Inhibition of eosinophil apoptosis by exposure to interleukin-5 (IL-5) is associated with the development of tissue eosinophilia and may contribute to the inflammation characteristic of asthma. Analysis of the signaling events associated with this process has been hampered by the inability to efficiently manipulate eosinophils by the introduction of active or inhibitory effector molecules. Evidence is provided, using a dominant-negative N17 H-Ras protein (dn-H-Ras) and MEK inhibitor U0126, that activation of the Ras-Raf-MEK-ERK pathway plays a determining role in the prolongation of eosinophil survival by IL-5. For these studies, a small region of the human immunodeficiency virus Tat protein, a protein transduction domain known to enter mammalian cells efficiently, was fused to the N-terminus of dn-H-Ras. The Tat-dn-H-Ras protein generated from this construct transduced isolated human blood eosinophils at more than 95% efficiency. When Tat-dn-H-Ras–transduced eosinophils were treated with IL-5, they exhibited a time- and dosage-dependent reduction in extracellular regulated kinase 1 and 2 activation and an inhibition of p90 Rsk1 phosphorylation and IL-5–mediated eosinophil survival in vitro. In contrast, Tat-dn-H-Ras did not inhibit CD11b up-regulation or STAT5 tyrosine phosphorylation. These data demonstrate that Tat dominant-negative protein transduction can serve as an important and novel tool in studying primary myeloid cell signal transduction in primary leukocytes and can implicate the Ras-Raf-MEK-ERK pathway in IL-5–initiated eosinophil survival.
Introduction
Asthma is a complex syndrome of variable airflow obstruction, bronchial hyperresponsiveness, and airway inflammation characterized by the infiltration of eosinophils and increased levels of interleukin-5 (IL-5). IL-5 is the principal eosinophil-active cytokine, and it affects the differentiation, recruitment, viability, and cytotoxic effector functions of the eosinophil. Several signaling pathways are implicated in IL-5–mediated events, including the activation of the tyrosine kinases Lyn, Syk, and Jak2,1-3phosphorylation of signal transducers and activators of transcription (STAT) factors,4-7 and the activation of the Ras-Raf-MEK-ERK pathway.3 8-10
Two lines of evidence suggest that IL-5 may activate extracellular signal-regulated kinase (ERK) 1 and ERK 2 through Ras-dependent pathways. First, in human peripheral blood eosinophils stimulated by IL-5, Ras is activated as measured by guanosine triphosphate loading.9,11 In addition, experiments in the cytokine-dependent myeloid cell line BaF3 recapitulated the observations in cytokine-stimulated eosinophils when a constitutively active mutant of Ras transfected into the BaF3 cells promoted ERK activation and suppressed apoptosis.12 These latter studies suggest that the Ras-Raf-MEK-ERK pathway may be important in IL-5–dependent suppression of apoptosis. Furthermore, a recent study has linked the survival of cerebellar neurons to the mitogen-activated protein (MAP) kinase pathway and reported that the ERK target protein, p90 ribosomal S6 kinase (p90 Rsk), could phosphorylate and inactivate the proapoptotic protein Bad and the cyclic adenosine monophosphate-responsive DNA-binding protein (CREB).13
To establish the importance of a protein such as Ras in a signaling pathway, one approach is to render the protein or pathway inoperative. Multiple methods achieve the inhibition of cellular processes, including the use of pharmacologic agents to inhibit enzyme activity, the use of antisense oligonucleotides to reduce the mass of a given protein in a cell, or the introduction of dominant-negative (dn) mutant proteins to counteract activity in wild-type cells. However, each of these methods has its drawbacks. The specificity of pharmacologic inhibitors is often suspicious, warranting the use of other methods to confirm results. Conversely, antisense oligonucleotides have several disadvantages attributable to the effects of these so-called nucleic acid drugs on molecules other than the intended target.14Furthermore, antisense approaches are unsatisfactory for the study of proteins that have low turnover rates, including many signal transduction molecules regulated by posttranslational modifications.14 Because of their unique specificity, a preferred choice for the inhibition of a signaling pathway is the expression of dn-mutant signaling proteins. Unfortunately, the expression of dn-signaling proteins in eosinophils has not been achieved, primarily because the eosinophil is a short-lived, terminally differentiated cell refractive to ordinary transfection methodologies. To facilitate the development of protocols for expressing mutant signaling molecules in peripheral blood eosinophils, we turned to the recently developed method of protein transduction.15,16 In the past decade, small regions of proteins that have the ability to cross biologic membranes efficiently—protein transduction domains—were identified. Such domains occur in a number of proteins including the Drosophila homeotic transcription factor antennaepedia, the herpes simplex virus type 1 transcription factor VP22, and the human immunodeficiency virus (HIV) Tat protein. The protein transduction domain of the HIV Tat protein is one of the most efficient domains at traversing cell membranes in a receptor-independent fashion.17-19 This characteristic is attributed to an 11-amino acid portion of the HIV Tat protein (YGRKKRRQRRR). When expressed as an N-terminal peptide linked to an unrelated protein, this sequence renders the fusion protein capable of entering cells in a time- and concentration-dependent fashion.20
In the studies described here, we linked the coding region for the Tat transduction motif to the gene coding for N17 dominant-negative H-Ras.21 22 The Ras mutant (N17) used in these studies contains an asparagine at position 17 that renders the mutant protein incapable of activating downstream targets and inhibits wild-type Ras function apparently by competing for upstream activators. When translated, the resultant protein was a fusion between the Tat transduction motif and dn-H-Ras. This Tat-dn-H-Ras fusion protein entered eosinophils with high efficiency and functioned as a dn-effector molecule. Using this construct, we provide evidence that IL-5–mediated Ras and ERK activation is necessary for IL-5–stimulated eosinophil viability and p90 Rsk1 activation. However, Ras activity is not required for the increased expression of the membrane receptor CD11b or the tyrosine phosphorylation of STAT5.
Materials and methods
Reagents
For eosinophil preparation, Percoll was purchased from Pharmacia (Piscataway, NJ), and anti-CD16–conjugated microbeads were purchased from Miltenyi Biotechnology (Auburn, CA). IL-5 was obtained from R&D Systems (Minneapolis, MN). Sigma Chemical y (St Louis, MO) was the source for phorbol myristate acetate (PMA). Protease inhibitor tablets (Complete) were obtained from Boehringer Mannheim (Indianapolis, IN). Annexin V was from Becton Dickinson (San Jose, CA). Immunoblotting reagents were purchased from a variety of suppliers, including Santa Cruz Biotechnology (Santa Cruz, CA; horseradish peroxidase-conjugated goat anti–rabbit IgG and anti–p90-Rsk1), Kierkegaard & Perry Laboratories (Gaithersburg, MD; Lumoglo chemiluminescence substrate reagents), Promega Life Sciences (Madison, WI; anti–active MAPK antisera), Upstate Biotechnology (Lake Placid, NY; anti-ERK1 and 2 -sera), Cell Signaling (Beverly, MA; anti–phospho-STAT5 antibody recognizing STAT5 phosphorylated on Tyr 694 and anti–phosphoro-p90 Rsk antibody recognizing p90 Rsk phosphorylated on Thr360/Ser364), Becton Dickinson (San Jose, CA; anti-CD11b), and Transduction Laboratories (Lexington, KY; anti-hemagglutinin [HA] and anti-Ras antibodies).
Cloning of dn-H-Ras in the TAT expression vector
The Tat-HA expression vector and Tat-HA–green fluorescence protein (Tat-GFP) were generously provided by Dowdy et al.15 The Tat-HA expression vector was cleaved withKpnI and XhoI (Promega). The N-terminal portion of the dn-N17 H-Ras gene (Upstate Biotechnology) was polymerase chain reaction amplified with the primers ATGGCCGGTACCATGACAGAATACAAGC and TGGCCGAGGTCTCGATGTAGGGGAT (each cycle was carried out at 95°C for 45 seconds, 60°C for 45 seconds, and 72°C for 1 minute, for a total of 25 cycles) using pfupolymerase (Stratagene, La Jolla, CA). The resultant polymerase chain reaction product was digested with KpnI and PpuMI (Promega). The C-terminal portion of the dn-H-Ras gene was cleaved from its expression plasmid with PpuMI andXhoI. The 2 fragments and the digested Tat plasmid were isolated by agarose gel electrophoresis followed by purification with the Gel Extract II kit (Qiagen, Valencia, CA). A 3-piece ligation was performed using T4 ligase (Promega). The construct Tat-HA-dn-H-Ras (Tat-dn-H-Ras) was confirmed by sequencing.
Protein purification
The polyhistidine-tagged Tat-dn-H-Ras gene was expressed in BL21 bacteria (Novagen, Madison, WI). After solubilization in purification buffer (8 M urea, 25 mM sodium phosphate, pH 8.0; Sigma), the bacteria solution was sonicated for 2 intervals of 30 seconds each (setting of 4 with a Branson Sonifier 250 sonicator) and centrifuged at 14 000g for 10 minutes. Supernatant containing the fusion protein was passed twice over a nickel-agarose column (Novagen), washed with 5 column volumes of purification buffer, and eluted with purification buffer containing 100 mM imidazole (Sigma). Urea and imidazole were removed from the resultant protein solution by dilution of the protein mixture 10-fold with 20 mM HEPES buffer and passage of the solution through a 10 000 mol wt cut-off filter (Sigma) using an Amicon filtration apparatus (Millipore, Bedford, MA); this procedure was repeated 3 times. To determine the concentration of the denatured protein, the molar extinction coefficient was calculated using Expasy Web-based software.23 Absorbance of the protein solution was measured at 280 nm, and this value was used to calculate a concentration according to Beer's law. Purity of Tat-dn-H-Ras and Tat-GFP was approximately 95%, as determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent Coomassie blue staining. Protein expression of the Tat-dn-H-Ras protein was confirmed by SDS-PAGE and immunoblotting as described above using anti-HA and anti-Ras antibodies.
Isolation of human eosinophils from peripheral blood
Blood donors included persons with and without atopy; eosinophil counts ranged from 3% to 12% of peripheral blood leukocytes. Eosinophils were purified from the heparinized peripheral blood of donors, as previously described.3 Briefly, a granulocyte mixture was obtained from the leukocyte buffy coat after centrifugation through a Percoll solution (density, 1.090 g/mL), and, after lysis of erythrocytes by hypotonic shock, the suspension was depleted of neutrophils by incubation with anti-CD16–conjugated microbeads and exposure to a magnetic field. Recovered eosinophils were resuspended at a concentration of 107 cells/mL in calcium- and magnesium-free Hanks balanced salt solution (HBSS) supplemented with 1% fetal calf serum (FCS). Eosinophil levels in these cell preparations were between 94% and 99%, as determined by microscopic examination of Wright-stained cytofuge preparations.
Eosinophil stimulation and preparation of cell lysates
Eosinophils were suspended in calcium- and magnesium-free HBSS with 1% FCS (107 cells/mL) and were preincubated for 30 minutes with Tat-dn-H-Ras followed by a 7-minute treatment with 1 nM IL-5. Next, the reactions were stopped by dilution with ice-cold buffer A (20 mM Tris, 137 mM NaCl, 1 mM EDTA, 0.1 mM sodium orthovanadate, 10 mM NaF, 10 mM β-glycerophosphate, and protease inhibitors, pH 7.4). Cells were pelleted, supernatants were discarded, and cell pellets were resuspended in buffer B (1% Triton-X-100, 0.25% deoxycholate, and 0.1% SDS in buffer A). Eosinophil lysates were centrifuged to remove the insoluble material, and supernatants were prepared for immunoblotting.
SDS-PAGE and immunoblotting
Eosinophil lysates were diluted with electrophoresis sample buffer, and the proteins were resolved on polyacrylamide slab gels. Lanes were loaded with samples that represented equal numbers of cells. Proteins were transferred to polyvinylidene difluoride membranes and immunoblotted using standard methods.24 The consistency of protein loading in all lanes was evaluated by staining of the polyvinylidene difluoride membrane with amido black after chemiluminography. In addition, samples were immunoblotted with anti-ERK 1 and -ERK 2 antisera to confirm that comparable protein mass was present in all lanes.
CD11b expression
Purified human blood eosinophils were resuspended in RPMI 1640 and 1% FCS.25 Tat-dn-H-Ras was added to a concentration of 100 nM, and IL-5 was added to a concentration of 100 pM. After 2 hours of incubation, the eosinophils were washed with FACS buffer (calcium and magnesium-free HBSS containing 1% bovine serum albumin and 0.1% azide) and labeled with anti-CD11b antibody or an isotype control for 1 hour at 4°C. After a wash with FACS buffer, the cells were incubated for 1 hour at 4°C with a fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibody (Becton Dickinson, San Jose, CA). The cells were then washed in FACS buffer and fixed overnight with 1% paraformaldehyde (Sigma). Fixed cells (10 000) were analyzed by flow cytometry on a FACScan (Becton Dickinson).
STAT5 phosphorylation
Human blood eosinophils were incubated with 100 nM Tat-dn-H-Ras for 30 minutes and then stimulated for 15 minutes with 300 pM IL-5. Cells were lysed using buffer B, and the resultant cell lysates were subjected to SDS-PAGE and immunoblotting with anti–phospho-STAT5 antibody. Parallel immunoblots probed with anti-STAT5 confirmed equivalent masses of target proteins in all eosinophil samples.
Cellular uptake of Tat-dn-H-Ras
Tat-dn-H-Ras was labeled with FITC (Pierce) according to manufacturer's instructions, and excess FITC was removed by passing the solution through a G-50 column (Pharmacia). Eosinophils resuspended in calcium- and magnesium-free HBSS, pH 6.8, containing 1% FCS were incubated with 100 nM FITC-Tat-dn-H-Ras for varying times at 37°C. The pH of the sheath fluid of the flow cytometer was adjusted to 6.8 to quench FITC fluorescence on the outside of the cell, and 10 000 cells were analyzed on a FACScan (Becton Dickinson).
Viability assay
Blood eosinophils were resuspended in RPMI 1640 containing 1% FCS to a concentration of 4 × 105 cells/mL. IL-5 was added to a concentration of 10 pM, and Tat-dn-H-Ras was added to a concentration of 100 nM. In some cases, the MAP ERK kinase (MEK) antagonist U0126 (Promega) was added to a concentration of 10 μM, or MEK antagonist PD98059 (Promega) was added to a concentration of 50 μM. Cells were incubated at 37°C with 5% CO2. Viability was determined by labeling cells with 5 mM propidium iodide and FITC-annexin V to determine apoptotic status (Sigma) followed by flow cytometry on a FACScan (Becton Dickinson).
Measurement of Ras activation
Human blood eosinophils were incubated with 100 nM Tat-dn-H-Ras for 30 minutes and then stimulated for 7 minutes with 1 nM IL-5. Cells were treated with lysis buffer (Upstate Biotechnology), and the resultant cell lysates were precleared using glutathione agarose beads (Pierce) and incubation at 4°C for 10 minutes, followed by bead removal by centrifugation. Resultant supernatants were incubated with glutathione agarose beads linked to a glutathione-s-transferase (GST) fusion of the Ras-binding domain of Raf (Upstate Biotechnology) for 30 minutes at 4°C. After 3 washes with lysis buffer, the mixture was boiled in sample buffer, separated by SDS-PAGE, and processed for immunoblot analysis using an anti-Ras antibody.
Results
Transduction of human eosinophils with Tat-dn-H-Ras
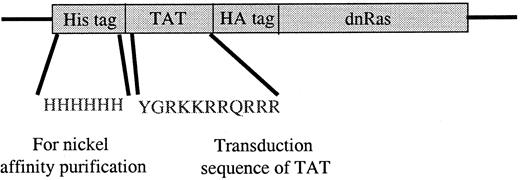
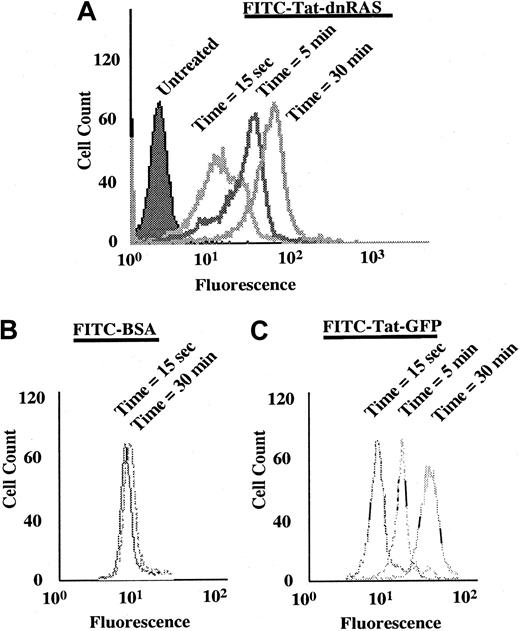
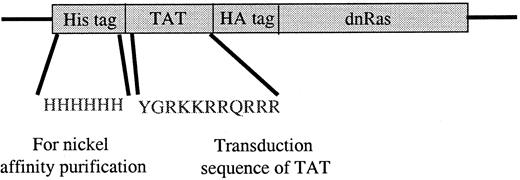
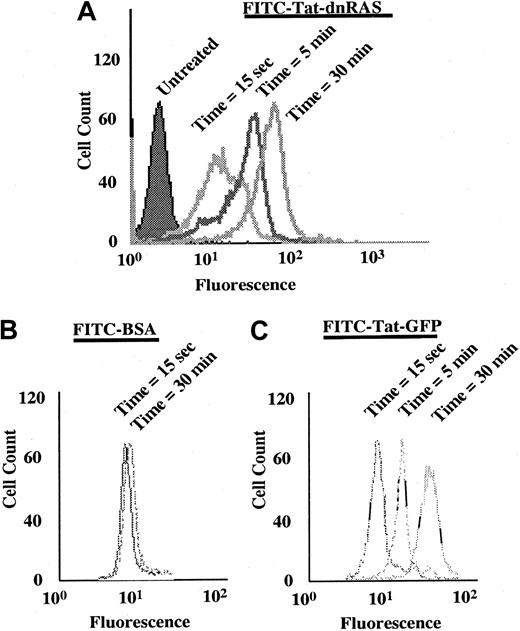
The Ras fusion protein consisted of 6 His residues, 11 amino acids of Tat, hemagglutinin (HA) tag, and N17 dominant-negative H-Ras (Tat-dn-H-Ras) (Figure 1). As described by Dowdy et al,15 proteins sized similarly to dn-H-Ras linked to the Tat peptide entered cells within 30 minutes in a concentration-dependent manner. To determine whether Tat-dn-H-Ras fusion protein enters human blood eosinophils, the protein was labeled with FITC. Eosinophils were incubated with 100 nM of the FITC-Tat-dn-H-Ras at a pH of 6.8, and samples were analyzed by flow cytometry every 5 minutes for 30 minutes (Figure2A) to determine cellular uptake of the labeled protein. FITC-labeled protein entering the cells was distinguished from that adhering to cell surfaces through an experiment at a pH of 6.8. At this pH, FITC-labeled proteins that enter the cell fluoresce, whereas those external to the cell have their fluorescence quenched.26 As seen in Figure 2, external FITC increased the fluorescence of the eosinophils at the 15-second time point. Subsequent increases in fluorescence from the 15-second time point are the result of FITC-labeled protein entering cells at later time points (5 and 30 minutes). Comparison was made of the time points at 15 seconds and 30 minutes, and more than 99% of the eosinophils had taken up the FITC-Tat-dn-H-Ras protein at the 30-minute time point. In contrast, incubation of eosinophils with FITC-bovine serum albumin not linked to the Tat peptide resulted in an increase in fluorescence from external FITC but did not result in an increase in eosinophil fluorescence over time (Figure 2B), demonstrating the Tat-linked fusion protein was entering the cells. Tat-GFP, used as a control in subsequent experiments, showed similar uptake kinetics (Figure 2C). Furthermore, the eosinophils were more than 98% viable during the course of the experiment, as indicated by propidium iodide staining (data not shown).
Tat-dominant-negative N 17 Ras construct.
The dominant-negative N17 H-Ras gene was cloned into the pTat-HA vector as described in “Materials and methods.” Six His residues precede the 11-amino acid Tat peptide. Another 10 amino acids of the HA tag and 6 Gly are immediately N-terminal to the N17 Ras protein. Tat-GFP also contains the HA tag (see “Materials and methods”).
Tat-dominant-negative N 17 Ras construct.
The dominant-negative N17 H-Ras gene was cloned into the pTat-HA vector as described in “Materials and methods.” Six His residues precede the 11-amino acid Tat peptide. Another 10 amino acids of the HA tag and 6 Gly are immediately N-terminal to the N17 Ras protein. Tat-GFP also contains the HA tag (see “Materials and methods”).
FITC-labeled Tat-dn-H-Ras enters eosinophils.
Tat-dn-H-Ras was labeled with FITC, as described in “Materials and methods.” Human blood eosinophils resuspended in pH 6.8 HBSS without calcium or magnesium and 1% fetal calf serum were incubated with 100 nM FITC-Tat-dn-H-Ras, and, at 5-minute intervals, aliquots were analyzed by flow cytometry on a FACScan (A). As a control for protein bound to the cellular exterior, FITC-labeled bovine serum albumin was incubated with eosinophils under identical conditions (B). Uptake of the control Tat-GFP protein is similar to that of Tat-dn-H-Ras (C). These histograms are representative of 3 independent experiments.
FITC-labeled Tat-dn-H-Ras enters eosinophils.
Tat-dn-H-Ras was labeled with FITC, as described in “Materials and methods.” Human blood eosinophils resuspended in pH 6.8 HBSS without calcium or magnesium and 1% fetal calf serum were incubated with 100 nM FITC-Tat-dn-H-Ras, and, at 5-minute intervals, aliquots were analyzed by flow cytometry on a FACScan (A). As a control for protein bound to the cellular exterior, FITC-labeled bovine serum albumin was incubated with eosinophils under identical conditions (B). Uptake of the control Tat-GFP protein is similar to that of Tat-dn-H-Ras (C). These histograms are representative of 3 independent experiments.
Activity of endogenous Ras in the presence of transduced Tat-dn-H-Ras
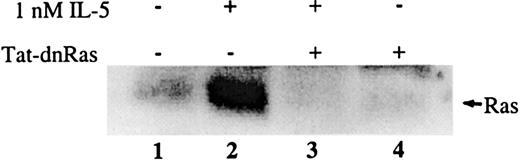
To determine whether the Tat-dn-H-Ras transduced into cells was capable of directly inhibiting the activation of endogenous Ras, blood eosinophils were incubated with or without 100 nM Tat-dn-H-Ras for 30 minutes at 37°C and were subsequently stimulated with 1 nM IL-5. Cell lysates generated from each sample were precleared with glutathione agarose beads to reduce nonspecific interactions. Next, the lysates were incubated with glutathione agarose beads linked to a GST fusion of the Ras-binding domain of Raf, which is known to bind only to the activated (guanosine triphosphate bound) form of Ras.9After a series of wash steps, the samples were separated by SDS-PAGE and immunoblotted. Little active Ras was detected in unstimulated eosinophils (Figure 3, lane 1), whereas the addition of IL-5 caused potent Ras activation (Figure 3, lane 2) as previously described.11 After pretreatment of eosinophils with Tat-dn-H-Ras, IL-5 did not stimulate Ras activation (Figure 3, lane 3), and no active Ras was detectable in eosinophils treated with Tat-dn-H-Ras alone (Figure 3, lane 4). Thus, Tat-dn-H-Ras is capable of efficiently inhibiting endogenous Ras activation in transduced eosinophils.
Transduced Tat-dn-H-Ras blocks activation of endogenous Ras on IL-5 stimulation.
Human blood eosinophils were incubated with 100 nM Tat-dn-H-Ras for 30 minutes and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were incubated with GST-Ras binding domain protein bound to glutathione agarose beads for 30 minutes at 4°C. After they were washed with lysis buffer, the mixture was boiled in sample buffer, separated by SDS-PAGE, and immunoblotted using anti-Ras antibody. Figure 3 is representative of 4 repetitions of this experiment.
Transduced Tat-dn-H-Ras blocks activation of endogenous Ras on IL-5 stimulation.
Human blood eosinophils were incubated with 100 nM Tat-dn-H-Ras for 30 minutes and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were incubated with GST-Ras binding domain protein bound to glutathione agarose beads for 30 minutes at 4°C. After they were washed with lysis buffer, the mixture was boiled in sample buffer, separated by SDS-PAGE, and immunoblotted using anti-Ras antibody. Figure 3 is representative of 4 repetitions of this experiment.
Dose-dependent effects of Tat-dn-H-Ras on IL-5–induced kinase ERK 1 and ERK 2 activation
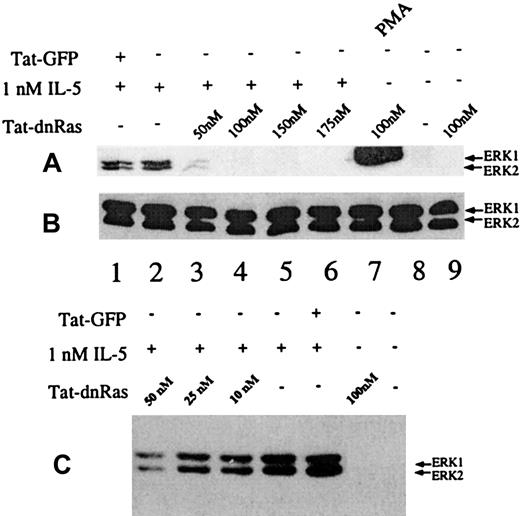
Incubation of peripheral blood eosinophils with IL-5 is known to stimulate multiple signal transduction molecules, including members of the Ras-Raf-MEK-ERK pathway.3,8,9,11,24 27 Although Tat-dn-H-Ras is taken up by eosinophils, it is important to demonstrate that Tat-dn-H-Ras was able to functionally inhibit signal transduction pathways downstream of Ras. To test this, various concentrations of Tat-dn-H-Ras were incubated with 106 eosinophils per treatment for 30 minutes at 37°C. The incubation time for the Tat-dn-H-Ras was based on studies described below. After the prescribed incubation period, the samples were stimulated with 1 nM IL-5 for 7 minutes to induce ERK 1 and ERK 2 activation. Resultant cell lysates were immunoblotted with activation-specific phospho-ERK antibodies (Figure 4A). These data demonstrate that Tat-dn-H-Ras maximally inhibits IL-5–mediated ERK activation at approximately 100 nM (Figure 4A, lanes 3-6). Concentrations less than 10 to 20 nM did not have a significant impact on IL-5–mediated ERK activation (Figure 4C). A 100-nM concentration of Tat-dn-H-Ras alone does not stimulate ERK 1 and ERK 2 activation (Figure 4A, lane 9). The activation of ERK 1 and ERK 2 by PMA is thought to be mediated through the activation of c-Raf-1 and, therefore, is not dependent on the activation of Ras. Consistent with this hypothesis, we observed that Tat-dn-H-Ras did not inhibit PMA-stimulated ERK activation in eosinophils (Figure 4, lane 7). Tat-GFP does not inhibit IL-5–mediated ERK 1 and ERK 2 activation at a concentration of 100 nM (Figure 4A, lane 1), and it was included as a control for the protein purification procedure and the effects of the Tat and HA fragments appended to the N-terminus of the GFP and the dn-H-Ras proteins (see “Materials and methods”).
Dosage of Tat-dn-H-Ras affects IL-5–stimulated ERK 1 and ERK 2 activation.
(A) Human blood eosinophils were incubated with various concentrations of Tat-dn-H-Ras for 30 minutes and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were resolved by SDS-PAGE and processed for immunoblot analysis using anti-active MAPK antibodies. (B) Cell lysates from panel A were also probed with anti-ERK 1 and ERK 2 antibodies to demonstrate equivalent masses of ERK 1 and ERK 2 in all samples. (C) The same experiment described for panel A was performed with concentrations of Tat-dn-H-Ras that were suboptimal for inhibiting ERK activation. These immunoblots are representative of 8 experiments.
Dosage of Tat-dn-H-Ras affects IL-5–stimulated ERK 1 and ERK 2 activation.
(A) Human blood eosinophils were incubated with various concentrations of Tat-dn-H-Ras for 30 minutes and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were resolved by SDS-PAGE and processed for immunoblot analysis using anti-active MAPK antibodies. (B) Cell lysates from panel A were also probed with anti-ERK 1 and ERK 2 antibodies to demonstrate equivalent masses of ERK 1 and ERK 2 in all samples. (C) The same experiment described for panel A was performed with concentrations of Tat-dn-H-Ras that were suboptimal for inhibiting ERK activation. These immunoblots are representative of 8 experiments.
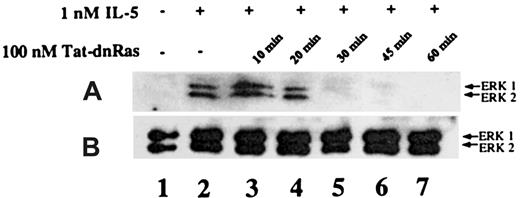
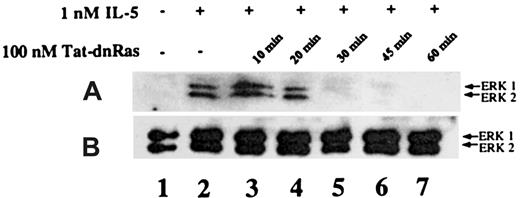
Time-dependent inhibition of IL-5–mediated ERK activation by Tat-dn-H-Ras
It has been reported that Tat fusion proteins could enter cells and effectively block biologic function within 30 minutes of exposure.15 To determine whether Tat-mediated protein transduction could be biologically active with similar kinetics, we assessed the incubation time required for Tat-dn-H-Ras to enter the cells and become effective at blocking IL-5–stimulated ERK 1 and ERK 2 activation. Eosinophils were incubated with 100 nM Tat-dn-H-Ras (optimal as determined in Figure 4) for 10, 20, 30, 45, and 60 minutes at 37°C (Figure 5, lanes 3-7). Samples from unstimulated and IL-5–stimulated samples are shown in lanes 1 and 2. After the specified time of incubation, the eosinophils were stimulated with 1 nM IL-5 for 7 minutes as described for the studies shown in Figure 4. Cell lysates obtained were immunoblotted with anti-active ERK antibodies. From these data, it is apparent that Tat-dn-H-Ras enters the cells within 20 minutes of incubation and remains effective in blocking ERK 1 and ERK 2 activation for at least 1 hour.
Time course of Tat-dn-H-Ras incubation as a function of inhibition of ERK 1 and ERK 2 activation.
(A) Human eosinophils were incubated with 100 nM Tat-dn-H-Ras for different time intervals and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were separated by SDS-PAGE and processed for immunoblot analysis using anti-active ERK 1 and ERK 2 antibodies. (B) Cell lysates from panel A were also probed with anti- ERK 1 and ERK 2 antibodies to demonstrate equivalent masses of protein in all samples. Figure 5 is representative of 3 repetitions of this experiment.
Time course of Tat-dn-H-Ras incubation as a function of inhibition of ERK 1 and ERK 2 activation.
(A) Human eosinophils were incubated with 100 nM Tat-dn-H-Ras for different time intervals and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were separated by SDS-PAGE and processed for immunoblot analysis using anti-active ERK 1 and ERK 2 antibodies. (B) Cell lysates from panel A were also probed with anti- ERK 1 and ERK 2 antibodies to demonstrate equivalent masses of protein in all samples. Figure 5 is representative of 3 repetitions of this experiment.
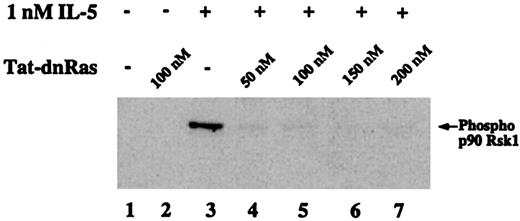
Effects of Tat-dn-H-Ras on IL-5–mediated p90 Rsk phosphorylation
The family of 90-kd ribosomal S6 kinases (p90 Rsk) are serine-threonine kinases that are direct substrates of the ERK kinases in several phenotypic contexts.28 However, the downstream effectors of the Ras-Raf-MEK-ERK pathway in eosinophils have not been identified. The following experiment was conducted to determine whether p90 Rsk is phosphorylated in eosinophils after IL-5 stimulation and whether that activation was Ras dependent. Eosinophils were incubated with 1 nM IL-5 for 7 minutes, cell lysates were prepared, and proteins were separated by SDS-PAGE. Immunoblotting with an antibody against p90 Rsk phosphorylated at Thr360/Ser364 was used to detect the phosphorylated forms of p90 Rsk (Figure6). Rsk phosphorylation was IL-5 dependent (Figure 6, lane 3) and addition of 50 nM, 100 nM, 150 nM, 200 nM Tat-dn-H-Ras (Figure 6, lanes 4 and 7) inhibited Rsk phosphorylation. No treatment (Figure 6, lane 1) or treatment with 100 nM Tat-dn-H-Ras alone (Figure 6, lane 2) showed no Rsk phosphorylation, whereas treatment with Tat-GFP did not have an effect on IL-5–mediated Rsk phosphorylation (data not shown). These data suggest that p90 Rsk is a downstream effector of the Ras-Raf-MEK-ERK pathway in IL-5–stimulated eosinophils.
Phosphorylation of the ERK substrate p90 Rsk1 is suppressed by Tat-dn-H-Ras.
Human blood eosinophils were incubated with 100 nM Tat-dn-H-Ras and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were separated by SDS-PAGE and processed for immunoblot analysis using anti–phospho-p90 Rsk1 antibody. Figure 6 is representative of 3 repetitions of this experiment.
Phosphorylation of the ERK substrate p90 Rsk1 is suppressed by Tat-dn-H-Ras.
Human blood eosinophils were incubated with 100 nM Tat-dn-H-Ras and then stimulated for 7 minutes with 1 nM IL-5. Resultant cell lysates were separated by SDS-PAGE and processed for immunoblot analysis using anti–phospho-p90 Rsk1 antibody. Figure 6 is representative of 3 repetitions of this experiment.
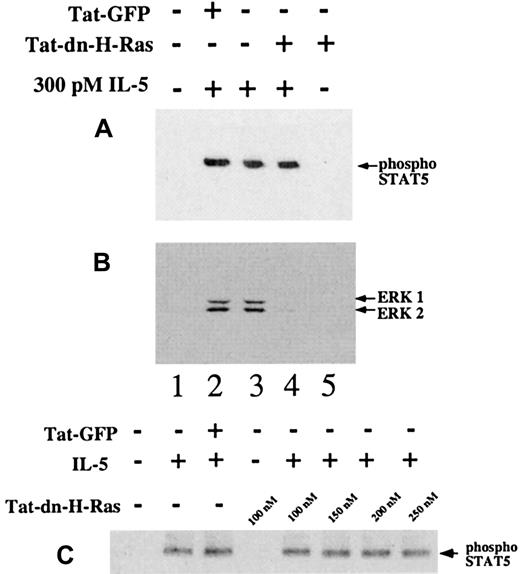
Status of STAT5 tyrosine phosphorylation in the presence of Tat-dn-H-Ras
To confirm that the effects of Tat-dn-H-Ras were confined to the Ras-ERK pathway, we tested the effects of Tat-dn-H-Ras on STAT5 tyrosine phosphorylation. Tyrosine phosphorylation of STAT5 is thought to be independent of Ras activation.3-5Cytoplasmic tyrosine kinase Jak2 is known to phosphorylate STAT5, thus triggering dimerization and subsequent translocation to the nucleus.29-31 Eosinophils were stimulated with IL-5 in the presence or absence of Tat-dn-H-Ras. Resultant cell lysates were immunoblotted for the presence of tyrosine phosphorylated STAT5 (Figure7A) or ERK 1 and ERK 2 activation (Figure7B). After IL-5 stimulation, 100 nM Tat-dn-H-Ras did not have a detectable effect on STAT5 tyrosine phosphorylation (Figure 7A, lanes 3 and 4), nor did concentrations up to 250 nM Tat-dn-H-Ras (Figure 7C). However, Tat-dn-H-Ras did block ERK 1 and ERK 2 activation (Figure 7B, lane 4) in these same cells. Furthermore, Tat-GFP had no effect on STAT or ERK activation (Figure 7B, lane 2). These data demonstrate that Tat-dn-H-Ras acts as a specific cell-permeable inhibitor and that protein transduction does not simply result in an overall specific decrease in eosinophil response to IL-5.
STAT5 phosphorylation after IL-5 stimulation is not blocked by Tat-dn-H-Ras.
Human blood eosinophils were incubated with 100 nM of Tat-dn-H-Ras or Tat-GFP for 30 minutes and then stimulated for 15 minutes with 300 pM IL-5. Resultant cell lysates were resolved by SDS-PAGE and processed for immunoblot analysis using either anti–phospho-STAT5 antibodies (A) or anti-active MAPK antibodies (B). The same experiment performed in panel A was performed with excessive concentrations of Tat-dn-H-Ras (C). An equal mass of protein in each sample was confirmed by immunoblotting with anti-STAT5 antibody (not shown). Data of 3 independent experiments are represented.
STAT5 phosphorylation after IL-5 stimulation is not blocked by Tat-dn-H-Ras.
Human blood eosinophils were incubated with 100 nM of Tat-dn-H-Ras or Tat-GFP for 30 minutes and then stimulated for 15 minutes with 300 pM IL-5. Resultant cell lysates were resolved by SDS-PAGE and processed for immunoblot analysis using either anti–phospho-STAT5 antibodies (A) or anti-active MAPK antibodies (B). The same experiment performed in panel A was performed with excessive concentrations of Tat-dn-H-Ras (C). An equal mass of protein in each sample was confirmed by immunoblotting with anti-STAT5 antibody (not shown). Data of 3 independent experiments are represented.
In addition to our STAT5 study, we evaluated the effects of Ras inhibition on other aspects of eosinophil biology such as IL-5–stimulated CD11b up-regulation. Constitutive expression of CD11b is augmented from cytoplasmic stores by stimulation with IL-5.32 Eosinophils were incubated with or without 1 nM IL-5 for 2 hours in the presence or absence of 100 nM Tat-dn-H-Ras or 100 nM Tat-GFP. Similar to findings in other reports,32 we observed that IL-5 up-regulated CD11b by 10.4% ± 1.6% (n = 3). Similarly, Tat-dn-H-Ras or 100 nM Tat-GFP treatment and subsequent IL-5 stimulation resulted in a CD11b increase of 10.1% ± 1.3% or 10.2% ± 1.8%, respectively, suggesting that Tat-dn-H-Ras does not inhibit IL-5–mediated CD11b up-regulation.
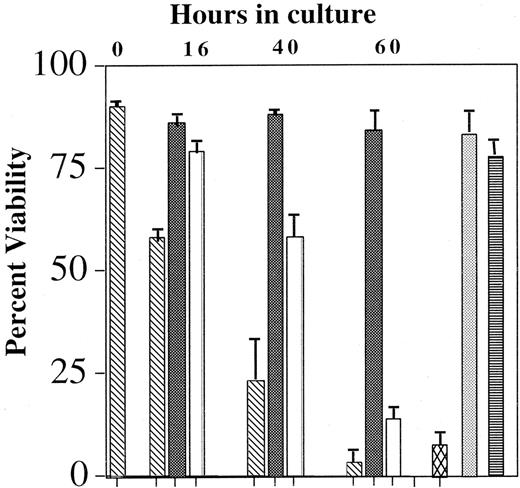
Effects of blocking the Ras-Raf-MEK-ERK pathway on IL-5–mediated eosinophil viability
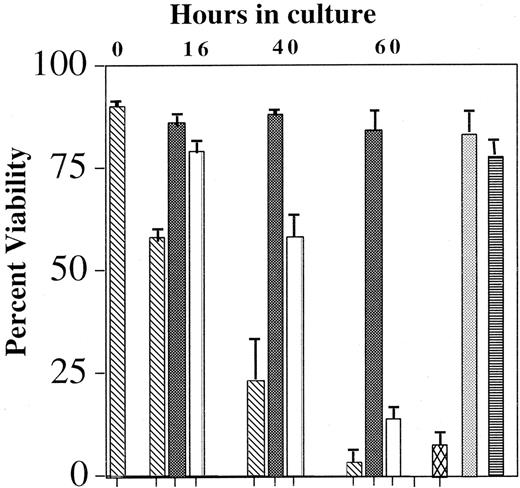
A multitude of signaling pathways has been implicated in IL-5–modulated eosinophil survival. Specific depletion of the tyrosine kinases Lyn,1,2 Syk,2 SHP-2,33and Raf-12 has resulted in an abrogation of the anti-apoptotic effects of IL-5. Similarly, inhibition of the Jak2 tyrosine kinase by AG490, the p38 MAP kinase by SB202589,34,35 and the PI-3 kinase by wortmanin9 also attenuates prolonged eosinophil viability in the presence of IL-5. However, the MEK 1 antagonist PD98059 failed to block the effects of IL-5–mediated survival. The reliability of studies using pharmacologic inhibitors is uncertain because of their nonspecific effects; therefore, the role of the Ras-Raf-MEK-ERK pathway in IL-5–mediated survival remains undefined. To determine whether the Ras-Raf-MEK-ERK pathway is pivotal in IL-5–mediated eosinophil survival, blood eosinophils were incubated with 10 pM IL-5 in the presence and absence of 100 nM Tat-dn-H-Ras or MEK 1/2 antagonist U012636 for up to 60 hours, and the effects of these treatments on cell viability were assessed by flow cytometry. Data summarized in Figure 8 show that Tat-dn-H-Ras or U0126 reduces the ability of IL-5 to suppress apoptosis in eosinophils. A comparable concentration of Tat-GFP protein did not inhibit IL-5–mediated survival during the same period of time, again revealing the selectivity of Tat-dn-H-Ras. In addition, as previously reported,37 the MEK 1 antagonist PD98059 did not inhibit IL-5–mediated survival. These data demonstrate for the first time that the entire Ras-Raf-MEK-ERK pathway is critical in regulating eosinophil survival and that this may be in part due to the activation not only of Raf-1 but of ERK 1 and ERK 2.
Effect of Tat-dn-H-Ras and MEK inhibitor U0126 on eosinophil viability.
Blood eosinophils were incubated with no IL-5 (▧), 10 pM IL-5 (▪), 100 nM Tat-dn-H-Ras (■), 100 nM Tat-GFP (░), 10 μM U0126 (▩), or 50 μM PD98059 (▤) for up to 60 hours. Viability was determined by FITC-annexin V and propidium iodide labeling of the cells and subsequent flow cytometry. Accumulated data of 3 independent experiments are represented.
Effect of Tat-dn-H-Ras and MEK inhibitor U0126 on eosinophil viability.
Blood eosinophils were incubated with no IL-5 (▧), 10 pM IL-5 (▪), 100 nM Tat-dn-H-Ras (■), 100 nM Tat-GFP (░), 10 μM U0126 (▩), or 50 μM PD98059 (▤) for up to 60 hours. Viability was determined by FITC-annexin V and propidium iodide labeling of the cells and subsequent flow cytometry. Accumulated data of 3 independent experiments are represented.
Discussion
Eosinophils are thought to be among the central effector cells involved in the development and course of asthma.38Unfortunately, this terminally differentiated cell type is not amenable to conventional methods of transfection commonly used as tools for elucidating molecular function. Furthermore, pharmacologic inhibitors and antisense oligonucleotides have drawbacks related to their specificity of inhibition. Therefore, we pursued protein transduction as an avenue for elucidating the contribution of specific effector molecules and signaling processes to cellular function. Protein transduction is the process by which a protein traverses the cellular membrane.15-18,39,40 This attribute is conferred by small, arginine-rich protein domains found in the transcription factors antennaepedia and VP22 and in the HIV Tat protein. The uptake of proteins fused to protein transduction domains does not appear to be mediated by normal cellular endocytosis, pinocytosis, or receptor-mediated processes.17 18
In this study, we show that human peripheral blood eosinophils can be transduced with a Tat fusion of the N17 dominant-negative H-Ras.21,22 The Ras mutant (N17) used in these studies contains an asparagine at position 17 that renders the mutant protein incapable of activating downstream targets and inhibits wild-type Ras function apparently by competing for upstream activators. Our laboratory and others have established that the Ras-Raf-MEK-ERK pathway plays a central role when eosinophils are stimulated with IL-5.3,9,24 27 Indeed, IL-5 is an important cytokine in eosinophil biology in that it stimulates bone marrow production of eosinophils, enhances their inflammatory properties, and prevents apoptosis.
We determined a role for Ras in the regulation of the MAP kinases ERK 1 and ERK 2 by demonstrating that Tat-dn-H-Ras can block IL-5–mediated ERK 1 and ERK 2 activation in a time- and dosage-dependent manner (Figures 4, 5) as assessed by anti–phospho-ERK immunoblotting. ERK family MAP kinases are important in a variety of eosinophil functions, including the promotion of LTC4 generation by IL-5 priming and chemotactic responses to eotaxin, but they have not been thought to be important in eosinophil viability.2,8The ERK family of kinases also mediates the phosphorylation of a number of targets including the p90 Rsk serine-threonine kinases and transcription factors Elk-1, c-fos, CREB, and c-jun.8,13 Indeed, we were able to show, for the first time in eosinophils, that p90 Rsk1 is phosphorylated in eosinophils on stimulation with IL-5 and that the blockade of the Ras-Raf-MEK-ERK pathway by Tat-dn-H-Ras blocked the phosphorylation event (Figure 6). The p90 Rsk family of kinases is also important in regulating apoptosis through Bad phosphorylation and in regulating transcription by phosphorylating a specific set of transcription factors related to cell cycle regulation.13 41 The immediate downstream effectors of active p90 Rsk in eosinophils are unknown and are the subject of current study.
Apoptosis is an important mechanism by which eosinophils are removed from the tissues.8,42,43 IL-5, IL-3, and granulocyte-macrophage colony-stimulating factor suppress this process, increasing the lifespan of eosinophils.44 Hence, the inhibition of apoptosis by these cytokines is considered an important mechanism in the development tissue eosinophilia in many allergic disorders. Previous studies using blood eosinophils have reported conflicting data on whether the Ras-Raf-MEK-ERK pathway is important in eosinophil survival. Based on the use of a chemical inhibitor of MEK 1 (PD98059), a previous study by Miike et al37 reported that ERK activity is not necessary for IL-5–stimulated blood eosinophil survival, and we have recapitulated those data (Figure 8). Another study has implicated c-Raf-1 as important in eosinophil-mediated survival but not in ERK activation.2 In the case of blood eosinophils, Tat-dn-H-Ras and the MEK 1 and 2 inhibitor U0126 restrain IL-5–mediated eosinophil survival, suggesting that the complete Ras-Raf-MEK-ERK pathway is necessary for the maintenance of viability in blood eosinophils by IL-5, not just c-Raf-1 (Figure 8). These data suggest a central role for Ras in mediating eosinophil viability after stimulation with IL-5. Indeed, active Ras is also capable of stimulating PI3K activation,45 and a role for PI3K in regulating eosinophil viability has been proposed through pharmacologic inhibition studies.9
Although several eosinophil responses to IL-5 stimulation were blocked by the presence of Tat-dn-H-Ras, 2 responses that may be potentiated by other signaling pathways were not blocked. Up-regulation of the adhesion molecule CD11b and STAT5 tyrosine phosphorylation were not inhibited in the presence of Tat-dn-H-Ras on IL-5 stimulation. In vitro CD11b up-regulation in eosinophils is probably analogous to that on neutrophils in which CD11b is rapidly mobilized from cytoplasmic stores instead of de novo protein synthesis.32 It is interesting to note that antisense blockade of Raf-1 prevents CD11b up-regulation,2 whereas blockade of Ras with Tat-dn-H-Ras does not. This suggests that Raf-1 may be stimulated by effectors other than Ras—such as protein kinases—as has been shown in other cell types.46
Eosinophils and their regulation by IL-5 are thought to provide one of the central mechanisms by which inflammatory processes contribute to the pathobiology of asthma. In the present report, we have identified Ras as an important mediator of primary myeloid cell survival in response to hematopoietic cytokines. Furthermore, the data provided in this study suggest that the introduction of dominant-negative signaling molecules into primary human myeloid cells may be achieved through the method of protein transduction. Subsequent studies of the contribution of specific signaling molecules to the inflammatory capabilities of the eosinophils are now possible.
We are grateful to Dr G. J. Wiepz for critical reading of the manuscript, and Dr Julie Sedgwick and Heather Gerbyshak for preparation of eosinophils.
Supported by National Institutes of Health grants HL56396 and AI34891 (P.J.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul J. Bertics, Department of Biomolecular Chemistry, 1300 University Ave, University of Wisconsin, Madison, WI 53706; e-mail: pbertics@facstaff.wisc.edu.