Previous studies have shown that the choice of envelope protein (pseudotype) can have a significant effect on the efficiency of retroviral gene transfer into hematopoietic stem cells. This study used a competitive repopulation assay in the dog model to evaluate oncoretroviral vectors carrying the envelope protein of the endogenous feline virus, RD114. CD34-enriched marrow cells were divided into equal aliquots and transduced with vectors produced by the RD114-pseudotype packaging cells FLYRD (LgGLSN and LNX) or by the gibbon ape leukemia virus (GALV)–pseudotype packaging cells PG13 (LNY). A total of 5 dogs were studied. One dog died because of infection before sustained engraftment could be achieved, and monitoring was discontinued after 9 months in another animal that had very low overall gene-marking levels. The 3 remaining animals are alive with follow-ups at 11, 22, and 23 months. Analyses of gene marking frequencies in peripheral blood and marrow by polymerase chain reaction revealed no significant differences between the RD114 and GALV-pseudotype vectors. The LgGLSN vector also contained the enhanced green fluorescent protein (GFP), enabling us to monitor proviral expression by flow cytometry. Up to 10% of peripheral blood cells expressed GFP shortly after transplantation and approximately 6% after the longest follow-up of 23 months. Flow cytometric analysis of hematopoietic subpopulations showed that most of the GFP-expressing cells were granulocytes, although GFP-positive lymphocytes and monocytes were also detected. In summary, these results show that RD114-pseudotype oncoretroviral vectors are able to transduce hematopoietic long-term repopulating cells and, thus, may be useful for human stem cell gene therapy.
Introduction
The hematopoietic compartment is a particularly attractive target for the introduction of therapeutic genes because of its accessibility, potential for massive expansion and self-renewal, and availability of well-established technologies for manipulation ex vivo. Retroviral vectors have been the most commonly employed means of gene transfer in clinical gene therapy trials because they have the potential to integrate into the host genome to provide durable transgene expression in target cells. However, although very high gene transfer rates were obtained in murine hematopoietic stem cells, similarly high transduction efficiencies have not been reproduced in large animal models or human trials.
In recent years modifications in transduction conditions have led to improved retrovirus-mediated gene transfer into hematopoietic repopulating cells in large animals such as nonhuman primates1-4 or dogs.5 6 These changes included use of fibronectin fragment CH-296, addition of cytokine combinations with activity in early hematopoietic cells, and use of the gibbon ape leukemia virus (GALV) pseudotype. Although initial gene transfer efficiencies of more than 20% were achieved, long-term levels usually declined to between 0.1% and 5%. For most clinical gene therapy applications, however, higher gene transfer levels will be required.
A key feature of retroviral transduction is the binding of viral envelope protein to specific cell surface proteins. This interaction determines the range of cells that can be transduced by a given viral vector. In addition, efficient transduction by retrovirus vectors requires high-level expression of the appropriate cell surface virus receptor.7-10 Previous studies in dogs and nonhuman primates have shown that the use of the GALV envelope (GALV-pseudotype) resulted in superior transduction of hematopoietic repopulating cells compared with the amphotropic pseudotype. This observation correlated well with the demonstration that the GALV receptor Pit1 is more abundantly expressed than the amphotropic receptor Pit2 in CD34+ hematopoietic cells.1 6
An alternative viral pseudotype based on the feline endogenous retrovirus RD114 has been described.11 Vectors bearing the RD114 envelope have been shown to transduce human hematopoietic cells at high efficiency.12-14 On the basis of these studies we wanted to investigate whether RD114-pseudotype vectors would improve gene transfer rates into hematopoietic stem cells compared with the widely used GALV-pseudotype vectors. To demonstrate the effect of titer, we used a low-titer–producing FLYRD clone, providing titers in the range of PG13 cells, as well as a high-titer–producing FLYRD clone, which produced up to 29-fold higher titers. This experimental design allowed us to compare high and low titers within one packaging system and between 2 different packaging cell lines. With the use of a canine competitive repopulation assay, we have directly examined whether RD114-pseudotype vectors would increase gene transfer rates.
Subjects, materials, and methods
Animals
Dogs were raised and housed at the Fred Hutchinson Cancer Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. All animals were provided with commercial chow and chlorinated tap water ad libitum. They were observed for disease for at least 2 months before study. All dogs were immunized for papillomavirus, leptospirosis, distemper, hepatitis, and parvovirus. Marrow draws were performed under general anesthesia. The animals received broad-spectrum antibiotics and recombinant canine granulocyte-colony stimulating factor (Amgen, Thousand Oaks, CA) after transplant until the absolute neutrophil count reached > 1000/μL. As preparation for transplantation, all animals received a single myeloablative dose of 920 cGy total body irradiation administered from 2 opposing 60Cobalt sources at 7 cGy/minute.
Retrovirus vectors and cell lines
The LgGLSN, LNX, and LNY vectors contain the bacterial neomycin phosphotransferase (neo) gene, which conveys G418 resistance. The 3 vectors were designed to incorporate slightly different lengths between the neo gene and the 3′ long terminal repeat (LTR). This design permitted the use of a single pair of primers to amplify characteristic products by polymerase chain reaction (PCR) and to distinguish cells genetically marked by the respective vectors. The LgGLSN vector contained in addition a gene encoding an enhanced green fluorescent protein (GFP; Green Lantern; GIBCO-BRL, Bethesda, MD) downstream of the 5′ LTR. In this vector, the Moloney murine leukemia virus proline tRNA primer binding site is replaced by a glutamine tRNA primer binding site from an endogenous mouse virus, designed to prevent binding of cellular proteins that causes repression of transcription from the retroviral LTR.15 Packaging cell lines were generated by using FLYRD (RD114) cells11 and PG13 (GALV) cells.16Stable producer clones were used for the LgGLSN/LNX/LNY comparisons.
Production of virus-containing medium
Packaging cells were plated at a density of 5 × 104/mL per 15-cm dish (Corning, Corning, NY). When cells reached 90% confluency, medium was replaced with fresh Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; GIBCO-BRL) and 1% Pen/Strep (GIBCO-BRL). After further incubation at 37°C for 12 to 16 hours, supernatant was collected and filtered through 0.45-μm pore size filters (Nalge Nunc International, Rochester, NY) before freezing and storing at −70°C.
Endpoint titer determination
On day 0, 5 × 104 D17 or HT1080 cells were plated in 6-well plates (Corning) in DMEM. After incubation for 24 hours, the medium was replaced by fresh DMEM supplemented with 10% FBS, 1% Pen/Strep, and Polybrene 4 μg/mL. The cells were then infected with 100 μL diluted vector supernatant (10−1 to 10−4 serial dilutions). After 24 hours of exposure to the virus, medium was removed, and the cells were trypsinized and split 1:10 in 4 mL DMEM containing G418 (0.5 mg/mL). At day 5, the G418-containing medium was renewed to maintain the G418 activity. At day 12, or when nontransduced control D17 cells were completely dead, colonies were stained by using Coomassie blue and enumerated by using an inverted microscope. The endpoint titer (colony forming unit [CFU]/mL) was calculated by averaging the colony numbers per well after multiplication by the dilution factor.
CD34 enrichment of bone marrow cells
The method used has been described previously.6 17Briefly, bone marrow mononuclear cells were separated by a Ficoll gradient and labeled with biotinylated monoclonal antibody 1H6 (immunoglobulin G1 anti-canine CD34) 40 μg/mL at 4°C for 30 minutes. The cells were washed twice, incubated with streptavidin-conjugated microbeads for 30 minutes at 4°C, washed, and then separated by using an immunomagnetic column technique according to the manufacturer's instructions (Miltenyi Biotec, Auburn, CA).
Transduction of enriched bone marrow cells
CD34-enriched cells were prestimulated for 24 hours in Iscoves modified Dulbecco medium supplemented with 12.5% horse serum (GIBCO-BRL), 12.5% FBS (GIBCO-BRL), 10−6 M hydrocortisone (Sigma Chemical, St Louis, MO), 10−4 M 2-mercaptoethanol (Sigma), 1% glutamine, 1% penicillin/streptomycin (GIBCO-BRL) in the presence of FLT3-ligand (FLT3-L; Immunex, Seattle, WA), canine stem cell factor, and canine granulocyte–colony-stimulating factor (Amgen) at a concentration of 50 ng/mL. After prestimulation, equal numbers of CD34-enriched cells were placed in 75-cm2 canted-neck flasks (Corning) coated with CH-296 (RetroNectin; Takara Shuzo, Japan) at a concentration of 2 μg/cm2. Fresh retrovirus-containing medium was replaced 4 times over a period of 48 hours, in addition to cytokines (50 ng/mL) and protamine sulfate (8 μg/mL). After transduction, nonadherent and adherent cells were pooled, counted, and infused intravenously into the animal within 24 hours of total body irradiation.
PCR analysis
Genomic DNA was prepared using proteinase K (Sigma) as described.6 Amplification conditions for LNY and LNX vectors have been described6 and were also used for LgGLSN. Briefly, 300 ng genomic DNA was amplified with SN 2257 and ASN 3210 primers by using 0.5 U Taq polymerase (GIBCO-BRL). Conditions were optimized to produce linear amplification in the range of intensity of the positive PCR samples: denaturation at 95°C, followed by 26 to 28 cycles (depending on gene marking levels) of 64°C annealing (1 minute), and 95°C denaturation (1 minute) with a final extension at 72°C for 7 minutes. For the detection of LN, LNX, and LNY, PCR was performed in the presence of 10 mCi/mL (370 MBq/mL)32P deoxycytidine triphosphate, and PCR products were separated on a 6% polyacrylamide gel. To correct for different amounts of DNA in the templates, the β-actin gene was amplified in duplicates of 100 ng and 300 ng genomic DNA with the following primers: actin-1, 5′ TCC TGT GGC ATC CAC GAA ACT 3′; and actin-2, 5′ GAA GCA TTT GCG GTG GAC GAT 3′. PCR conditions for the amplification of β-actin were the same as for the neo gene, except that only 24 cycles were performed. Phosphor image analysis was used to quantify PCR signal intensities by comparing PCR signals in DNA from peripheral blood with PCR signals in DNA from a dilution series of single-vector copy HT1080/LN, -LNX, and -LNY cells. Calculated gene transfer percentages were corrected for the number of experimental arms and assume that peripheral blood cells contain an average of one copy of the corresponding vector per cell. For the analysis of hematopoietic subpopulations, DNA from T cells, granulocytes, and monocytes was isolated. Cell separations were performed by using monoclonal antibodies and a fluorescence-activated cell sorter (FACSVantage; Becton Dickinson, San Jose, CA). Granulocytes were purified by using the monoclonal antibody DM5,18 which recognizes a canine myeloid antigen. CD3+ T cells were purified by using canine-specific anti-CD3 antibody, CA17.2A12 (provided by Dr Peter Moore, University of California, Davis, CA), and monocytes were isolated by using a human anti-CD14 antibody (DAKO, Carpinteria, CA).
Flow cytometric analysis
Flow cytometric quantification of at least 20 000 propidium iodide (1 μg/mL) excluding forward and right-angle light scatter-gated events was performed on a FACSCalibur (Becton Dickinson). Analysis of flow cytometric data was performed with the use of CELLQuest v3.1f software with gating to exclude ≤ 0.01% control cells in the relevant region. Monoclonal antibodies conjugated to phycoerythrin, which had been shown to bind canine CD markers CD3 and CD14, were used to study canine T cells and monocytes, and DM5 was used to examine dog granulocytes. Gene transfer levels were corrected for the number of experimental arms used in the different dogs.
Northern blot analysis
RNA was prepared from 1 to 3 × 107 NIH3T3, FLYRD, HT 1080, and D17 cells by using Trizol (GIBCO-BRL). For Northern blot analysis, 10 μg total RNA per sample was separated in a 1.2% agarose gel containing 2.2 M formaldehyde and transferred to a nitrocellulose filter (Hybond-N+, Buckinghamshire, England). Membranes were hybridized at 65°C to RDR19 or Pit1 probes that had been radiolabeled with γ-32P dCTPs, using a random priming kit (Random Primed DNA Labeling Kit; Boehringer-Mannheim, Germany). Both probes had identical specific activities of 1 × 106 cpm/μg DNA. Filters were exposed to phosphor image screens for 72 hours and analyzed using Image Quant software (Molecular Dynamics, Sunnyvale, CA). After analysis filters were stripped using 0.5% sodium dodecyl sulfate and reprobed using the second probe.
Detection of helper virus
After transplantation, peripheral blood mononuclear cell DNA was assayed for the presence of recombinant helper virus genomes by envelope-specific PCR by using identical conditions to those used for amplification of the neo gene. Positive control log dilutions of DNA from PG13/LN packaging cells into normal dog DNA were quantified concurrently. The sequences of the primers used to amplify the GALV envelope gene have been described previously.1The sequences of the primers used to amplify the RD114 envelope gene were as follows: Rde324F, 5′ CCA GGA CAT CCA CAA ACT AGC C 3′; Rde792R, 5′ GTC TTG CCT CCG CAA GTT ACC 3′. The detection limit using this approach was approximately 1 copy per 10 000 cells. PCR analyses of peripheral blood samples for the presence of GALV and RD114 envelope sequences were negative in all animals, indicating the absence of helper-virus infection.
Results
CD34 enrichment and engraftment of transduced cells
We employed a competitive repopulation assay to study gene transfer efficiency into canine repopulating cells by using RD114-pseudotype retrovirus vectors. Five dogs were transplanted with transduced CD34-enriched autologous marrow cells to compare gene transfer efficiencies with LgGLSN (FLYRD) and LNY (PG13). In the first 3 animals a RD114-pseudotype vector (LNX) with a lower titer as compared with LgGLSN (RD114) was also included to determine the influence of titer and multiplicity of infection (MOI) on gene transfer.
Cell yield, purity, and recovery after CD34 enrichment are summarized in Table 1. After transplantation, one animal died because of infection before sustained leukocyte engraftment was achieved (E668). In the 4 surviving animals a stable absolute neutrophil count of > 500/μL was reached at a median of 17 days (range, 10-23 days), and a platelet count of > 50 000/μL was reached at a median of 58 days (range, 43-76 days). Neither the purity of enriched CD34 cells nor total cell number was predictive of the duration of thrombocytopenia. Monitoring was discontinued after 9 months in the animal with lowest overall transduction rate (E737). The remaining 3 dogs are now at 11, 22, and 23 months after transplant and healthy.
Ex vivo gene transfer efficiency in hematopoietic progenitors
CD34-enriched bone marrow cells were prestimulated and divided into 3 equal fractions. Then each fraction was transduced with the different vectors. In 3 dogs (E668, E523, and E695), the different fractions were transduced with LNY (PG13), LNX (FLYRD), and LgGLSN (FLYRD). In 2 additional dogs (E737 and E760), cells were divided into only 2 fractions and transduced with LNY and LgGLSN. Vector titers were measured on D17 cells. Because of the differences in titers, mean MOIs for the transduction of CD34-enriched cells were 1.2 (0.2-1.7), 3.8 (2.1-5), and 25.6 (3.8-37) for LNY, LNX, and LgGLSN, respectively (Table 2).
In all instances cells transduced with the GALV-pseudotyped virus expanded more (on average 1.62-fold) than the cells transduced with the RD114-pseudotype virus (on average 1.22-fold), although that difference was not statistically significant.
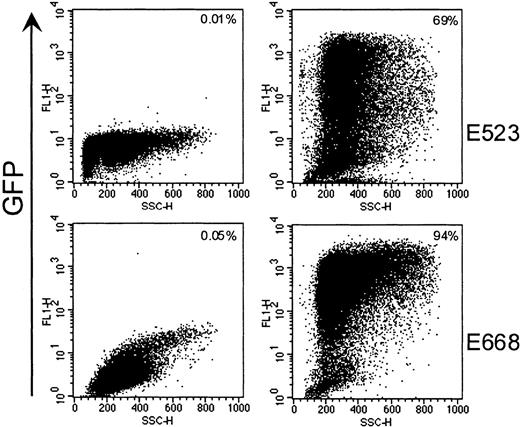
Expression rates were determined by flow cytometry in fractions transduced with GFP-containing vector LgGLSN (FLYRD). Gene transfer efficiency in these CD34-enriched cells was between 69% and 94% (Figure 1, Table 1). To exclude pseudotransduction (ie, GFP protein transfer from the virus-containing medium) as a possible explanation for the high gene transfer rates by flow cytometry, we transduced canine D17 cells with vector-containing medium from FLYRD/LgGLSN and measured GFP expression over time. After an initial increase during the first 2 days following transduction, GFP expression levels remained constant for more than 14 days (data not shown), suggesting that the initial gene transfer rates as determined by flow cytometry were indeed from integrated virus and not because of pseudotransduction.
High gene transfer efficiency into canine CD34+ cells by using a RD114-pseudotype vector.
Representative flow-cytometric analysis of CD34-enriched cells after transduction with RD114-pseudotype vector LgGLSN and before infusion into dogs E523 and E668. The left panel depicts mock-transduced CD34+ cells, and the right panel shows gene transfer efficiencies after transduction by LgGLSN.
High gene transfer efficiency into canine CD34+ cells by using a RD114-pseudotype vector.
Representative flow-cytometric analysis of CD34-enriched cells after transduction with RD114-pseudotype vector LgGLSN and before infusion into dogs E523 and E668. The left panel depicts mock-transduced CD34+ cells, and the right panel shows gene transfer efficiencies after transduction by LgGLSN.
In vivo comparison between the GALV- and RD114-pseudotype vectors
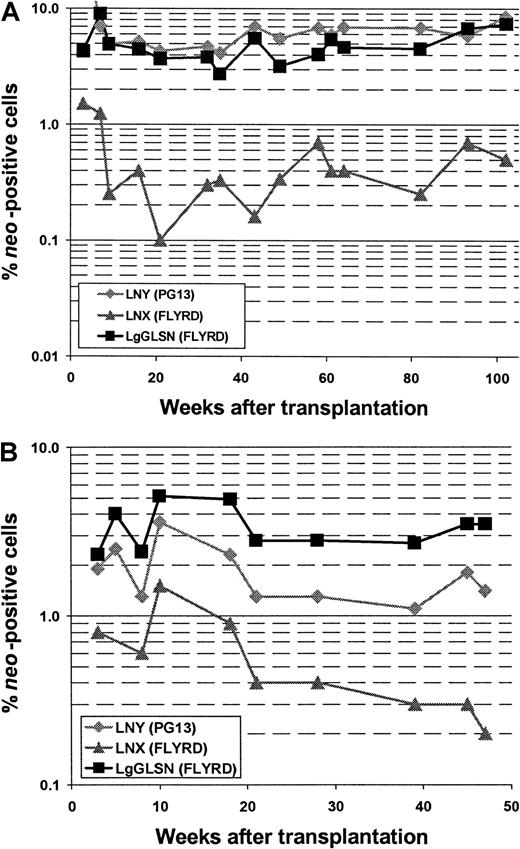
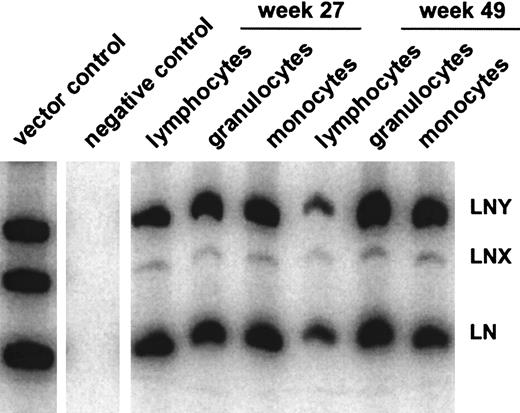
In all 5 animals gene transfer efficiencies were compared between the high-titer RD114-pseudotype vector LgGLSN and the GALV-pseudotype vector LNY. In 3 of the 5 animals, a low-titer RD114-pseudotype vector (LNX) was included; the titer of this vector was similar to the titer of the GALV-pseudotype vector. Gene transfer rates after transplantation were measured by flow cytometry (for LgGLSN only) and by PCR (for all vectors used) of peripheral blood leukocytes. In contrast to the high-titer vector, the low-titer RD114-pseudotype vector performed significantly worse compared with the GALV-pseudotype vector even though both vectors had similar titers on D17 cells. These results suggest that higher titer viruses will be required for efficient transduction using RD114-pseudotype vectors produced by FLYRD. The use of the low-titer RD114-psuedotype vector LNX also resulted in significantly inferior gene transfer compared with the high-titer RD114-pseudotype vector LgGLSN (P < .0002), indicating that vector concentration is important for efficient gene transfer into hematopoietic repopulating cells using FLYRD-derived vectors. Representative analyses of 2 dogs (E523 and E695) are shown in Figure 2. In E695 and E760 the use of LgGLSN led to slightly higher gene transfer levels compared with the PG13-derived vector. In E737, the overall gene transfer rate was low, and only LgGLSN was detected between 0.1% and 0.01%. In E668 the gene transfer levels for LNY were higher than for LgGLSN with very short follow-up, and in E523 no difference between the 2 vectors was detected. To demonstrate successful gene transfer into primitive multipotential hematopoietic cells, peripheral blood cells from 3 dogs were enriched using fluorescence-activated cell sorting for granulocytes (DM-5), T lymphocytes (CD 3), and monocytes (CD14). The presence of all 3 vectors was demonstrated by PCR in granulocytes, T lymphocytes, and monocytes in all 3 dogs. Figure3 shows the PCR results from E523 at 27 and 49 weeks after transplant. In summary, 4 of 5 dogs demonstrated at least equivalent gene transfer efficiency using the high-titer RD114-pseudotype vector compared with the GALV-pseudotype vector.
Comparison between RD114- and GALV-pseudotype vectors in a competitive repopulation assay in dogs.
Detection of vector sequences in peripheral blood cells from dogs E523 (A) and E695 (B) transplanted with CD34-enriched marrow cells transduced with either RD114-pseudotype vectors produced from a high-titer clone of a human packaging cell line (LgGLSN), by a low-titer clone of the same human packaging cell line (LNX), or by GALV-pseudotype vectors produced by a mouse packaging cell line (LNY). Percentage of vector-positive DNA as measured by phosphor image analysis of signal intensities for LNY, LNX, and LgGLSN corrected for the amount of DNA and the number of experimental arms.
Comparison between RD114- and GALV-pseudotype vectors in a competitive repopulation assay in dogs.
Detection of vector sequences in peripheral blood cells from dogs E523 (A) and E695 (B) transplanted with CD34-enriched marrow cells transduced with either RD114-pseudotype vectors produced from a high-titer clone of a human packaging cell line (LgGLSN), by a low-titer clone of the same human packaging cell line (LNX), or by GALV-pseudotype vectors produced by a mouse packaging cell line (LNY). Percentage of vector-positive DNA as measured by phosphor image analysis of signal intensities for LNY, LNX, and LgGLSN corrected for the amount of DNA and the number of experimental arms.
Gene transfer rates in hematopoietic subpopulations.
Detection of vector sequences in different hematopoietic lineages in peripheral blood from dog E523 at 27 and 49 weeks after transplantation with transduced CD34-enriched bone marrow cells.
Gene transfer rates in hematopoietic subpopulations.
Detection of vector sequences in different hematopoietic lineages in peripheral blood from dog E523 at 27 and 49 weeks after transplantation with transduced CD34-enriched bone marrow cells.
Analysis of GFP expression in peripheral blood cells after transplantation
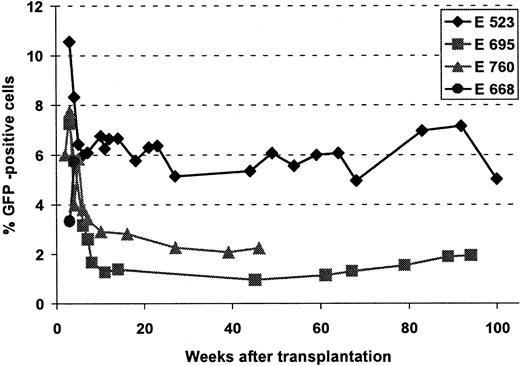
Because one of the RD114-pseudotype vectors (LgGLSN) contained the GFP gene, expression from integrated vector could be measured by flow cytometry. Figure 4 provides a summary of GFP expression in peripheral blood samples over time in 4 of the 5 animals. In the fifth dog, E737, gene transfer was very low and GFP expression could not be detected. In the other 4 dogs initial expression levels between 4% and 10% were achieved. However, within the first 10 to 15 weeks after transplantation expression declined to a baseline level between 1% and 7%, which has been sustained for 1 to almost 2 years in 3 dogs.
Gene transfer into long-term repopulating cells, using a RD114-pseudotype vector.
Flow cytometric analysis of GFP-expressing cells in peripheral blood at different time points after transplantation. Dog E668 died at 35 days; therefore, only short-term follow-up is available; in E737 gene transfer level was very low and expression of GFP was never detectable.
Gene transfer into long-term repopulating cells, using a RD114-pseudotype vector.
Flow cytometric analysis of GFP-expressing cells in peripheral blood at different time points after transplantation. Dog E668 died at 35 days; therefore, only short-term follow-up is available; in E737 gene transfer level was very low and expression of GFP was never detectable.
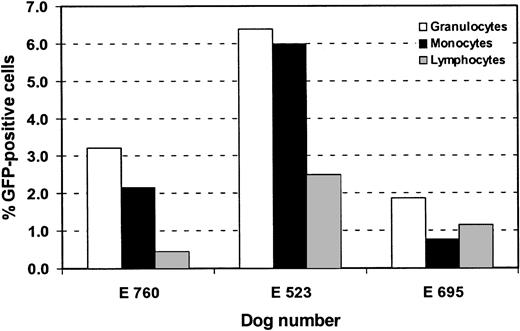
To assess gene expression in specific hematopoietic lineages, peripheral blood cells in long-term expressing dogs were labeled with monoclonal antibodies against granulocytes (DM-5), T lymphocytes (CD3), and monocytes (CD14) and were analyzed by flow cytometry at different time points. Figure 5 summarizes the results in the 3 dogs. Sustained GFP expression was detected in all subsets examined. Very similar patterns of GFP expression in subpopulations were seen at different time points after transplantation (data not shown). The percentage of GFP-expressing cells was generally higher in granulocytes (up to 8.1%) than in T lymphocytes (up to 3.5%) or monocytes (up to 5.7%). Two animals were examined for GFP expression in platelets and were found to have between 1.2% and 1.3% GFP-positive platelets at 9 and 21 months after transplant. We were not able to detect GFP in erythrocytes.
Retroviral gene expression in hematopoietic subpopulations, using a RD114-pseudotype vector.
Flow cytometric analysis of GFP expression in granulocytes (DM5), monocytes (CD14), and T lymphocytes (CD3). Bars represent percentage of GFP-positive cells in these subpopulations. Analysis was done at different time points after transplantation. E760 was analyzed at week 10, E523 at week 49, and E695 at week 61.
Retroviral gene expression in hematopoietic subpopulations, using a RD114-pseudotype vector.
Flow cytometric analysis of GFP expression in granulocytes (DM5), monocytes (CD14), and T lymphocytes (CD3). Bars represent percentage of GFP-positive cells in these subpopulations. Analysis was done at different time points after transplantation. E760 was analyzed at week 10, E523 at week 49, and E695 at week 61.
Northern analysis of retroviral receptor expression
To determine whether titer differences observed between the RD114- and the GALV-pseudotype vectors could be a consequence of retroviral receptor expression and to compare receptor levels in cells of human and canine origin, we analyzed messenger RNA levels of GALV and RD114 receptor on different cell lines. Expression levels of Pit1 were comparable to the RD114 receptor (RDR) in D17 cells, which was in contrast to human cell lines (HT1080, FLYRD), in which Pit1 expression was more abundant than RDR expression (data not shown). These findings indicate that the titer differences observed between RD114- and GALV-pseudotype vectors on D17 and HT1080 cells were not because of differential expression of Pit1 and RDR.
Discussion
The low gene transfer efficiency into hematopoietic long-term repopulating cells has been a major obstacle in the application of stem cell–based gene therapy. A significant barrier to efficient transduction with the widely used GALV-pseudotype or amphotropic vectors has been the relatively low-level expression of retroviral receptors Pit1 and Pit2 on hematopoietic stem/progenitor cells.1 10 In this study we used a competitive repopulation assay in the dog to evaluate an alternative viral pseudotype based on the feline endogenous retrovirus RD114. We have shown that RD114-pseudotype vectors produced by FLYRD were able to transduce long-term repopulating cells in lethally irradiated dogs. GFP-positive peripheral blood granulocytes, lymphocytes, monocytes, and platelets have been detected for almost 2 years to date, indicating that RD114-pseudotype vectors can transduce true multipotential hematopoietic stem cells.
In all 5 dogs studied we compared transduction efficiency of RD114-pseudotype vectors with that of the GALV-pseudotype vector. All 3 vectors used in this study contained the neo gene, and one of the RD114-pseudotype vectors (LgGLSN) also contained the GFP gene. Thus, the comparison between the RD114-pseudotype vectors and GALV-pseudotype vector was based on PCR analysis of the differentneo signatures in the different vectors. The high-titer RD114-pseudotype vector resulted in a marginally superior gene transfer efficiency in long-term repopulating cells. However, the overall gene transfer levels were not markedly different from dogs that received CD34-enriched cells transduced by GALV-pseudotype vectors in this study or in previous studies.5 6
Although we achieved long-term marking of, and gene expression in, hematopoietic stem cells using RD114-pseudotype vectors, the in vivo gene transfer levels after hematopoietic reconstitution were significantly lower than transduction rates in CD34+ cells as determined by flow cytometric analysis before infusion into the animal. It is unlikely that the high gene transfer levels demonstrated before infusion were due to pseudotransduction because we demonstrated constant GFP expression levels over an extended period of time in cells transduced by RD114-pseudotype vectors. A more likely explanation for this discrepancy is the recently described effect of the conditioned medium generated from the RD114-pseudotype FLYRD packaging cells on hematopoietic progenitor/stem cells. Kelly et al13 have shown that conditioned medium from HT1080 cells, which is the parental cell line used to generate FLYRD cells, led to the differentiation of primitive hematopoietic cells and subsequent loss of engraftment in the nonobese diabetic/severe combined immunodeficiency model. We did not analyze the phenotype of our transduced CD34-enriched cells; however, the number of culture colony-forming units recovered after transduction from the different experimental arms was consistently lower after transduction with LgGLSN (FLYRD) compared with the GALV-pseudotype vector LNY (data not shown), suggesting the loss of more primitive cells under these culture conditions. This finding would also suggest that engraftment of RD114-transduced cells, and thus gene transfer levels in the dog, could be considerably improved by using modified transduction conditions, including the avoidance of exposure to the HT1080-conditioned medium as described by Kelly et al.13 Alternatively, other human packaging cells could be used to generate RD114-pseudotype vectors. There is evidence that vectors produced by human 293 cells may be superior to vectors produced by murine packaging cells, which have been shown to produce inhibitory factors.20-22
In 3 of the dogs we included a third experimental arm by using a RD114-pseudotype vector from a low-titer FLYRD clone. The titer differences between the high-titer FLYRD/LgGLSN and the low-titer FLYRD/LNX cells were up to 29-fold. This finding resulted in a marked difference in the gene transfer rates in vivo between the 2 vectors, suggesting that a higher MOI facilitated the transduction of canine repopulating cells. It is important to note that, although titers for the PG13-derived LNY vector were similar to the low-titer RD114-pseudotype vector, transduction efficiency in vivo was substantially higher with the PG13-derived vectors. These data suggest that, although titers and MOI are important, comparisons can only be made within one packaging system. Moreover, high-titer vector appears to be required for efficient transduction of hematopoietic-repopulating cells by RD114-pseudotype vectors, although we have not ruled out the possibility that the low-titer RD114 producer cell line was generating an increased amount of noninfectious particles that could have competed with the same receptor and thereby interfered with transduction. The titer differences observed on D17 and HT1080 cells between the high-titer RD114-pseudotype and the GALV-pseudotype vector were not due to differences in the corresponding receptor expression, because expression of Pit1 and RDR were comparable in D17 cells, and Pit1 was even more abundant in HT1080 cells than RDR.
After an initial decline in GFP-positive cells, we have observed stable persistence of gene-modified cells for almost 2 years at last assessment. The initial decline was most likely due to transduced progenitor cells and not due to gene silencing because we saw a similar decline when peripheral blood cells were analyzed for the presence of the transgene by PCR. The possible development of an immune response against GFP-containing cells is an unlikely explanation for the initial decline because GFP-positive cells remained detectable at stable levels for almost 2 years.
In conclusion, our data show stable transduction of canine hematopoietic repopulating cells by using RD114-pseudotype oncoretroviral vectors. The fact that we were able to detect gene-modified cells in multiple lineages for almost 2 years to date suggests that the RD114-pseudotype was able to mediate transduction of a true multipotential hematopoietic stem cell. In light of high gene transfer rates observed in human progenitor cells and nonobese diabetic/severe combined immunodeficiency repopulating cells and the more abundant expression of the RD114 receptor on human cells as compared with dog cells, the application of RD114-pseudotype vectors offers a promising alternative in human stem cell gene therapy.
We thank Eric Bell, Alix Smith, Lori Ausburn, the technicians of the Canine Shared Resource, the hematology and pathology laboratories for their technical assistance, and Bonnie Larson and Helen Crawford for assistance with the preparation of the manuscript. We also thank Drs Peter McSweeney, Brenda Sandmaier, and Peter Moore for providing us with canine antibodies used in this study.
Supported in part by grants HL36444, DK47754, and DK56465 from the National Institutes of Health. M.G. and P.A.H. are supported by the German Krebshilfe. J.E.J.R. was supported by Fellowship DRG081 of the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation. H.P.K. is a Markey Molecular Medicine Investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hans-Peter Kiem, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N D1-100, PO Box 19024, Seattle, WA 98109-1024; e-mail: hkiem@fhcrc.org.