and SRE4 have been described.19 The pEgr-1(SRE5M)LUC was provided by W. Aird (Harvard University, Boston, MA). The pcDNA3 (Invitrogen, San Diego, CA) was used as a control plasmid for transfections. Plasmids expressing dominant-negative versions of Ras, Raf1, MEK1, ERK1, and ERK2 have been described.43 In addition, we used plasmid expressing dominant-negative MEK kinase-1 (MEKK1). The control plasmid pFA-CMV expresses the GAL4 DNA-binding domain alone and was obtained from Stratagene Cloning Systems. The pFA2–Elk-1 (pGAL4–Elk-1TA) expresses the GAL4 DNA-binding domain fused with the transactivation domain of Elk-1 (Stratagene Cloning Systems). The reporter plasmid pFR-LUC (pGAL4-LUC) contains 5 copies of the GAL4 binding site upstream of a minimal promoter that drives expression of the firefly luciferase reporter gene.
Transfections
THP-1 cells were transfected using DEAE-dextran.33After transfection, cells were incubated in complete media for 46 hours at 37°C before stimulating with LPS (10 μg/mL) for 5 hours at 37°C. In some experiments, cells were preincubated with PD98059 (25 or 50 μM) for 30 minutes at 37°C before the addition of LPS. Cell lysates were assayed for luciferase activity as described in the manufacturer's protocol (Promega) using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA). In selected experiments, cells were cotransfected with pRLTK, which expressesRenilla luciferase (Promega). Renilla luciferase was measured according to the manufacturer's protocol (Promega) and used to normalize the activity of the firefly luciferase. Variability of the transfection was found to be less than 10%.
Data analysis
The number of experiments analyzed is indicated in each figure. Band intensities were quantified by densitometric analyses using a Personal Densitometer (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software. Statistical analysis was performed using the unpaired Student t test, and differences were determined to be statistically significant at P < .05.
Results
LPS induction of TF expression requires the MEK-ERK1/2 kinase pathway
LPS induces TF and TNF-α expression in human monocytes and THP-1 monocytic cells.27,33,44,45 A previous study showed that activation of the MEK-ERK1/2 pathway was required for LPS induction of TNF-α in human monocytes.6 Therefore, we used TNF-α as a positive control in this study. To determine if the MEK-ERK1/2 pathway is required for LPS induction of TF expression in monocytes, we measured LPS induction of TF expression in the presence and absence of the MEK1/2 inhibitor PD98059.46 LPS induction of TF and TNF-α in PBMCs was inhibited in a dose-dependent manner by PD98059 (Figure 1A,B). Similar results were observed using THP-1 cells (Figures 2A and 3A). Therefore, we used THP-1 cells as a model of human monocytes to determine how LPS activation of the MEK-ERK1/2 pathway regulates TF and TNF-α expression. In THP-1 cells, PD98059 inhibited LPS-induced TF protein expression (Figure 2B) and TF mRNA expression at both 2 and 4 hours (Figure 2C). The larger TF transcript represents a partially spliced form that is observed in monocytic cells.47 48 LPS-induced TNF-α mRNA expression was also inhibited by PD98059 at 2 and 4 hours (Figure3B). Finally, we examined the effect of PD98059 on LPS-induced TF and TNF-α promoter activity. PD98059 (25 μM) inhibited LPS-induced TF and TNF-α promoter activities by 70% (Figure 2D) and 47% (Figure 3C), respectively. These results indicate that the MEK-ERK1/2 MAPK pathway plays a major role in the LPS induction of the TF and TNF-α genes in monocytic cells.
LPS induction of TF and TNF-α expression in PBMCs is inhibited by the MEK inhibitor PD98059.
PBMCs were preincubated by various doses of PD98059 (5, 10, 20 μM) for 30 minutes prior to stimulation with LPS (100 ng/mL) for 5 hours. (A) TF activity was determined in triplicate using a one-step clotting assay. (B) TNF-α antigen in cell supernatants was determined in duplicate by ELISA. These data represents the mean ± SD from 3 independent experiments.
LPS induction of TF and TNF-α expression in PBMCs is inhibited by the MEK inhibitor PD98059.
PBMCs were preincubated by various doses of PD98059 (5, 10, 20 μM) for 30 minutes prior to stimulation with LPS (100 ng/mL) for 5 hours. (A) TF activity was determined in triplicate using a one-step clotting assay. (B) TNF-α antigen in cell supernatants was determined in duplicate by ELISA. These data represents the mean ± SD from 3 independent experiments.
LPS induction of TF expression in THP-1 cells is inhibited by PD98059.
(A) THP-1 cells were preincubated with various doses of PD98059 (0.1, 0.5, 5, 10, 20 μM) prior to stimulation with LPS (10 μg/mL) for 5 hours. TF activity was determined in triplicate using a one-step clotting assay. Data (mean ± SD) from 3 independent experiments are shown. (B) TF antigen was detected by Western blotting in LPS-stimulated (5 hours) THP-1 cells with or without preincubation with PD98059 (25 μM). (C) Total RNA was extracted from THP-1 cells stimulated with LPS for 2 or 4 hours at 37°C after preincubation for 30 minutes with or without PD98059 (50 μM). TF mRNA levels were determined by Northern blotting. The membrane was reprobed with a G3PDH probe to assess RNA loading. (D) The pTF-LUC (3 μg) was transiently transfected into THP-1 cells. After 46 hours, transfected cells were preincubated in the presence or absence of PD98059 (25 μM) for 30 minutes prior to incubation with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are from 3 independent experiments.
LPS induction of TF expression in THP-1 cells is inhibited by PD98059.
(A) THP-1 cells were preincubated with various doses of PD98059 (0.1, 0.5, 5, 10, 20 μM) prior to stimulation with LPS (10 μg/mL) for 5 hours. TF activity was determined in triplicate using a one-step clotting assay. Data (mean ± SD) from 3 independent experiments are shown. (B) TF antigen was detected by Western blotting in LPS-stimulated (5 hours) THP-1 cells with or without preincubation with PD98059 (25 μM). (C) Total RNA was extracted from THP-1 cells stimulated with LPS for 2 or 4 hours at 37°C after preincubation for 30 minutes with or without PD98059 (50 μM). TF mRNA levels were determined by Northern blotting. The membrane was reprobed with a G3PDH probe to assess RNA loading. (D) The pTF-LUC (3 μg) was transiently transfected into THP-1 cells. After 46 hours, transfected cells were preincubated in the presence or absence of PD98059 (25 μM) for 30 minutes prior to incubation with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are from 3 independent experiments.
LPS induction of TNF-α expression in THP-1 cells is inhibited by PD98059.
(A) THP-1 cells were preincubated with various doses of PD98059 (0.1, 0.5, 5, 10, 20 μM) prior to stimulation with LPS (10 μg/mL) for 5 hours. TNF-α antigen in cell supernatants was determined in duplicate by ELISA. Data (mean ± SD) from 5 experiments are shown. (B) Total RNA was extracted from THP-1 cells stimulated with LPS for 2 or 4 hours at 37°C after preincubation for 30 minutes with or without PD98059 (50 μM). TNF-α mRNA levels were determined by Northern blotting. The membrane was reprobed with a G3PDH probe to assess RNA loading. (C) The pTNF-LUC (3 μg) was transiently transfected into THP-1 cells. After 46 hours, transfected cells were preincubated in the presence or absence of PD98059 (25 μM) for 30 minutes prior to incubation with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are from 3 independent experiments.
LPS induction of TNF-α expression in THP-1 cells is inhibited by PD98059.
(A) THP-1 cells were preincubated with various doses of PD98059 (0.1, 0.5, 5, 10, 20 μM) prior to stimulation with LPS (10 μg/mL) for 5 hours. TNF-α antigen in cell supernatants was determined in duplicate by ELISA. Data (mean ± SD) from 5 experiments are shown. (B) Total RNA was extracted from THP-1 cells stimulated with LPS for 2 or 4 hours at 37°C after preincubation for 30 minutes with or without PD98059 (50 μM). TNF-α mRNA levels were determined by Northern blotting. The membrane was reprobed with a G3PDH probe to assess RNA loading. (C) The pTNF-LUC (3 μg) was transiently transfected into THP-1 cells. After 46 hours, transfected cells were preincubated in the presence or absence of PD98059 (25 μM) for 30 minutes prior to incubation with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are from 3 independent experiments.
Does inhibition of the MEK-ERK1/2 pathway reduce nuclear activity of NF-κB and AP-1?
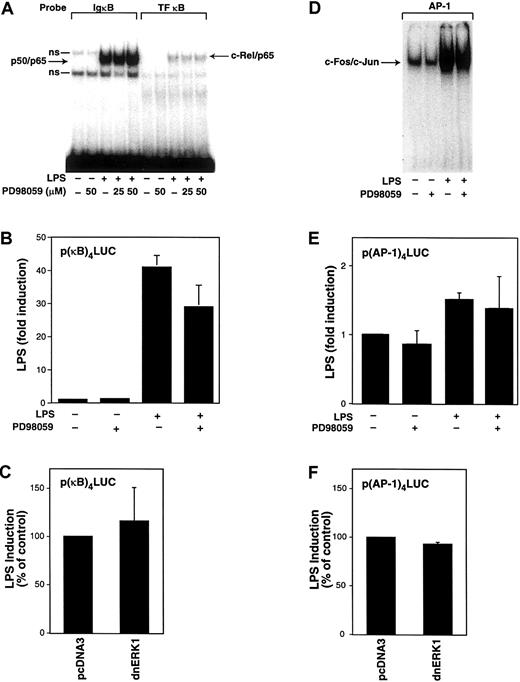
We and others have shown that NF-κB/Rel and AP-1 proteins play a major role in the LPS induction of TF and TNF-α gene expression in human monocytic cells.27,33,40,49 Therefore, we determined the effect of PD98059 on LPS-stimulated nuclear binding of NF-κB and AP-1 as well as LPS-induced κB- and AP-1–dependent transcription in THP-1 cells. Nuclear translocation was examined by electrophoretic mobility shift assays (EMSAs) using oligonucleotides that contain a prototypic κB site (IgκB), a κB site from the human TF promoter (TF-κB) that binds c-Rel/p65, or an AP-1 site that binds c-Fos/c-Jun.34 No nuclear NF-κ/Rel complexes were detected in unstimulated cells (Figure4A). LPS induced nuclear translocation, and binding of both p50/p65 and c-Rel/p65 was not affected by PD98059 (Figure 4A). Next, we determined the effect of PD98059 on LPS-induced κB-dependent transcription. THP-1 cells were transiently transfected with a plasmid containing a κB-dependent luciferase reporter gene. PD98059 weakly inhibited (29%) LPS-induced, κB-dependent transcription using a reporter plasmid containing a multimerized κB site (Figure 4B). Inhibition of κB-dependent transcription by PD98059 may in part contribute to the reduction of LPS-induced TF and TNF-α expression by PD98059. However, the magnitude of this inhibition (29%) did not appear to account for the stronger inhibition (48%-70%) of PD98059 on LPS-induced TF and TNF-α promoter expression. Cotransfection of the κB-dependent reporter plasmid with a plasmid expressing a dominant-negative ERK1 mutant did not affect LPS-induced, κB-dependent transcription (Figure 4C). Similarly, PD98059 did not affect the LPS-induced increase in AP-1 binding or AP-1–dependent transcription (Figure 4D,E). In addition, expression of a dominant-negative ERK1 mutant did not affect LPS-induced, AP-1–dependent transcription (Figure 4F). These studies indicate that inhibition of the MEK-ERK1/2 pathway does not reduce LPS induction of TF expression by affecting either LPS-induced NF-κB or AP-1 activity.
Effect of PD98059 on LPS-induced nuclear binding of NF-κB and AP-1 and κB- and AP-1–dependent transcription in THP-1 cells.
(A) Nuclear extracts were prepared from THP-1 cells preincubated with or without PD98059 (50 μM) for 30 minutes before stimulation with LPS (10 μg/mL) for 2 hours. EMSAs were used to measure binding of p50/p65 and c-Rel/p65 to oligonucleotides containing κB sites from the murine IgκB and human TF genes. Two nonspecific (ns) complexes were observed binding to the IgκB oligonucleotide. (B) The p(κB)4-LUC (3 μg) was transfected into THP-1 cells. After transfection, cells were preincubated in the presence or absence of PD98059 (50 μM) for 30 minutes before incubation with or without LPS for 5 hours at 37°C. (C) The p(κB)4-LUC was cotransfected with either vector control (pcDNA3) or a plasmid expressing a dominant-negative ERK1 mutant. (D) EMSAs were used to measure c-Fos/c-Jun binding to an AP-1 site using nuclear extracts from cells stimulated with LPS (1 hour) with or without PD98059. (E) LPS-induced, AP-1–dependent transcription was measured in cells transfected with p(AP-1)4-LUC in the presence and absence of PD98059. (F) LPS-induced, AP-1–dependent transcription was measured in the presence and absence of a plasmid expressing a dominant-negative ERK1 mutant. Transfection data are shown from 6 independent transfection experiments and are expressed as mean ± SD.
Effect of PD98059 on LPS-induced nuclear binding of NF-κB and AP-1 and κB- and AP-1–dependent transcription in THP-1 cells.
(A) Nuclear extracts were prepared from THP-1 cells preincubated with or without PD98059 (50 μM) for 30 minutes before stimulation with LPS (10 μg/mL) for 2 hours. EMSAs were used to measure binding of p50/p65 and c-Rel/p65 to oligonucleotides containing κB sites from the murine IgκB and human TF genes. Two nonspecific (ns) complexes were observed binding to the IgκB oligonucleotide. (B) The p(κB)4-LUC (3 μg) was transfected into THP-1 cells. After transfection, cells were preincubated in the presence or absence of PD98059 (50 μM) for 30 minutes before incubation with or without LPS for 5 hours at 37°C. (C) The p(κB)4-LUC was cotransfected with either vector control (pcDNA3) or a plasmid expressing a dominant-negative ERK1 mutant. (D) EMSAs were used to measure c-Fos/c-Jun binding to an AP-1 site using nuclear extracts from cells stimulated with LPS (1 hour) with or without PD98059. (E) LPS-induced, AP-1–dependent transcription was measured in cells transfected with p(AP-1)4-LUC in the presence and absence of PD98059. (F) LPS-induced, AP-1–dependent transcription was measured in the presence and absence of a plasmid expressing a dominant-negative ERK1 mutant. Transfection data are shown from 6 independent transfection experiments and are expressed as mean ± SD.
Egr-1 is required for maximal induction of TF and TNF-α expression in LPS-stimulated monocytic cells
Previous work in our laboratory and others suggests that the transcription factor Egr-1 is required for maximal induction of TF expression in LPS-stimulated human monocytic cells.33-35For instance, LPS induced the TF proximal promoter (−111) that contained 3 Egr-1 sites but lacked AP-1 and κB sites.33In addition, DNase I footprinting mapped LPS-inducible footprints to an Egr-1 site in the TF promoter.35 However, the role of Egr-1 in TF gene expression in monocytic cells has not been examined directly.
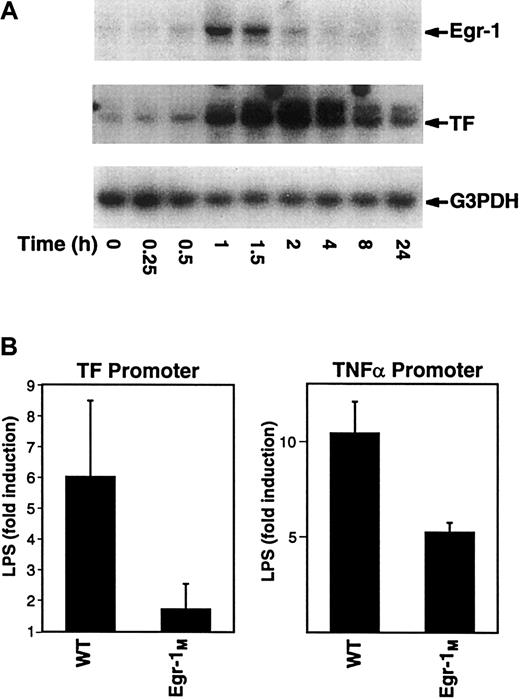
In this study, we analyzed the kinetics of induction of Egr-1 and TF mRNA expression in LPS-stimulated THP-1. Maximal levels of Egr-1 mRNA were observed at 1 hour and preceded the maximal induction of TF mRNA at 2 hours (Figure 5A). Similarly, LPS induction of Egr-1 mRNA in mouse peritoneal macrophages preceded induction of TF mRNA (data not shown). Next, we analyzed the effect of mutation of Egr-1 sites in the TF and TNF-α promoters on inducibility by LPS. Mutation of the Egr-1 sites in the TF and TNF-α promoter reduced LPS induction by 72% and 50%, respectively (Figure5B). Taken together, these results indicate that Egr-1 contributes to LPS induction of TF and TNF-α gene expression in monocytic cells.
Regulation of TF and TNF-α by Egr-1.
(A) Total RNA was isolated from THP-1 cells stimulated with LPS (0-24 hours). Levels of Egr-1, TF, and G3PDH mRNA were determined by Northern blotting. (B) THP-1 cells were transfected with plasmids containing either the wild-type TF promoter (wt) or an Egr-1 mutant (Egr-1M) as well as the wild-type TNF-α promoter (wt) or an Egr-1M. Transfected cells were incubated with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and the results were expressed as fold induction. Data (mean ± SD) are from at least 3 independent experiments.
Regulation of TF and TNF-α by Egr-1.
(A) Total RNA was isolated from THP-1 cells stimulated with LPS (0-24 hours). Levels of Egr-1, TF, and G3PDH mRNA were determined by Northern blotting. (B) THP-1 cells were transfected with plasmids containing either the wild-type TF promoter (wt) or an Egr-1 mutant (Egr-1M) as well as the wild-type TNF-α promoter (wt) or an Egr-1M. Transfected cells were incubated with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and the results were expressed as fold induction. Data (mean ± SD) are from at least 3 independent experiments.
LPS induces Egr-1 expression in human monocytes and THP-1 monocytic cells
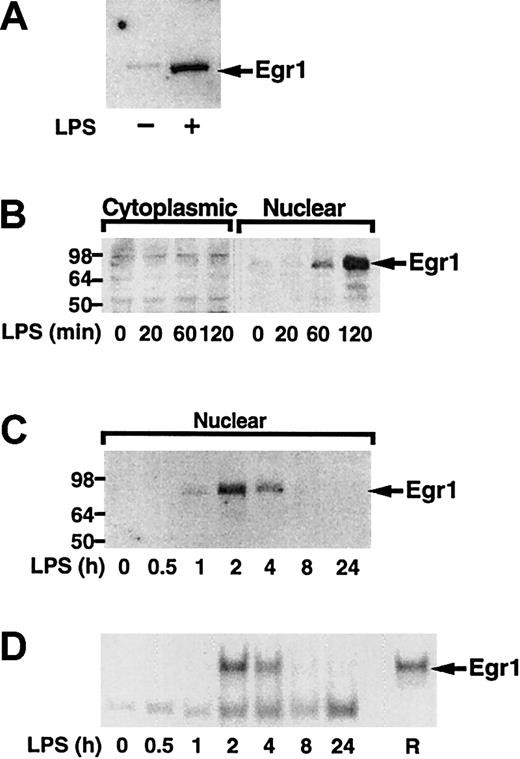
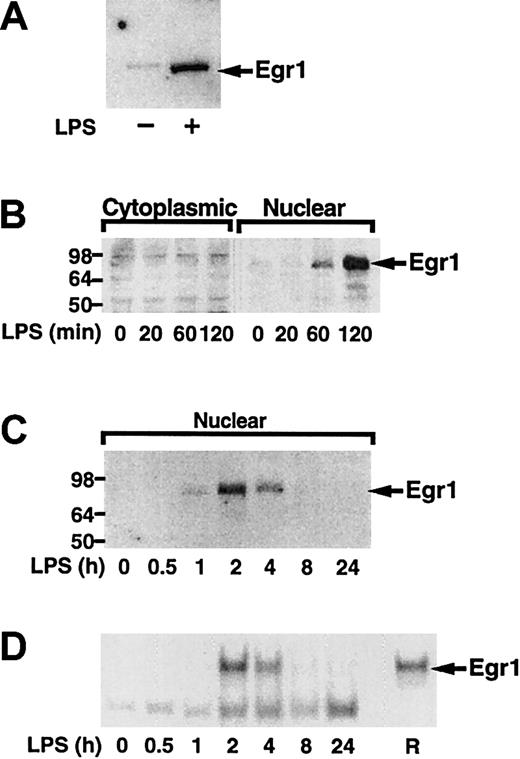
Next, we analyzed if LPS induced Egr-1 expression in human monocytes and THP-1 cells. We found that LPS induced Egr-1 protein in both monocytes and THP-1 cells (Figure6A,B). Time course experiments using THP-1 cells demonstrated that LPS transiently induced Egr-1 expression with maximal levels in the nucleus at 2 hours (Figure 6C). The newly synthesized Egr-1 protein was functional and bound to an oligonucleotide containing a prototypic Egr-1 sequence (Figure 6D). The Egr-1 complex was competed with an oligonucleotide containing an Egr-1 site and was supershifted with an anti–Egr-1 antibody (data not shown). Sp1 binding was not affected by stimulation with LPS (data not shown). These results confirm that LPS induced Egr-1 expression in both adherent monocytes and THP-1 cells.
LPS induction of Egr-1 expression in human monocytes and THP-1 cells.
(A) Nuclear extracts were prepared from adherent human monocytes with or without LPS (100 ng/mL) stimulation for 2 hours. Proteins were separated by SDS-PAGE and Egr-1 detected by Western blotting using anti–Egr-1 antibody. (B) THP-1 cells were stimulated with LPS (10 μg/mL) for various times (0-2 hours), and cytoplasmic and nuclear extracts were prepared. Egr-1 was detected by Western blotting. (C) THP-1 cells were stimulated with LPS for various times (0-24 hours). Egr-1 was detected in nuclear extracts by Western blotting. (D) EMSAs were performed using nuclear extracts and a radiolabeled oligonucleotide containing an Egr-1 site. Recombinant human Egr-1 (R) was used as a control.
LPS induction of Egr-1 expression in human monocytes and THP-1 cells.
(A) Nuclear extracts were prepared from adherent human monocytes with or without LPS (100 ng/mL) stimulation for 2 hours. Proteins were separated by SDS-PAGE and Egr-1 detected by Western blotting using anti–Egr-1 antibody. (B) THP-1 cells were stimulated with LPS (10 μg/mL) for various times (0-2 hours), and cytoplasmic and nuclear extracts were prepared. Egr-1 was detected by Western blotting. (C) THP-1 cells were stimulated with LPS for various times (0-24 hours). Egr-1 was detected in nuclear extracts by Western blotting. (D) EMSAs were performed using nuclear extracts and a radiolabeled oligonucleotide containing an Egr-1 site. Recombinant human Egr-1 (R) was used as a control.
LPS induction of Egr-1 expression is mediated by the Ras-Raf-MEK-ERK1/2 pathway
To determine if the MEK-ERK1/2 pathway was required for LPS induction of Egr-1, we examined the effect of the MEK-specific inhibitor PD98059 on LPS-induced Egr-1 expression in THP-1 cells. PD98059 inhibited LPS induction of Egr-1 protein and mRNA (Figure7A,B), and LPS induction of the Egr-1 promoter was also strongly inhibited by PD98059 (Figure 7C). In contrast, another MAPK inhibitor, SB203850, which specifically inhibits p38, had no inhibitory effect on LPS-mediated Egr-1 induction (data not shown). These results indicate that the MEK-ERK1/2 pathway mediates LPS induction of the Egr-1. To complement the experiments using a pharmacologic inhibitor, we determined the effect of dominant-negative mutants of Ras, Raf-1, ERK1, and ERK2 on LPS-induced Egr-1 promoter activity in THP-1 cells. Cotransfection with plasmids expressing dominant-negative versions of Ras, Raf-1, ERK1, or ERK2 reduced LPS induction of Egr-1 promoter activity more than 75% (Figure 7D). In contrast, expression of dominant-negative MEKK-1, an upstream activator of JNK and p38 MAPK, had no significant effect on LPS-induced Egr-1 promoter activity. These results demonstrate the specificity of the Ras-Raf-MEK-ERK1/2 pathway in mediating LPS induction of Egr-1 gene expression in monocytic cells.
LPS induction of Egr-1 expression is mediated by the Ras-Raf-MEK-ERK pathway.
(A) THP-1 cells were preincubated with PD98059 (25 μM) for 30 minutes prior to stimulation with LPS for 2 hours. Nuclear extracts were analyzed by Western blotting using an anti–Egr-1 antibody. (B) Total RNA was extracted from THP-1 cells that were unstimulated or stimulated with LPS (10 μg/mL) for 1 hour at 37°C with or without PD98059 (25 μM). Egr-1 mRNA levels were determined by Northern blotting using a radiolabeled human Egr-1 cDNA probe. The membrane was reprobed with a G3PDH probe as a measure of loading. (C) THP-1 cells were transiently transfected with pEgr-1–LUC (3 μg). After transfection, cells were preincubated in the presence or absence of PD98059 (25 μM) for 30 minutes before incubation with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as fold induction (n = 3). (D) THP-1 cells were cotransfected with pEgr-1–LUC (1.5 μg) and either pcDNA3 (5.5 μg) or plasmids expressing a dominant-negative version of Ras, Raf-1, ERK1, ERK2, or MEKK-1. Transfected cells were treated with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as percentage of control induction (n = 3).
LPS induction of Egr-1 expression is mediated by the Ras-Raf-MEK-ERK pathway.
(A) THP-1 cells were preincubated with PD98059 (25 μM) for 30 minutes prior to stimulation with LPS for 2 hours. Nuclear extracts were analyzed by Western blotting using an anti–Egr-1 antibody. (B) Total RNA was extracted from THP-1 cells that were unstimulated or stimulated with LPS (10 μg/mL) for 1 hour at 37°C with or without PD98059 (25 μM). Egr-1 mRNA levels were determined by Northern blotting using a radiolabeled human Egr-1 cDNA probe. The membrane was reprobed with a G3PDH probe as a measure of loading. (C) THP-1 cells were transiently transfected with pEgr-1–LUC (3 μg). After transfection, cells were preincubated in the presence or absence of PD98059 (25 μM) for 30 minutes before incubation with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as fold induction (n = 3). (D) THP-1 cells were cotransfected with pEgr-1–LUC (1.5 μg) and either pcDNA3 (5.5 μg) or plasmids expressing a dominant-negative version of Ras, Raf-1, ERK1, ERK2, or MEKK-1. Transfected cells were treated with or without LPS for 5 hours at 37°C. Luciferase activity in cell lysates was determined, and results were expressed as percentage of control induction (n = 3).
Localization of the LPS response element in the Egr-1 promoter
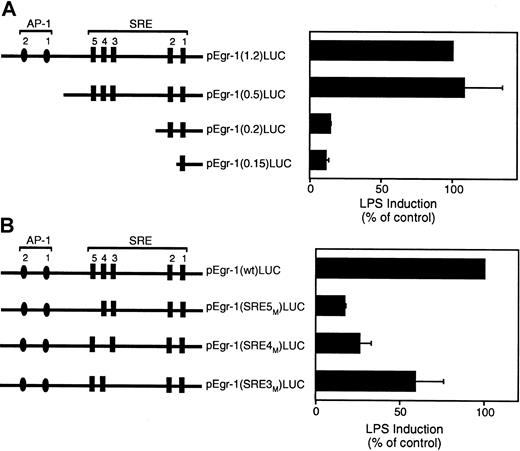
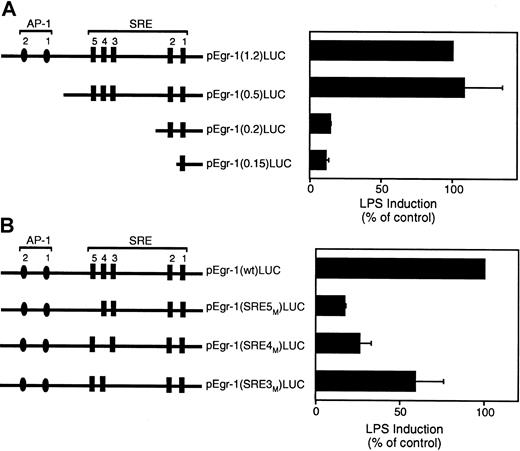
The Egr-1 promoter contains 2 distal AP-1 binding sites and 5 SREs (Figure 8A). We used a series of luciferase reporter plasmids containing 5′ deletions of the Egr-1 promoter to identify the cis-acting LPS response element in the Egr-1 promoter.21 Deletion of a region containing 2 distal AP-1 sites had no effect on LPS induction of the Egr-1 promoter (Figure 8A). In contrast, deletion of a region containing the distal SRE sites 3 to 5 dramatically decreased Egr-1 promoter activity (Figure8A). Deletion of a region containing the 2 proximal SRE sites showed no further decrease in promoter activity. These results suggest that SRE sites 3 to 5 mediate LPS induction of the Egr-1 promoter. We therefore examined the effect of individual mutations of the SRE3, SRE4, or SRE5 sites in the Egr-1 promoter on LPS-inducible promoter activity. Mutation of SRE5 showed the strongest inhibition although each of the 3 SRE mutations reduced LPS induction of the Egr-1 promoter (Figure 8B). These results indicate that SRE sites 3 to 5 mediate LPS induction of the Egr-1 promoter in THP-1 cells.
LPS induction of the Egr-1 promoter is mediated by SRE3, SRE4, and SRE5.
(A) LPS induction of a 5′ deletion series of the Egr-1 promoter. (B) LPS induction of the wild-type Egr-1 promoter and mutants of SRE3, SRE4, and SRE5. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are shown for 3 independent experiments.
LPS induction of the Egr-1 promoter is mediated by SRE3, SRE4, and SRE5.
(A) LPS induction of a 5′ deletion series of the Egr-1 promoter. (B) LPS induction of the wild-type Egr-1 promoter and mutants of SRE3, SRE4, and SRE5. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are shown for 3 independent experiments.
LPS induces formation of an Elk-1/SRF complex at SRE sites
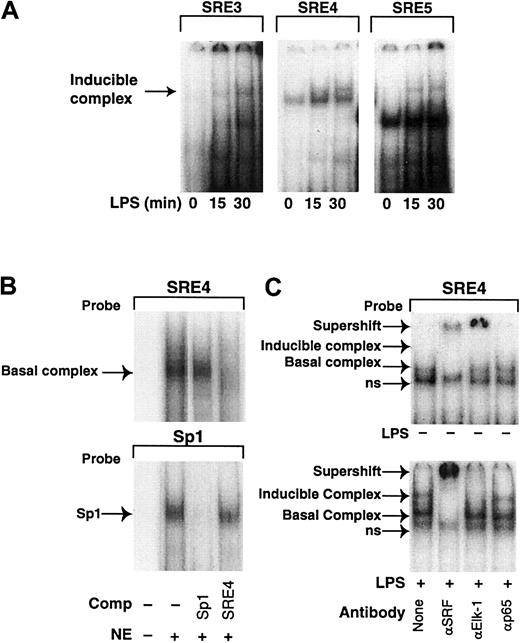
SREs are known to bind multicomponent complexes, which contain SRF and TCFs, such as Elk-1, Sap-1, and Fli-1.19,50Phosphorylation of SRF by various signaling pathways has been shown to increase SRF binding to the SRE.51 TCFs are also targets of MAPK pathways, and phosphorylation-dependent activation of different TCFs has been shown to contribute to Egr-1 gene regulation in cell-type and stimulus-specific manner.52 53 To analyze transcription factors that bind to the SRE sites 3 to 5 in the Egr-1 promoter in monocytic cells, we performed EMSAs with oligonucleotides spanning SRE3, SRE4, and SRE5. An LPS-inducible protein-DNA complex with the same mobility was observed using oligonucleotides spanning each of the SRE sites 3 to 5 (Figure 9A). Additional constitutive protein-DNA complexes were observed with the SRE4 and SRE5.
LPS increases binding of nuclear proteins to SRE3, SRE4, and SRE5 in the Egr-1 promoter.
(A) Nuclear extracts were prepared from THP-1 cells stimulated with LPS for 0 to 30 minutes. An inducible complex was observed in EMSAs using radiolabeled oligonucleotides spanning SRE3, SRE4, and SRE5. (B) Nuclear extracts from unstimulated THP-1 cells were incubated with radiolabeled oligonucleotides containing SRE4 or an Sp1 site. A 50 × excess of unlabeled oligonucleotide was used as a competitor. (C) Nuclear extracts were prepared from THP-1 cells with or without LPS (10 μg/mL) for 30 minutes. Nuclear extracts from unstimulated (upper panel) or LPS-stimulated (lower panel) cells were incubated with SRE4. Supershift experiments were performed with antibodies against SRF, Elk-1, and p65.
LPS increases binding of nuclear proteins to SRE3, SRE4, and SRE5 in the Egr-1 promoter.
(A) Nuclear extracts were prepared from THP-1 cells stimulated with LPS for 0 to 30 minutes. An inducible complex was observed in EMSAs using radiolabeled oligonucleotides spanning SRE3, SRE4, and SRE5. (B) Nuclear extracts from unstimulated THP-1 cells were incubated with radiolabeled oligonucleotides containing SRE4 or an Sp1 site. A 50 × excess of unlabeled oligonucleotide was used as a competitor. (C) Nuclear extracts were prepared from THP-1 cells with or without LPS (10 μg/mL) for 30 minutes. Nuclear extracts from unstimulated (upper panel) or LPS-stimulated (lower panel) cells were incubated with SRE4. Supershift experiments were performed with antibodies against SRF, Elk-1, and p65.
We performed a detailed characterization of the protein-DNA complexes formed using SRE4. The SRE 4 site bound a protein-DNA complex present in unstimulated THP-1 cells that was competed by an oligonucleotide containing SRE4 but not by an Sp1 site (Figure 9B, upper panel). A similar complex was observed using an oligonucleotide containing the c-fos SRE (data not shown). Sp1 was used as a control and bound to an oligonucleotide containing an Sp1 site. The Sp1 complex was competed with an oligonucleotide containing an Sp1 site but not by an oligonucleotide containing SRE4 (Figure 9B, lower panel).
The protein-DNA complexes that bound to SRE4 in unstimulated and LPS-stimulated THP-1 cells could be further separated by performing the electrophoresis at 4°C. The basal complex observed in unstimulated cells could be separated into 2 distinct complexes (Figure 9C, upper panel). The slower migrating basal complex was supershifted with anti-SRF antibody but not by an anti–Elk-1 or an anti-p65 antibody, indicating that it contained SRF but not Elk-1. A faster-migrating nonspecific complex was not recognized by any of the antibodies. Three distinct complexes were observed using nuclear extracts from LPS-stimulated cells that included an inducible complex migrating more slowly than the basal complex. The inducible complex was supershifted with an anti-SRF antibody and an anti–Elk-1 antibody but not by an anti-p65 antibody, indicating that it was a complex composed of SRF and Elk-1 (Figure 9C, lower panel). The LPS-inducible complex observed with SRE5 was also supershifted with antibodies to SRF and Elk-1 (data not shown). LPS also induces activation of JNK1 and phosphorylation of Sap-1.37 However, the LPS-inducible complex was not supershifted with an anti–Sap-1 antibody (data not shown). These results indicate that LPS stimulation of THP-1 cells specifically induced the binding of an Elk-1/SRF complex to the Egr-1 promoter.
LPS transiently induces phosphorylation of ERK1/2 and Elk-1
Several studies have shown that LPS rapidly induces ERK1/2 phosphorylation in monocytes and macrophages.6,7,54,55 We examined the time course of ERK1/2 phosphorylation in LPS-stimulated THP-1 monocytic cells. LPS stimulation transiently increased ERK1/2 phosphorylation with a peak at 15 minutes and a return to baseline at 25 minutes (Figure 10A). An antibody against total ERK1/2 was used to demonstrate equal loading of the samples. ERK1/2 has been shown to phosphorylate the transcription factor Elk-1 in fibroblast and HeLa cells.24 25 We therefore determined if LPS stimulation of THP-1 monocytic cells induced phosphorylation of Elk-1. We observed a transient phosphorylation of Elk-1 that peaked at 15 minutes in LPS-stimulated THP-1 cells (Figure 10B). The kinetics of LPS-induced assembly of the Elk-1/SRF complex was also evaluated by EMSAs. LPS transiently increased protein binding to SRE4 with maximal level at 15 minutes (Figure 10C). LPS stimulation did not change Sp1 binding. PD98059 abolished formation of the LPS-induced complex (Figure 10D). These data demonstrated that the kinetics of LPS-induced Elk-1 phosphorylation was identical to the increase in protein binding to SRE4.
LPS induces phosphorylation of Elk-1 in THP-1 cells.
(A) THP-1 cells were stimulated with LPS (10 μ/mL) for 0 to 25 minutes. Cytoplasmic extracts were prepared, and proteins were separated by SDS-PAGE. Western blot analysis was performed using antibodies against either phosphorylated or nonphosphorylated ERK1/2. (B) THP-1 cells were stimulated with LPS (10 μg/mL) for 0 to 60 minutes. Nuclear extracts were prepared, and proteins were separated by SDS-PAGE. Western blot analysis was performed using antibodies against phosphorylated Elk-1. (C) Nuclear extracts from LPS-stimulated THP-1 cells (0-60 minutes) were incubated with oligonucleotides containing SRE4 or Sp1 and protein-DNA complexes analyzed by EMSAs. (D) THP-1 cells were LPS-stimulated (15 minutes) with or without PD98059. Nuclear extracts were incubated with SRE4, and complexes were analyzed by EMSAs.
LPS induces phosphorylation of Elk-1 in THP-1 cells.
(A) THP-1 cells were stimulated with LPS (10 μ/mL) for 0 to 25 minutes. Cytoplasmic extracts were prepared, and proteins were separated by SDS-PAGE. Western blot analysis was performed using antibodies against either phosphorylated or nonphosphorylated ERK1/2. (B) THP-1 cells were stimulated with LPS (10 μg/mL) for 0 to 60 minutes. Nuclear extracts were prepared, and proteins were separated by SDS-PAGE. Western blot analysis was performed using antibodies against phosphorylated Elk-1. (C) Nuclear extracts from LPS-stimulated THP-1 cells (0-60 minutes) were incubated with oligonucleotides containing SRE4 or Sp1 and protein-DNA complexes analyzed by EMSAs. (D) THP-1 cells were LPS-stimulated (15 minutes) with or without PD98059. Nuclear extracts were incubated with SRE4, and complexes were analyzed by EMSAs.
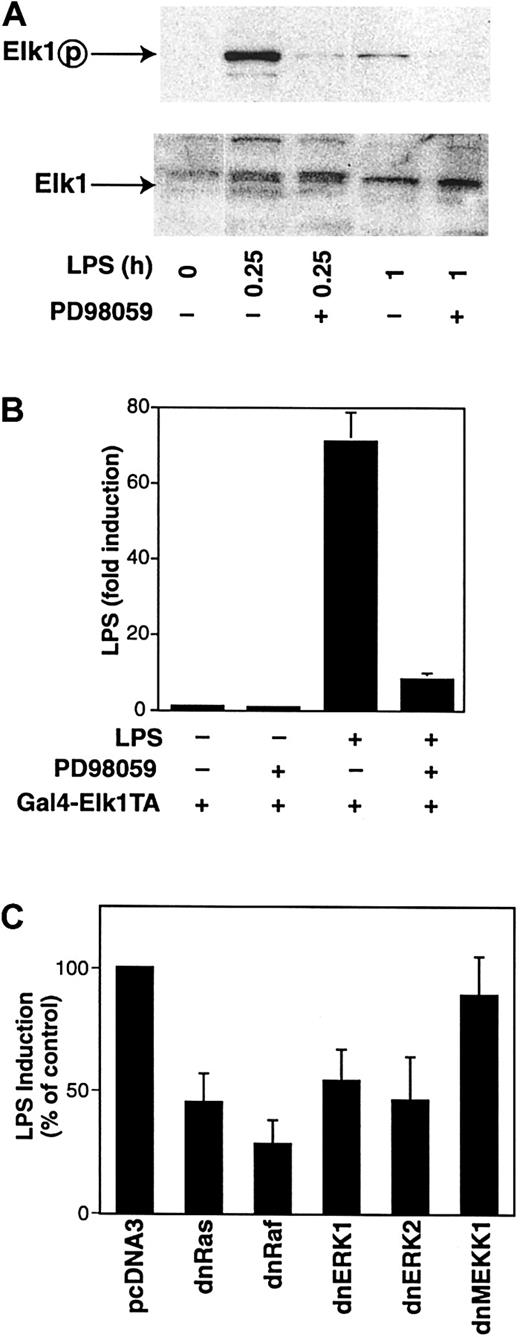
LPS activation of the MEK-ERK1/2 pathway mediates phosphorylation of Elk-1 and Elk-1–dependent transcription
We finally confirmed if LPS stimulation of THP-1 cells induces ERK1/2-mediated activation of endogenous Elk-1. First, we showed that LPS-induced phosphorylation of Elk-1 was inhibited by PD98059 (Figure11A). To analyze if LPS induction of the MEK-ERK1/2 pathway can lead to functional activation of Elk-1, THP-1 cells were cotransfected with the reporter plasmid pGAL4-LUC and pGAL4-Elk-1-TA, which expresses a chimeric protein that contains the GAL4 DNA-binding domain fused to the transactivation domain of Elk-1. LPS induced Elk-1–dependent transcription 71-fold, which was strongly inhibited by PD98059 (Figure 11B). No induction was observed using a plasmid that expresses only pGAL4-binding domain (not shown). Next, we determined the effect of dominant-negative versions of Ras, Raf-1, ERK1, and ERK2 on LPS activation of GAL4–Elk-1–TA. Cotransfection with plasmids expressing dominant-negative versions of Ras, Raf-1, ERK1, or ERK2 reduced LPS induction of Elk-1–dependent transcription by 54% to 72% (Figure 11C). In contrast, expression of dominant-negative MEKK-1 had no significant effect on LPS-induced Elk-1–dependent transcription. These results indicate that LPS stimulation of monocytic cells leads to phosphorylation and activation of Elk-1 via the Ras-Raf1-MEK-ERK1/2 pathway.
LPS induces Elk-1–dependent transcription.
(A) Nuclear extracts were prepared from LPS-stimulated THP-1 cells with or without PD98059 (25 μM). Western blot analysis was performed using antibodies against either phosphorylated or nonphosphorylated Elk-1. (B) The pGAL4-LUC (3 μg) and pGAL4–Elk-1TA (3 μg) were cotransfected into THP-1 cells. Transfected cells were preincubated in the presence or absence with PD98059 (25 μM) for 30 minutes prior to incubation with or without LPS for 5 hours. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are shown for 3 independent experiments. (C) THP-1 cells were cotransfected with pGAL4–Elk-1TA (4.0 μg) and pGAL4-LUC (1.5 μg) together with either pcDNA3 (4.5 μg) or plasmids expressing a dominant-negative version of Ras, Raf-1, ERK1, ERK2, or MEKK-1. Transfected cells were treated with or without LPS for 5 hours. Luciferase activity in cell lysates was determined, and results were expressed as percentage of the control plasmid (pcDNA3).
LPS induces Elk-1–dependent transcription.
(A) Nuclear extracts were prepared from LPS-stimulated THP-1 cells with or without PD98059 (25 μM). Western blot analysis was performed using antibodies against either phosphorylated or nonphosphorylated Elk-1. (B) The pGAL4-LUC (3 μg) and pGAL4–Elk-1TA (3 μg) were cotransfected into THP-1 cells. Transfected cells were preincubated in the presence or absence with PD98059 (25 μM) for 30 minutes prior to incubation with or without LPS for 5 hours. Luciferase activity in cell lysates was determined, and results were expressed as fold induction. Data (mean ± SD) are shown for 3 independent experiments. (C) THP-1 cells were cotransfected with pGAL4–Elk-1TA (4.0 μg) and pGAL4-LUC (1.5 μg) together with either pcDNA3 (4.5 μg) or plasmids expressing a dominant-negative version of Ras, Raf-1, ERK1, ERK2, or MEKK-1. Transfected cells were treated with or without LPS for 5 hours. Luciferase activity in cell lysates was determined, and results were expressed as percentage of the control plasmid (pcDNA3).
Discussion
In this study, we used the MEK1/2 inhibitor PD98059 to show that LPS activation of the MEK-ERK1/2 pathway was required for the maximal expression of TF and TNF-α in human monocytes and THP-1 monocytic cells. A similar study using PD98059 and Ro 09-2210 demonstrated a role of the MEK-ERK1/2 pathway in LPS induction of TNF-α expression in human monocytes.6 We extended these observations by determining the mechanism by which the MEK-ERK1/2 pathway contributes to TF and TNF-α gene expression. We showed that LPS activation of the MEK-ERK1/2 pathway induced Egr-1 expression, which was required for maximal expression of TF and TNF-α. Despite an early study showing LPS induction of Egr-1 in murine macrophages,20 there have been no studies on the transcription factors that regulate LPS induction of Egr-1 in human monocytic cells. We showed that LPS induction of the Egr-1 promoter was mediated by 3 SRE sites that bind a complex containing SRF and Elk-1. We showed for the first time that LPS stimulation of monocytic cells induced phosphorylation of Elk-1 via the MEK-ERK1/2 pathway.
The TF and TNF-α genes are regulated by NF-κB/Rel proteins in monocytic cells.27,33 In addition, mutational studies have shown a role for Egr-1 in TNF-α gene expression.27 Other studies suggest that Egr-1 also regulates TF gene expression in monocytic cells.33-35 Inhibition of MEK1/2 with PD98059 reduced LPS induction of TF and TNF-α promoter activity by 70% and 47%, respectively. The inhibition of TF and TNF-α expression did not appear to be due to a reduction in the activity of NF-κB/Rel and AP-1 proteins because PD98059 had a minimal effect on LPS induction of reporter plasmids containing an enhancer with either multimerized κB or AP-1 sites. In addition, a dominant-negative ERK1 mutant did not affect LPS-induced κB- or AP-1–dependent transcription. In contrast, PD98059 dramatically inhibited LPS induction of Egr-1 gene expression. Kinetic experiments showed that LPS induction of Egr-1 preceded the maximal levels of TF expression, which is consistent with Egr-1 regulating TF gene expression. In addition, mutation of the Egr-1 sites in the TF and TNF-α promoters reduced the level of LPS induction. Mutation of the Egr-1 sites in the TF promoter reduced the level of LPS induction 72%, which is similar to the effect of PD98059 (70%). Mutation of the Egr-1 site in the TNF-α promoter reduced the level of LPS induction by 50%, which is similar to the effect of PD98059 (47%). These data suggest that the effect of inhibition of the MEK-ERK1/2 pathway on LPS-induced TF and TNF-α expression is primarily due to inhibition of Egr-1 expression. PD98059 (25 μM) was a more potent inhibitor of LPS-induced TF promoter activity (70%) and protein expression compared with the inhibition of TNF-α promoter activity (47%) and protein expression in both monocytes and THP-1 cells. The difference in sensitivity to PD98059 may reflect the fact that the TF promoter contains 3 Egr-1 sites whereas the TNF-α promoter contains a single Egr-1 site.
We demonstrated that LPS rapidly induced Egr-1 expression in human monocytes and THP-1 cells. Inhibition of the Ras-Raf1-MEK-ERK1/2 pathway dramatically reduced LPS induction of Egr-1 promoter activity. Analysis of a 5′ deletion series of the Egr-1 promoter localized thecis-acting LPS response element to a region containing SRE sites 3 to 5. Mutation of each of the SRE sites 3 to 5 reduced LPS induction of the Egr-1 promoter, indicating that these sites mediated LPS induction. However, these studies do not exclude the possibility that SRE1 and SRE2 are necessary for the function of these upstream elements. EMSAs demonstrated that SRE3, SRE4, and SRE5 bound an LPS-inducible complex. Supershift experiments indicated that this complex was composed of SRF and Elk-1. We did not observe Sap-1 in the complex. We found that LPS stimulation of cells induced a rapid MEK1/2-dependent phosphorylation of Elk-1 that peaked at 15 minutes and corresponded with the maximal levels of an LPS-inducible protein-DNA complex. Finally, expression of dominant-negative mutants of the Ras-ERK1/2 pathway and the use of PD98059 demonstrated that LPS stimulation of monocytic cells induced Elk-1–dependent transcription via the Ras-Raf1-MEK-ERK1/2 pathway. Dominant-negative MEKK1 did not reduce LPS induction of either the Egr-1 promoter or the Elk-1–dependent reporter gene, suggesting that the JNK MAPK pathway does not play a significant role in LPS induction of Egr-1 in monocytic cells.
A model of LPS induction of TF in monocytic cells is shown in Figure12. The current study demonstrated that LPS stimulation of monocytic cells leads to the rapid MEK1/2-dependent phosphorylation of Elk-1. An LPS-inducible complex consisting of Elk-1 and SRF mediated induction of Egr-1 gene expression. We propose that newly synthesized Egr-1 binds to the TF promoter and contributes to maximal induction of TF expression in LPS-stimulated cells. LPS stimulation of monocytic cells also activated the JNK MAPK pathway. The JNK pathway phosphorylates and activates AP-1 proteins bound to the TF promoter.56 Finally, LPS stimulation activates IKKβ and induces nuclear translocation c-Rel/p65. Egr-1 and p65 have been shown to synergistically induce gene expression.57 Thus, assembly of a multiprotein complex on the TF promoter containing AP-1, c-Rel/p65, and Egr-1 is required for maximal induction of TF in LPS-stimulated monocytic cells.
LPS induction of the TF gene in monocytes.
Binding of LPS to the CD14 and TLR4/MD2 complex activates several intracellular signaling pathways that include ERK1/2, JNK, and NF-κB. The ERK1/2 pathway rapidly phosphorylates Elk-1 in an Elk-1/SRF complex that is bound to the Egr-1 promoter. We propose that newly synthesized Egr-1 binds to the TF promoter and, in association with AP-1 and c-Rel/p65, induces TF gene expression. JNK phosphorylates and activates AP-1 proteins. IKKβ phosphorylates inhibitor proteins, which allows nuclear translocation of c-Rel/p65. E indicates Egr-1; F, c-Fos; I, IκBs; J, c-Jun; R, c-Rel; 65, p65.
LPS induction of the TF gene in monocytes.
Binding of LPS to the CD14 and TLR4/MD2 complex activates several intracellular signaling pathways that include ERK1/2, JNK, and NF-κB. The ERK1/2 pathway rapidly phosphorylates Elk-1 in an Elk-1/SRF complex that is bound to the Egr-1 promoter. We propose that newly synthesized Egr-1 binds to the TF promoter and, in association with AP-1 and c-Rel/p65, induces TF gene expression. JNK phosphorylates and activates AP-1 proteins. IKKβ phosphorylates inhibitor proteins, which allows nuclear translocation of c-Rel/p65. E indicates Egr-1; F, c-Fos; I, IκBs; J, c-Jun; R, c-Rel; 65, p65.
We thank R. Clarkson and W. Aird for mutated versions of the Egr-1 promoter, and J. Robertson and B. Parker for preparing the manuscript.
Supported by grants from the National Institutes of Health (HL48872) to N.M. and a postdoctoral fellowship from the American Heart Association (M.A.O.).
M.G. and M.A.O. contributed equally to the study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Nigel Mackman, Dept of Immunology, C-204, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail: nmackman@scripps.edu.