Abstract
Chromosomal translocation t(6;14)(p21.1;q32.3) has been reported as a rare but recurrent event not only in myeloma and plasma cell leukemia but also in diffuse large B-cell non-Hodgkin lymphoma (B-NHL) (diffuse large B-cell lymphoma [DLBCL]) and splenic lymphoma with villous lymphocytes (SLVL); however, the nature of the target gene(s) has not been determined. This study identified t(6;14)(p21.1;q32.3) in 3 cases of transformed extranodal marginal zone B-NHL, in 1 case of SLVL, and in 1 case of a low-grade B-cell lymphoproliferative disorder. In a sixth case, a CD5+ DLBCL, the translocation was identified by molecular cloning in the absence of cytogenetically detectable change. Two chromosomal translocation breakpoints were cloned by using long-distance inverse polymerase chain reaction methods. Comparison with the genomic sequence for chromosome 6p21.1 showed breakpoints approximately 59 and 73.5 kilobases 5′ of the cyclin D3 (CCND3) gene with no other identifiable transcribed sequences in the intervening region. Although Southern blotting with derived genomic 6p21.1 probes failed to detect other rearrangements, fluorescent in situ hybridization assays, using BAC (bacterial artificial chromosome) clones spanning and flanking theCCND3 locus, along with probes for IGHconfirmed localization of 6p21.1 breakpoints within the same region, as well as fusion of the CCND3 and IGH loci. Furthermore, in all cases, high-level expression of CCND3was demonstrated at RNA and/or protein levels by Northern and Western blotting and by immunohistochemistry. These data implicateCCND3 as a dominant oncogene in the pathogenesis and transformation in several histologic subtypes of mature B-cell malignancies with t(6;14)(p21.1;q32.3) and suggest thatCCND3 overexpression seen in about 10% of DLBCL cases may have a genetic basis.
Introduction
The lymphomas and leukemias of mature B cells are a biologically and histologically heterogeneous group of malignancies.1 Their molecular pathogenesis for the most part remains unknown, although cloning of chromosomal translocations targeted to the immunoglobulin (IG) loci continues to allow the identification of novel dominant oncogenes and to define new pathogenic mechanisms.2 These translocations result in the deregulated expression of genes involved in several pathways, including the control of programmed cell death (apoptosis) and proliferation.
IG translocations that involve genes controlling progression through the cell cycle have been described. Cyclin D1 (CCND1) on chromosome 11q13 is involved in the t(11;14)(q13;q32.3). This translocation is found in nearly all cases of mantle cell lymphoma but also in some cases of myeloma, in marginal zone malignancies, such as splenic lymphoma with villous lymphocytes (SLVL), and in B-cell prolymphocytic leukemia.3-6 The cyclin-dependent kinase gene (CDK6) in chromosome band 7q22 is involved in t(7;14)(q22;q32) or more commonly in t(2;7)(p12;q22); these translocations appear to be specific for a subset of SLVL.7,8 The involvement of the cell-cycle regulatory genes in marginal zone B-cell malignancies was unanticipated, because they are characteristically indolent diseases. Whether these genes might have functions in B-cell differentiation other than cell-cycle regulation in mature B cells is not yet clear; cyclin D3 expression, for example, is associated with differentiation of some cell types.9
Cytogenetic abnormalities of chromosome band 6p21 reported in mature B-cell malignancies are various and include translocations, amplifications, and deletions.10,11 Such translocations in diffuse large B-cell lymphoma (DLBCL) often involve the BCL6gene on chromosome band 3q27 and may juxtapose BCL6 with either the histone H4 gene or the PIM1oncogene.12,13 The t(6;14)(p12∼p21;q32) has been previously reported in a variety of B-cell malignancies, principally in myeloma and plasma cell leukemia but also in DLBCL and splenic marginal zone lymphomas (SMZLs).14-18 Here we report the recurrent involvement of a third cell-cycle regulatory gene, cyclin D3/CCND3, in 6 cases of mature B-cell malignancy, including one case of CD5+ DLBCL, through its direct involvement in t(6;14)(p21.1;q32.3).
Patients, materials, and methods
Patient clinical characteristics
Cytogenetics
Long-distance inverse polymerase chain reaction
Long-distance inverse polymerase chain reaction (LDI-PCR) and DNA sequencing of IGHJ rearrangements was performed as previously described.22 Primers designed to amplify illegitimate IG switch rearrangements observed with a 3′ Sγ probe on Southern blot were used. In case 1 from Table 1, DNA was digested to completion with SphI and ligated at low concentration, followed by nested PCR using γR1 primer (5′-GACCAGTGGACACTGTTCTCAGATGG-3′) and γF4 primer (5′-CACGCAGAAGAGCCTCTCCCTGT-3′) and γR2 primer (5′-CCTCCAAGGCCCTTTTCTTCTGTG-3′) and γF5 primer (5′-CCCAGCATGGAAATAAAGCACCC-3′). In case 2 from Table 1, DNA was digested with Taq1 and amplified first with primers γR1 and γF1 (5′-TCCCTGAGGTGGCACCGATG-3′) and then in a second round of amplification with primers γR2 and γF2 (5′-CCAGAGCTGAGGCCAAGCTAGAG-3′). The sizes of the PCR products obtained (1.8 and 2.5 kilobases [kb], respectively) corresponded to the sizes of the rearranged fragments seen on Southern blot (allowing for the position of the LDI-PCR primers), indicating that neither fragment arose because of PCR artifact.
Radiation hybrid mapping
Radiation hybrid (RH) mapping was performed with DNA primers 1060 (5′-TCACCAGAGTTTACCCCTGT-3′) and 1061 (5′-TGCATATACATCCATCCATGC-3′) derived from the breakpoint sequence of the cloned LDI-PCR fragment from case 1 by means of the Stanford G3 Panel (Research Genetics, Huntsville, AL). Map localization was calculated on the Stanford Human Genome Center RH server.47
Bacterial artificial chromosome and plasmid artificial chromosome screening and sequencing
Bacterial artificial chromosome (BAC) and plasmid artificial chromosome (PAC) clones derived from the chromosome band 6p21 contig Chr_6ctg648 were screened by PCR, using primers 1060/1061. Positive clones were rescreened with a second primer pair derived from the same breakpoint sequence (1077, 5′-CATTGTATGGATGTATATGC-3′; 1078, 5′-TACAAAAGACCTGCTCAAGG-3′). Screening of a gridded PAC library was also performed by hybridization, using a single-copy probe derived from the breakpoint sequence.23 Positive clones were rehybridized twice with the breakpoint clone and with aCCND3 expressed sequence tag (EST) clone spanning the open reading frame to show colocalization of the cloned breakpoint withCCND3. The complete genomic sequence of the region around the CCND3 gene derived from clones RP11-533O20 and RP5-973N23 was obtained at the Sanger Centre, using methods previously described.24 The accession numbers for these finished sequences are AL513008 and AL160163, respectively.
Fluorescence in situ hybridization studies
Fluorescence in situ hybridization (FISH) for the detection of breakpoints in the IGH locus was performed, using the LSIIGH Break-apart probe (Vysis, Downers Grove, IL). For the detection of breaks affecting the CCND3 locus, 2 different interphase FISH assays were developed. The first involved the use of probes closely flanking the breakpoint cloned from the index patient 1; these should colocalize in normal cells but should be separated in cells carrying a CCND3 translocation. Pooled PAC clones RP1-321B9 and RP1-139D8 were chosen as centromeric and PAC RP5-973N23 and BAC RP11-298J23 as telomeric probes for this approach. The second assay aimed to identify fusion of chromosomal regions 6p21 and 14q32.3 by means of differently labeled probes spanning the CCND3and IGH loci, respectively. These probes should be separated in normal cells but fused on both der(6)t(6;14)(p21.1;q32.3) and der(14)t(6;14)(p21.1;q32.3) by a t(6;14)(p21.1;q32.3) affectingIGH and CCND3. The CCND3 probes were PAC RP5-973N23 containing the CCND3 gene as well as the flanking clones RP1-321B9, RP1-139D8, and RP11-298J23. A pool of cosmids for the IGVH and IGCα regions served as probe for the IGH locus.25 PAC and BAC DNA was prepared by using the Perfectprep Plasmid Maxi Kit (Eppendorf, Cologne, Germany) and labeled with Spectrumgreen-dUTP and Spectrumorange-dUTP (Vysis, Downers Grove, IL), respectively, using random-primed DNA labeling (Life Technologies, Eggenstein, Germany). Each PAC/BAC clone (200 ng) was coprecipitated with 5 μg Cot1 DNA (Life Technologies) and dissolved in 10 μL hybridization buffer containing 50% formamide, 10% dextran sulfate, and 2× sodium chloride/sodium citrate. FISH was performed on cells left from cytogenetic analysis or on cytospin preparations of cells stored in dimethyl sulfoxide. Preparation of slides, pretreatment with pepsin, and simultaneous denaturation and hybridization were performed as described.20 Experiments were evaluated by using a fluorescence microscope (Zeiss, Göttingen, Germany) equipped with appropriate filter sets (AHF, Tübingen, Germany) and were documented using the ISIS program (MetaSystems, Altlussheim, Germany).
Southern, Northern, and Western blots
High molecular weight DNA obtained from all cases was Southern blotted and probed with the chromosome band 6p21 breakpoint clone derived from patient 1 and IGH probes using previously described methods and probes.26,27 Northern and Western blotting were performed as described.27 For Northern blotting, a 2.0-kb complementary DNA (cDNA) spanning theCCND3 open-reading frame from EST clone, IMAGE No. 2 015 357 was used. The cyclin D3 monoclonal antibody (Mab) was clone DCS22 (Oncogene Research Products, Boston, MA) and the p27Kip1 MAb was clone DCS72-F6 (Neomarkers, Freemont, CA).
Immunohistochemistry
Paraffin section and cytospin immunohistochemistry were performed by using routine methods. The cyclin D3 Mab was identical to that used for Western blots (clone DCS22). Immunostains were performed on an automated immunostainer (Ventana, Tucson, AZ) by using a streptavidin technique. The primary antibody was used at a 1:20 dilution with microwave antigen retrieval and citrate buffer, pH 6.0. Reactive tonsil was used as positive control and the negative control omitted the primary antibody.
Results
Molecular cloning of t(6;14)(p21.1;q32.3)
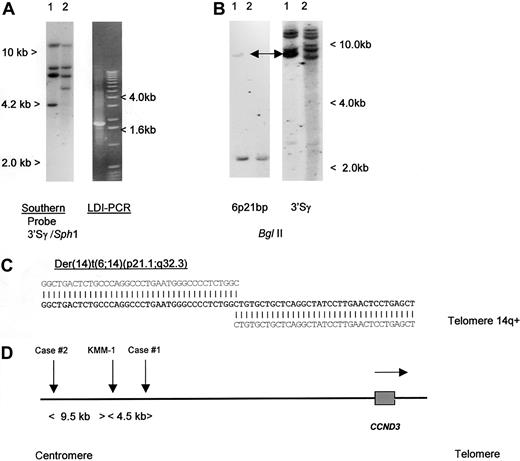
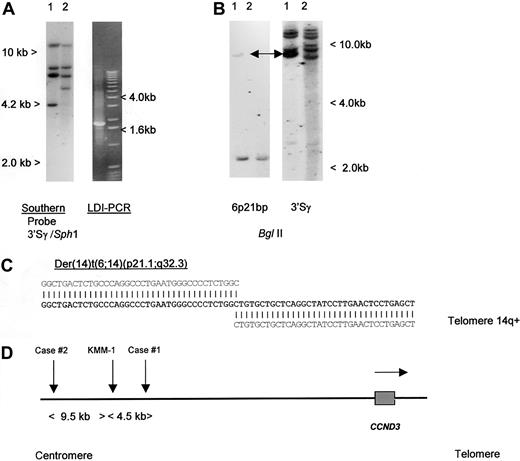
To detect IGH translocations in the absence of cytogenetic data, we have developed LDI-PCR methods to clone rearrangements involving the JH and S region segments of the IGH locus22 (T.S., T.G.W., M.J.S.D., unpublished data, December 2000). Translocations targeted to the S regions may also be detected by Southern blot as the lack of comigration of 5′ and 3′ rearranged IGHSfragments.26 We initially studied a case of aggressive CD5+ DLBCL in which cytogenetic studies had shown a complex karyotype with multiple structural abnormalities, including del(6p) but no cytogenetically obvious t(6;14)(p21.1;q32.3) (Table 1, patient 1).19 No suitable cytogenetic material was available from this case to permit more detailed FISH analysis on metaphase preparations. Analysis of the configuration of the IGH loci was performed, using both Southern blot and LDI-PCR methods. Southern blot showed 2 clonal IGHJ rearrangements; both were cloned by LDI-PCR and sequenced and showed productive VDJ andDJ rearrangements (data not shown). Further analysis of Southern blot with a panel of IGHS probes, including 5′ and 3′ Sγ probes showed an illegitimate 3′ Sγ fragment of 4.2 kb in SphI digestion (Figure1A). This fragment was cloned by using 3′ Sγ LDI-PCR and sequenced (Accession No. AF388309); the sequence is partially shown in Figure 1C. Beyond the Sγ2 sequence, all homology with the IGH locus was lost. Because the size of the amplified fragment corresponded to the size anticipated from the Southern blotting experiments, it was unlikely to have been derived from an LDI-PCR artifact. This finding was confirmed by showing comigration of the rearranged illegitimate IGHS fragment with the rearranged fragment seen on Southern blot with the derived genomic probe (Figure 1B). At the time of cloning, this sequence had no homology to any of the genomic or EST sequences on the public databases.
Molecular cloning of illegitimate 3′ switch γ rearrangement from a case of CD5+ DLBCL.
(A) Left- hand panel: Southern blot using an IGH 3′Sγ probe. Tumor DNA (lane 1) showed a 3′Sγ rearranged fragment of 4.2 kb that did not comigrate with a 5′Sγ probe (data not shown). Lane 2 contained germline control DNA. The 4.2-kb-SphI rearranged fragment was amplified by LDI-PCR. Right-hand panel: The LDI-PCR target for IGH 3′Sγ yielded a 1.8-kb product that corresponded to the 4.2-kb fragment seen on Southern blot. The LDI-PCR product was subcloned and sequenced. (B) Left-hand panel: Southern blot of tumor and normal DNA digested with BglII and hybridized with chromosome 6p21 breakpoint probe, showing single rearrangement in the tumor DNA. Comigrating rearranged bands are shown with an arrow. Right-hand panel: Same filter rehybridized with IGH 3′Sγ, showing comigration of rearranged 6p21 and IGH sequences (arrows). (C) Sequence of the der(14)t(6;14)(p21;q32) breakpoint from case 1 (Accession No. AF388309). Bold letters denote breakpoint sequence; above, IGHSγ2 sequence (gi 1 066 109 gb U39 934.1 HSU39 934); and below, 6p21.1 BAC clone RP11-533O20, which contained both the breakpoint andCCND3. (D) Ideogram to show the localization of chromosome 6p21.1 breakpoints. All 3 cloned breakpoints fell 5′ (centromeric) of the cyclin D3/CCND3 gene, in regions containing repetitive DNA sequences. Within the BAC clone RP11-533O20, the breakpoints were located at nucleotides 79567 (case 1, AF388309), 84057 (myeloma cell line KMM-1, AF364073),29 and 93510 (case 2, AF388310). Horizontal arrow denotes transcriptional orientation ofCCND3.
Molecular cloning of illegitimate 3′ switch γ rearrangement from a case of CD5+ DLBCL.
(A) Left- hand panel: Southern blot using an IGH 3′Sγ probe. Tumor DNA (lane 1) showed a 3′Sγ rearranged fragment of 4.2 kb that did not comigrate with a 5′Sγ probe (data not shown). Lane 2 contained germline control DNA. The 4.2-kb-SphI rearranged fragment was amplified by LDI-PCR. Right-hand panel: The LDI-PCR target for IGH 3′Sγ yielded a 1.8-kb product that corresponded to the 4.2-kb fragment seen on Southern blot. The LDI-PCR product was subcloned and sequenced. (B) Left-hand panel: Southern blot of tumor and normal DNA digested with BglII and hybridized with chromosome 6p21 breakpoint probe, showing single rearrangement in the tumor DNA. Comigrating rearranged bands are shown with an arrow. Right-hand panel: Same filter rehybridized with IGH 3′Sγ, showing comigration of rearranged 6p21 and IGH sequences (arrows). (C) Sequence of the der(14)t(6;14)(p21;q32) breakpoint from case 1 (Accession No. AF388309). Bold letters denote breakpoint sequence; above, IGHSγ2 sequence (gi 1 066 109 gb U39 934.1 HSU39 934); and below, 6p21.1 BAC clone RP11-533O20, which contained both the breakpoint andCCND3. (D) Ideogram to show the localization of chromosome 6p21.1 breakpoints. All 3 cloned breakpoints fell 5′ (centromeric) of the cyclin D3/CCND3 gene, in regions containing repetitive DNA sequences. Within the BAC clone RP11-533O20, the breakpoints were located at nucleotides 79567 (case 1, AF388309), 84057 (myeloma cell line KMM-1, AF364073),29 and 93510 (case 2, AF388310). Horizontal arrow denotes transcriptional orientation ofCCND3.
Breakpoints of t(6;14)(p21.1;q32.3) lie 5′ of theCCND3 gene
To define the chromosomal localization of this novel sequence, RH mapping, BAC library screening using PCR primers, and screening of a gridded PAC library using a derived single-copy probe were performed. RH mapping of a sequence-tagged site derived from the LDI-PCR fragment placed the breakpoint on the short arm of chromosome 6 with SHGC-11207 being the most closely linked marker (distance 8cR, LOD-Score 14.09). The EST SHGC-11207 located in the interval between D6D1616 and D6S427 (59.6-73.9 cM) at 2422 cR10000 (F) represented the CCND3gene.49CCND3 has been cytogenetically assigned to the chromosomal region 6p12∼21.1.28 Database searches using the BLAST algorithm50 revealed PAC clone RP5-973N23 to contain CCND3. This PAC clone is part of the contig Chr_6ctg6 established by the Sanger Center.48 BAC and PAC clones from Chr_6ctg6 covering more than 1 Mb around theCCND3 locus were screened by PCR and revealed only the one BAC clone (RP11-533O20) containing the translocation breakpoint. This clone contained the EST marker stSG8750 specific for the 3′ end ofCCND3. Furthermore, one of the isolated PAC clones hybridized not only with the translocation breakpoint probe but also with a CCND3 cDNA probe confirming that the 2 sequences were immediately adjacent. Restriction mapping with rare-cutting restriction enzymes indicated that the breakpoint clone was about 40 kb distant from the CCND3 gene (data not shown). To determine the precise relationship of the breakpoint to CCND3, the complete sequences of overlapping clones RP11-533O20 and RP5-973N23 were determined24 (and GenBank Accession No. AL513008 andAL160163). These data showed that the breakpoint in patient 1 lay approximately 59 kb 5′ of the CCND3 coding sequences. Analysis of the intervening DNA sequence between the breakpoint andCCND3 failed to detect any other obvious transcribed DNA sequences.51
To determine if other cases with cytogenetically defined t(6;14)(p21.1;q32.3) also had breaks within the same region, Southern blotting was initially performed, using a genomic probe derived from the breakpoint of the index patient. No additional breaks were detected, suggesting that 6p21.1 breaks were dispersed. LDI-PCR cloning and sequencing of case 2, Table 1, was therefore undertaken. As with case 1, the breakpoint on chromosome 14 fell within IGHS, but in this case in Sγ1. Comparison of the derived sequence (Accession No. AF388310) with that of RP11-533O20 showed the breakpoint to lie 73.5 kb 5′ of CCND3. During the preparation of this article, another group reported the cloning of the same chromosomal translocation from a myeloma cell line KMM-1.29 The breakpoint in this cell line (GenBank Accession No. AF364072 and AF364073) mapped 4490 base pair (bp) centromeric of that from case 1 reported here, suggesting some possible clustering. A schema of the translocation breakpoints is shown in Figure 1D; all breakpoints were 5′ of CCND3 and resulted in juxtaposition with IGHS sequences. None of the 6p21.1 breakpoint sequences contained identifiable IG recombination signal sequences, and all were adjacent to Alu repetitive sequences.
CCND3 is the target gene of t(6;14)(p21.1;q32.3)
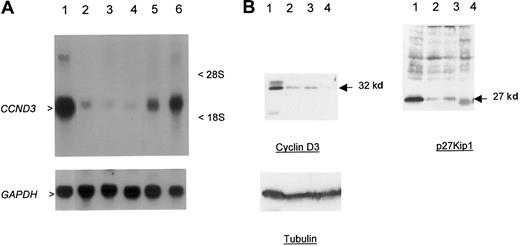
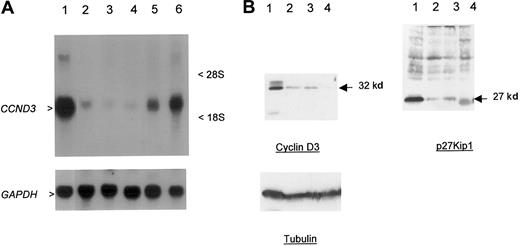
The close physical linkage of the breakpoints to CCND3, the lack of any obvious transcribed sequences in the intervening region, and the previous demonstration of involvement of other G1 cell-cycle regulatory proteins in B-cell malignancies suggested thatCCND3 was the probable target gene of t(6;14)(p21.1;q32.3). To confirm this, Northern and Western blotting was performed in cases 1 and 2 in which there was suitable material available (Figure2A-B). Marked overexpression ofCCND3 RNA in case 1 and cyclin D3 protein in both cases were seen in comparison to other B-cell malignancies and lymphoid cell lines lacking cytogenetic abnormalities of chromosome band 6p21. Immunohistochemistry showed that all the neoplastic lymphoid cells expressed cyclin D3 (Figure 3A-B). Both cases also showed high-level expression of p27Kip1. Together, these data indicated that the illegitimate IGH Sγ rearrangement in both cases represented a t(6;14)(p21.1;q32.3), with breakpoints adjacent to CCND3, resulting in deregulated expression of cyclin D3.
Overexpression of cyclin D3 in a case of CD5+ DLBCL.
(A) Northern blot; 30 μg total RNA was used per lane. Upper panel probe CCND3 mRNA, lower panel GAPDH as loading control. RNA from patient 1 (lane 1) showed overexpression ofCCND3 in comparison to DLBCL cell line MD903 with t(3;14)(q27;q32) (lane 2), DLBCL cell line CTB-1 with t(3;14)(q27;q32) (lane 3), mantle-cell NHL cell line Granta 519 with t(11;14)(q13;q32) (lane 4), transformed SLVL cell line Karpas 1718 (lane 5), and B-ALL (FAB L3) cell line KHM-10B with t(8;22)(q24;q11) (lane 6). Arrowheads indicate 2.0-kb CCND3 transcript. (B) Western blot analysis of CCND3 protein. Total cell extracts (30 μg) from patient 1 were subjected to Western blot and probed with Mabs to cyclin D3, p27Kip1, and tubulin (lower panel) as a loading control. Lane 1, patient material; lane 2, HUT78 T-NHL cell line; lane 3, CEMO cell line; and lane 4, RPMI8226. Arrow indicates 32-kd cyclin D3 and 27-kd p27Kip1 proteins.
Overexpression of cyclin D3 in a case of CD5+ DLBCL.
(A) Northern blot; 30 μg total RNA was used per lane. Upper panel probe CCND3 mRNA, lower panel GAPDH as loading control. RNA from patient 1 (lane 1) showed overexpression ofCCND3 in comparison to DLBCL cell line MD903 with t(3;14)(q27;q32) (lane 2), DLBCL cell line CTB-1 with t(3;14)(q27;q32) (lane 3), mantle-cell NHL cell line Granta 519 with t(11;14)(q13;q32) (lane 4), transformed SLVL cell line Karpas 1718 (lane 5), and B-ALL (FAB L3) cell line KHM-10B with t(8;22)(q24;q11) (lane 6). Arrowheads indicate 2.0-kb CCND3 transcript. (B) Western blot analysis of CCND3 protein. Total cell extracts (30 μg) from patient 1 were subjected to Western blot and probed with Mabs to cyclin D3, p27Kip1, and tubulin (lower panel) as a loading control. Lane 1, patient material; lane 2, HUT78 T-NHL cell line; lane 3, CEMO cell line; and lane 4, RPMI8226. Arrow indicates 32-kd cyclin D3 and 27-kd p27Kip1 proteins.
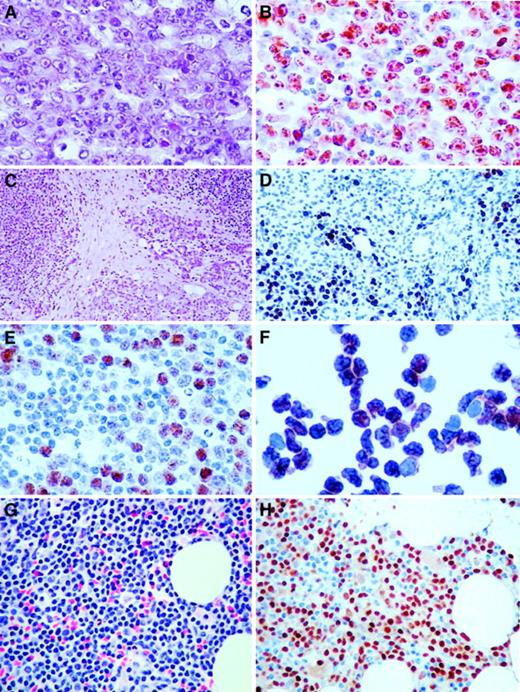
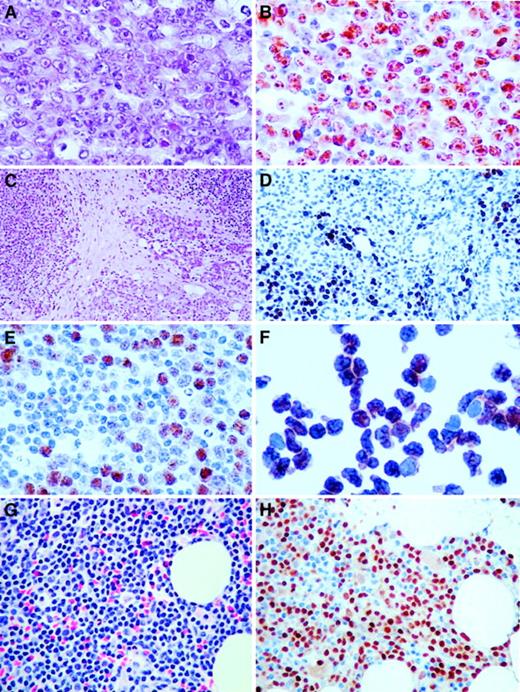
Immunohistochemistry of cases with t(6;14)(p21.1;q32.3).
(A) Hematoxylin and eosin (H&E) section of a lymph node biopsy of the index case (Table 1, case 1), showing a diffuse infiltrate of large neoplastic B cells including centroblasts and some immunoblasts (original magnification × 400). (B) Same section as A stained with antibody to cyclin D3, showing the majority of the tumor cells with strong nuclear positivity (original magnification × 400). (C) H&E section of case 5 (original magnification × 100), showing background parotid gland and extranodal marginal zone B-cell lymphoma of MALT type. (D) Cyclin D3 staining of a focal area of large B-cell transformation in a background of parotid gland and low-grade extranodal marginal zone B-cell lymphoma of MALT type. Note salivary gland duct epithelium without cyclin D3 staining (original magnification × 200). (E) Same section as D stained with antibody to cyclin D3, showing focus of large neoplastic B cells with nuclear staining (original magnification × 300). (F) Cytospin of cells in the peripheral blood of case 3 stained with antibody to cyclin D3, showing intense nuclear staining of virtually all cells (original magnification × 400). (G) H&E section of bone marrow biopsy (Table 1, case 3), showing infiltrate of small B cells (original magnification × 400). (H) Same section of bone marrow stained with antibody to cyclin D3, showing intense nuclear staining of the small neoplastic B cells (original magnification × 400).
Immunohistochemistry of cases with t(6;14)(p21.1;q32.3).
(A) Hematoxylin and eosin (H&E) section of a lymph node biopsy of the index case (Table 1, case 1), showing a diffuse infiltrate of large neoplastic B cells including centroblasts and some immunoblasts (original magnification × 400). (B) Same section as A stained with antibody to cyclin D3, showing the majority of the tumor cells with strong nuclear positivity (original magnification × 400). (C) H&E section of case 5 (original magnification × 100), showing background parotid gland and extranodal marginal zone B-cell lymphoma of MALT type. (D) Cyclin D3 staining of a focal area of large B-cell transformation in a background of parotid gland and low-grade extranodal marginal zone B-cell lymphoma of MALT type. Note salivary gland duct epithelium without cyclin D3 staining (original magnification × 200). (E) Same section as D stained with antibody to cyclin D3, showing focus of large neoplastic B cells with nuclear staining (original magnification × 300). (F) Cytospin of cells in the peripheral blood of case 3 stained with antibody to cyclin D3, showing intense nuclear staining of virtually all cells (original magnification × 400). (G) H&E section of bone marrow biopsy (Table 1, case 3), showing infiltrate of small B cells (original magnification × 400). (H) Same section of bone marrow stained with antibody to cyclin D3, showing intense nuclear staining of the small neoplastic B cells (original magnification × 400).
In other cases of t(6;14)(p21.1;q32.3) in which suitable material for blotting experiments was not available, expression of cyclin D3 protein was determined by using immunohistochemistry. Previous immunophenotypic studies on the expression of cyclin D3 in both normal and malignant B cells have demonstrated low-level expression usually in only a subpopulation of cells, which are thought to represent proliferating cells or blasts.30-33 With the exception of case 4, in which the translocation occurred in only a subpopulation of malignant cells, all cases with t(6;14)(p21.1;q32.3) showed high levels of expression of cyclin D3 in all the malignant cells (Figure 3). These data suggest the involvement of CCND3/cyclin D3 in these cases also; this finding was confirmed by the use of FISH assays described below. A series of lymphoid malignancies with cytogenetic abnormalities of chromosome band 6p21 other than t(6;14)(p21.1;q32.3), which were shown not to involve CCND3 by FISH, were also investigated for cyclin D3 expression by Western blot. All showed low levels of cyclin D3 expression, confirming the involvement of other genes in this region in B-cell malignancies (data not shown).
t(6;14)(p21.1;q32.3) is a recurrent abnormality in mature B-cell malignancies
t(6;14)(p21.1;q32.3) has been reported previously, mainly in cases of myeloma or plasma cell leukemia, although an association with SLVL has recently been reported.17 Material from these 3 cases of SLVL was not available for study. The same translocation was, however, identified in 4 additional cases from our cytogenetic databases; additional investigation of these cases was therefore undertaken. Morphologic review of all cases was performed by a hematopathologist (R.D.G.). A summary of the clinical details of all patients is shown in Table 1. In all but 2 patients, the karyotypes were very complex with multiple structural abnormalities. There were no other consistent associated cytogenetic abnormalities, but notably 3 of the 6 cases exhibited other chromosomal translocations involving 14q32, although the partner chromosomes were variable. In one case of SLVL (Table 1, case 3), t(6;14)(p21.1;q32.3) was the only detectable cytogenetic abnormality, suggesting that it may have been a primary event in the pathogenesis of the disease. This patient presented with a lymphocytosis of 30 × 109/L only, with the immunophenotype consistent with SLVL but with no lymphadenopathy or splenomegaly. However, in a case of SMZL with local transformation to DLBCL (Table 1, case 4), t(6;14)(p21.1;q32.3) was found only in the lymph nodal sample containing transformed cells and not in the splenic SMZL sample. Associated high-level expression of cyclin D3 was restricted to the subpopulation of large cells in the lymph node alone, suggesting that in this instance the translocation was a secondary event associated with transformation to high-grade disease. Two other cases (Table 1, cases 5 and 6) involved the salivary gland and shared identical histologic features. Both biopsies of the parotid gland showed a background histology compatible with a mucosa-associated lymphoid tissue (MALT) lymphoma, including reactive lymphoid follicles, lympho-epithelial lesions, and infiltrates of marginal zone B cells. Additionally, both cases had foci of large transformed cells consistent with diffuse large B-cell transformation; in both cases, the large transformed cells were strongly cyclin D3+ (Figure 3C-D). In case 2, an initial diagnosis of B-cell chronic lymphocytic leukemia (B-CLL) was made on the basis of the lymphocytosis and the concurrent presence of a t(14;19)(q32;q13), a translocation that is found predominantly, but not exclusively, in B-CLL.34 The bone marrow morphology in this case was, however, more suggestive of a lymphoplasmacytic lymphoma, including prominent infiltration of the squash preparations with mast cells. Immunophenotyping of peripheral blood lymphocytes showed expression of HLA-DR, CD19, CD20, CD22, CD38, FMC7, and sIgMκ but no expression of CD5, CD10, CD23, CD11c, or CD25. No lymph node biopsy was performed, and the case was classified as a low-grade B-cell lymphoproliferative disorder, not further classifiable.
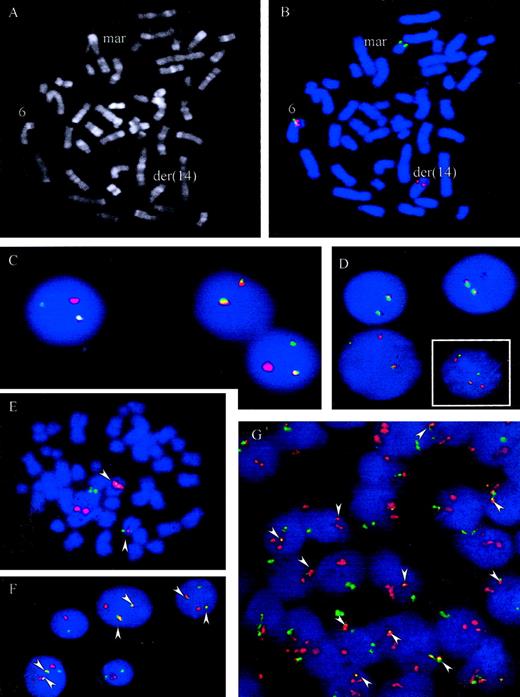
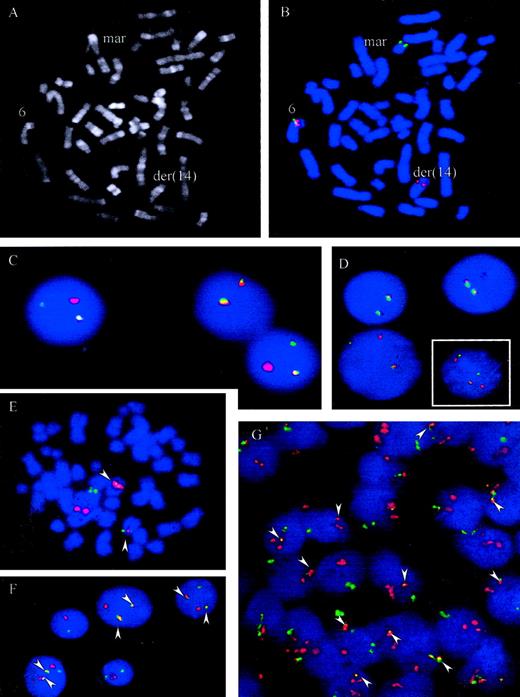
t(6;14)(p21.1;q32.3) juxtaposes IGH andCCND3
A metaphase spread from case 5 is shown in Figure4A. To confirm recurrent involvement ofCCND3 and juxtaposition with IGH in these cases, 3 different interphase FISH assays were applied. Using the LSIIGH assay (Vysis), which includes differentially labeled probes hybridizing to the VH region and centromeric of theCH region, breakpoints within the IGH locus were confirmed in all 6 cases. Some of cases displayed complex signal patterns for this probe in metaphase and interphase cells, indicating different IGH-associated translocations as well as gains and losses of derivative chromosomes. To study involvement of theCCND3 locus in interphase nuclei a “CCND3break-apart” assay was developed that applied differentially labeled probes flanking the breakpoint determined in case 1. The signals derived from these probes, which consisted of pooled BAC and PAC clones, should colocalize in cells with intact CCND3locus but should be separated in cells carrying a translocation with breakpoint in the CCND3 locus (Figure 4B-D). By means of this assay, all 6 cases showed a signal pattern indicative of disruption of the CCND3 locus.
FISH assays for the detection of t(6;14)(p21.1;q32.3).
(A-B) R-banding and FISH analysis of patient 5, Table 1. FISH was performed with chromosome 6p21 probes hybridizing immediately centromeric (green) and telomeric (red) of the CCND3 gene. Signals were observed on the intact chromosome 6 (colocalization of red and green signals) and on the der(14)t(6;14)(p21.1;q32.3) containing the telomeric 6p21 signal (red). The green signal (centromeric 6p21) was on a marker chromosome whose constitution could not be determined by morphology. (C-D, large picture) FISH with probes flanking the CCND3 locus in cases 2 and 4. An intact CCND3 locus is indicated by a colocalized red/green signal. The split indicates the break event. (D, small figure) FISH in case 6 shows a complexly aberrant nucleus with 2 intact CCND3 loci (fused signals) as well as a split of the signals, indicating the breakpoint in the CCND3locus. (E-G) FISH with probes spanning the IGH andCCND3 loci in cases 1 and 3, Table 1. As both IGH(green) and CCND3 (red) probe pools span the recurrent breakpoint regions 2 fusion signals (arrowheads) are to be expected, indicating the der(6)t(6;14)(p21.1;q32.3) and der(14)t(6;14)(p21.1;q32.3) in addition to each isolated signal indicating the intact IGH and CCND3 loci. This expected pattern is clearly seen in the aberrant metaphase (E) and the tumor cell nuclei (F) of case 3. In case 1 (G) the pattern is more complex, but fusion of IGH and CCND3 can be seen in most of the cells (arrowheads). On the basis of FISH analyses with differentially labeled probes flanking the CCND3 andIGH loci, respectively, the lack of one fusion signal and the supernumerary CCND3 signals detectable in many cells in this case is most likely due to a gain of one intact copy of the CCND3 locus and loss of the der(6)t(6;14)(p21.1;q32.3) (data not shown).
FISH assays for the detection of t(6;14)(p21.1;q32.3).
(A-B) R-banding and FISH analysis of patient 5, Table 1. FISH was performed with chromosome 6p21 probes hybridizing immediately centromeric (green) and telomeric (red) of the CCND3 gene. Signals were observed on the intact chromosome 6 (colocalization of red and green signals) and on the der(14)t(6;14)(p21.1;q32.3) containing the telomeric 6p21 signal (red). The green signal (centromeric 6p21) was on a marker chromosome whose constitution could not be determined by morphology. (C-D, large picture) FISH with probes flanking the CCND3 locus in cases 2 and 4. An intact CCND3 locus is indicated by a colocalized red/green signal. The split indicates the break event. (D, small figure) FISH in case 6 shows a complexly aberrant nucleus with 2 intact CCND3 loci (fused signals) as well as a split of the signals, indicating the breakpoint in the CCND3locus. (E-G) FISH with probes spanning the IGH andCCND3 loci in cases 1 and 3, Table 1. As both IGH(green) and CCND3 (red) probe pools span the recurrent breakpoint regions 2 fusion signals (arrowheads) are to be expected, indicating the der(6)t(6;14)(p21.1;q32.3) and der(14)t(6;14)(p21.1;q32.3) in addition to each isolated signal indicating the intact IGH and CCND3 loci. This expected pattern is clearly seen in the aberrant metaphase (E) and the tumor cell nuclei (F) of case 3. In case 1 (G) the pattern is more complex, but fusion of IGH and CCND3 can be seen in most of the cells (arrowheads). On the basis of FISH analyses with differentially labeled probes flanking the CCND3 andIGH loci, respectively, the lack of one fusion signal and the supernumerary CCND3 signals detectable in many cells in this case is most likely due to a gain of one intact copy of the CCND3 locus and loss of the der(6)t(6;14)(p21.1;q32.3) (data not shown).
To demonstrate juxtaposition of the CCND3 and IGHloci, differentially labeled probe sets spanning the CCND3and IGH loci, respectively, were applied in an interphase FISH assay. In this assay, a simple t(6;14)(p21.1;q32.3) in a diploid cell should result in a split of each one CCND3 andIGH signal, resulting in a total of 3 CCND3 andIGH signals. Moreover, each one of the different signals should be colocalized on the der(6)t(6;14)(p21.1;q32.3) and der(14)t(6;14)(p21.1;q32.3), leading to a signal pattern of 2 fusion signal plus each one CCND3 and IGH signal, the latter both indicating the normal chromosomes 6 and 14, respectively (Figure 4E-F). In all 5 cases with suitable material for this FISH assay available, fusion of IGH and CCND3 loci could be confirmed. Variant signal patterns were observed frequently (Figure 4 and data not shown). Review of the original cytogenetics of the index case (case 1), in the light of the above findings, failed to detect a t(6;14)(p21.1;q32.3); the case exhibited a highly complex karyotype19 (L. Harder, unpublished observations, December, 2000). Nevertheless, FISH not only unambiguously confirmed breakpoints in the CCND3 and IGH loci as well as IGH/CCND3 fusion in this case but also provided evidence for a gain of one intact copy of the CCND3 locus and loss of the der(6)t(6;14)(p21.1;q32.3) (Figure 4G and data not shown).
Discussion
In this study, we demonstrate the recurrent involvement of the cyclin D3 gene in t(6;14)(p21.1;q32.3) of mature B-cell lymphoproliferative disorders; comparable results have been obtained by others in myeloma.29 In our series, the translocation was observed in patients with SLVL, MALT lymphoma at transformation to DLBCL, and in DLBCL. In the 2 cases cloned here, the IGHbreakpoint occurred in the IG switch regions, whereas the breakpoints on chromosome 6p21.1 occurred some distance 5′ of theCCND3 gene. Thus, both the anatomy of this translocation and its presence in several histologic subtypes of B-cell malignancy appear to be similar to the t(11;14)(q13;q32.3) involving the cyclin D1, which is found predominantly in mantle cell lymphoma but also in SLVL, B-cell prolymphocytic leukemia, and myeloma.
The expression of all the D family of cyclins in both normal and malignant B-cell populations has been extensively studied by immunohistochemistry and at the RNA level30-33,35-38; the 2 may not necessarily parallel each other as cyclin D3 messenger RNA (mRNA) is subject to extensive posttranscriptional control.39 They comprise a group of molecules necessary for progression through the G1 phase of the cell cycle and are thought to have largely overlapping and possibly redundant roles.
Cyclin D3 expression in reactive lymph nodes has been observed in the blast cells of germinal centers and in interfollicular areas. cyclin D3+ mantle and marginal zone cells wereBCL2−, indicating that they represent activated blast cells and suggesting a link between B-cell proliferation and cyclin D3 expression.31,33 In B-cell malignancies, cyclin D3 is absent from cases of mantle cell B-NHL that overexpress cyclin D130,32,36; the causes for this lack of expression are not known. Otherwise, cyclin D3 RNA and protein expression appear to be ubiquitous, although within subgroups of disease, the levels of expression can vary substantially and correlate with proliferative activity; usually only a subpopulation of cells is cyclin D3+ on immunohistochemistry. In DLBCL for example, most cases exhibit less than 10% of cells positive for cyclin D3.30-33 Similarly in B-CLL, the frequency of cyclin D3+ cells is usually low (about 10% of cells staining), whereas higher levels of expression have been reported in DLBCL and Burkitt lymphoma.30,32 In one series, there were 6 of 74 cases of DLBCL in which more than 90% of malignant cells expressed cyclin D3, indicating constitutive expression.32 Cyclin D3 expression correlated with high-level expression of p27Kip1, a cell-cycle inhibitory molecule. It was suggested that the high levels of cyclin D3 complexed with, and sequestered p27Kip1, allowing unhindered proliferation, although the causes for the high-level expression of both proteins were not established.
A genetic cause for cyclin D3 overexpression has long been suspected but not previously demonstrated.40 Involvement of cyclin D2 in a translocation with the immunoglobulin λ light chain locus has been shown in a case of Richter transformation of B-CLL.41Although such translocations are rare, these data are of some interest because cyclin D2 is crucial for the development of CD5+ B cells and also plays an important role in mediating proliferative signaling.42 The data reported here show deregulated expression of cyclin D3 in B-NHL arising as a consequence of t(6;14)(p21.1;q32.3) and suggest that cyclin D3 may also act as a dominant oncogene. Among B-NHL, it seems that t(6;14)(p21.1;q32.3) may be associated with lymphomas of marginal zone origin.17Whether cases of DLBCL with the same translocation represent transformation of low-grade marginal zone lymphoma cannot be determined.18 In one case of SMZL reported here, the chromosomal translocation occurred in only a fraction of the cells in a lymph node specimen, whereas a concurrent spleen specimen had a clone with trisomy 12 and add(12)(q22) as the sole chromosome abnormalities. In this instance therefore, t(6;14)(p21.1;q32.3) was likely to have been a secondary event, perhaps associated with early histologic transformation.
Whether t(6;14)(p21.1;q32.3) is the only mechanism underlying high-level cyclin D3 expression in B-cell malignancies remains to be determined. High-level expression of cyclin D3 occurs in DLBCL at a frequency much higher than that reported to date for t(6;14)(p21.1;q32.3), although the complexity of the cytogenetic abnormalities may mask the translocation. Amplifications of 6p21 involving CCND3 and resulting in high-level expression have been described in other malignancies.43 Comparable amplifications of this region have been observed in comparative genomic hybridization studies of follicular lymphoma, although whether this involved CCND3 was not clear.11 Alternatively, more local alterations within the CCND3 gene may occur. As with CCND1 in mantle cell lymphoma, small deletions within the 3′ untranslated regions of the CCND3 gene might allow changes in the levels of expression through increased mRNA stability.44,45 Finally, it is possible that high-level expression of cyclin D3, like p27Kip1, will be a useful prognostic marker, not only in DLBCL but also in other subtypes of B-cell malignancy.46
We thank all members of the chromosome 6 project group, in particular Sarah Sims and Sophie Williams, and the Human Genome Resource Centre, Hinxton Hall, United Kingdom, for EST, BAC, PAC, and cosmid clones. We thank Ms Claudia Becher, Dorit Schuster, Hiromi Ogata, and Marta Salido for skillful technical assistance. We thank Professor Hiroaki Mitsuya, Drs Shima Uneda, Hiroyuki Hata (Internal Medicine II, Kumamoto University), and Drs Kenichi Iyama and Jyunji Tsuruta (Department of Pathology, Kumamoto University Hospital) for their assistance in reporting the index case. We thank Dr Luisa Dı́az Fernández for kindly providing one of the cases.
Supported by the Bud Flanagan Leukaemia Trust, the Kay Kendall Leukaemia Fund, the Lady Tata Memorial Trust, The Daiwa Anglo-Japanese Foundation, The Human Resources Support Foundation under Kumamoto City Municipal Centennial Commemorative Project, the Deutsche Krebshilfe (grant 10-1556-Schl4), and the IZKF, Kiel. R.D.G. and D.E.H. were supported by grant 5 U01 CA84967-02 from the National Institutes of Health. The Sanger Centre is funded by the Wellcome Trust.
R.S. and M.J.S.D. are co–senior authors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Martin J. S. Dyer, Department of Haematology, University of Leicester, Robert Kilpatrick Clinical Sciences Building, Leicester Royal Infirmary, PO Box 65, Leicester, LE2 7LX United Kingdom; e-mail: mjsd1@le.ac.uk.