Correct developmental regulation of β-like globin gene expression is achieved by preferential transcription of a gene at a given developmental stage, silencing of other β-like gene promoters, and competition among these promoters for interaction with the locus control region (LCR). Several evolutionarily conserved DNA elements in the promoters of the β-like genes and LCR have been studied in detail, and the role of their binding factors has been investigated. However, the β-globin promoter includes additional evolutionarily conserved sequences of unknown function. The present study examined the properties of a 21-base pair (bp) promoter-conserved sequence (PCS) located at positions −115 to −136 bp relative to the transcription start site of the β-globin gene. A helicaselike transcription factor (HLTF) belonging to the SWI2/SNF2 family of proteins binds to the PCS and a partly homologous sequence in the enhancer region of the LCR hypersensitive site 2 (HS2). Elevation of the level of HLTF in K562 erythroleukemic cells increases β-promoter activity in transient transfection experiments, and mutations in the PCS that remove HLTF-binding regions abolish this effect, suggesting that HLTF is an activator of β-globin transcription. Overexpression of HLTF in K562 cells does not affect the endogenous levels of γ- and ε-globin message, but it markedly activates β-globin transcription. In conclusion, this study reports a transcription factor belonging to the SWI2/SNF2 family, which preferentially activates chromosomal β-globin gene transcription and which has not previously been implicated in globin gene regulation.
Introduction
The expression of globin genes is controlled at several levels in vivo. Temporal switching from embryonic to fetal and then to adult globin is a much-discussed subject.1,2 In addition, globin production becomes a progressively larger part of total protein synthesis as erythroid precursors divide and mature. The levels of production of the α- and non–α-globin chains are precisely balanced, although there is no evident feedback regulation of α-globin transcription by β-globin gene activity or vice-versa.3 Thus, the balanced activity of the α- and β-like promoters appears to be intrinsic to the structure ofcis-acting elements, even though the promoters and related enhancers are considerably different in structure, and the genes are differently expressed during development.4 5
The β-like globin promoters contain several transcription factor binding sites such as GATA, CACCC, β-DRE, and stage-selecting factor binding sites.6-11 Several of the factors binding to these sites are restricted to a limited number of hematopoietic lineages, including erythrocyte precursors, and some act specifically on one or more of the globin genes. Negative as well as positive regulation of globin genes is crucial for their correct developmental regulation. Globin genes are turned off by 2 general mechanisms: autonomous gene silencing involving sequences located in the proximal and distal promoters and competition between genes for interaction with the locus control region (LCR).12 Silencers have been identified in the β-globin cluster, particularly upstream of the embryonic globin gene.13-16 In the case of adult β-globin, an upstream promoter element between −500 and −250 base pair (bp) region that harbors binding sites for BP1 and BP2 proteins has been shown to participate in the negative regulation of transcription.17-19
β-Globin transcription is also modulated by LCRs.20 The β-globin LCR is located 50 Kb upstream from the adult β-globin gene. It contains 4 regions that exhibit erythroid-specific DNase hypersensitivity in vivo (HS1, HS2, HS3, and HS4) in order proceeding upstream from the β-globin gene cluster. Among these, HS2 harbors typical enhancer activity in transfected cells.21-23 The NF-E2 site of HS2 is critical for this enhancing function.24,25 A basic leucine zipper protein called p45 NF-E2 and several of its related factors in association with the p18 Mafs bind to the NF-E2 DNA sequence,26-28 but they may not be the only factors mediating its activity.29-31
Although a variety of transcription factors have been identified that bind to the LCR and promoters of the β-like genes, the overall molecular mechanism of globin gene switching is not fully understood. Identification of any additional transcription factors that normally act on the LCR or globin gene promoters is an essential step toward understanding the molecular mechanisms involved in globin gene switching. Phylogenetic footprints at different regions of the β-locus have proved to be useful in identifying the regulatory elements and their binding factors based on the consensus binding motifs.32-35 However, there are several conserved regions of unknown function on the β-locus that do not match with the consensus binding sites for the known DNA-binding proteins.
In the present study we have concentrated on a 21-bp (−115 to −136 bp) sequence on the β-promoter (the PCS) that is phylogenetically conserved. A helicaselike transcription factor (HLTF) with DNA-dependent adenosine triphosphatase (ATPase) activity and homology to the SWI2/SNF2 family of proteins binds to this sequence and its homologous counterpart at the NF-E2 site of the HS2. Mutational analysis of PCS coupled with transient transcription studies indicates that HLTF is an activator of the β-globin transcription. Finally, we show that increased expression of HLTF activates transcription of the endogenous β-globin gene in K562 cells.
Materials and methods
Cell growth and nuclear extracts
K562 cells were grown on RPMI 1640 (with 300 mg/L glutamine) supplemented with 10% fetal bovine serum and 1× antibiotics and antimycotic (100 U/mL ampicillin, 100 U/mL streptomycin, and 0.25 μg/mL amphotericin). Nuclear extracts were prepared as described previously.36 About 14 to 18 mg nuclear extracts at 10 μg/μL concentration were obtained from 109 cells and stored in 20-μL aliquots at −80°C.
Antibodies and Western blotting
The rabbit polyclonal Rush-1 and Rush-2 antibodies were kind gifts from Beverly Chilton's laboratory (Texas Technical University Health Science Center, Lubbock, TX).37 We refer to these antibodies as peptide antibody A and antibody B, respectively. Affinity purified and lyophilized antibodies were dissolved in 10 mM Tris-HCl pH 8.0 and stored at −80°C. Antibody A was raised against amino acids 8 to 38 (which is identical to the HLTF peptide at the same position), and antibody B was raised against amino acids 370 to 387 (that has 65% identity and 88% similarity with the HLTF peptide at the same position) of the Rush-1α protein, a rabbit homologue of human HLTF.37 Antibodies against p18 Maf and Sin3A and His tag were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). For Western blots, 50 to 70 μg nuclear or total protein extract per lane was resolved on a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a Hybond nylon blotting membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The membranes were blocked with 5% milk and 0.05% Tween 20 in phosphate-buffered saline (PBS) and probed with anti-HLTF antibodies A and B or anti-His antibody for 1 hour at room temperature in PBS containing 1% nonfat dry milk and 0.05% Tween 20 at 1:2000 dilution. Horseradish peroxidase–tagged rabbit anti-immunoglobulin G in mouse was used as a secondary antibody at 1:200 000 dilution. The blots were developed by the Super Signal West Femto kit supplied by Pierce Company (Rockford, IL).
Transfections
A lipofectin-based transfection protocol supplied by Gibco BRL (Rockville, MD) was used. For each transfection, 3 × 106K562 cells, 5 μg pGL3 plasmid construct, and 20 μL lipofectin were used. Transfection was allowed to continue for 5 hours after which time cells were allowed to recover for 40 hours in RPMI medium. In case of transient transfections, cells were harvested at the end of recovery, washed twice with 2 mL PBS, and used to prepare cell-free extract as per the protocol supplied with Luciferase assay kit from Promega. The same assay kit was used to assay luciferase activity in the cell-free extracts. The photon emission in each assay was measured for 10 seconds on a Lumat LB9501 luminometer (EG&G Wallace, Gaithersburg, MD). The luciferase activities represent the averages of 3 to 8 separate experiments, most of which were done in duplicate. To generate a pool of stably transfected cells, G418 (neomycin) was added at the end of the 40-hour recovery period at a concentration of 0.8 mg/mL, after which time the cells were incubated in a standard CO2incubator at 37°C for 3 to 4 weeks or until the neomycin-resistant cells start growing actively. Every 3 days, the cells were fed with fresh medium and G418. Two preparations of DNA were used in separate transfection experiments for each construct.
Statistical analysis
Pairwise comparisons of luciferase activities of all the transient transfections in each experiment were done by Studentt test. Unless otherwise indicated, only statistically significant results are discussed in the text. Only those differences withP values < .05 were taken as significant.
Electrophoretic mobility shift assays
The double-stranded DNA (100 ng) was labeled by a polynucleotide kinase reaction using [γ-32P] adenosine triphosphate (ATP). Aliquots (20 μL) of the nuclear extract were used in the electrophoretic mobility shift assay (EMSA). The binding buffer contained 10 mM HEPES pH 7.9, 50 mM KCl, 10% glycerol, and 0.02% NP40. A typical 20-μL binding reaction contained 1 μg poly dIdC, 1 mM dithiothreitol, 2 ng 32P DNA probe (∼100 000 cpm), and 5 μg nuclear extract in the binding buffer. Binding took place at room temperature for 10 minutes. The mixture was loaded on a 5% polyacrylamide gel and run for 2.5 hours at 200 volts (25 mAmp) in 0.5× 45 mM Tris-Borate and 1 mM EDTA ph 8.0 (TBE).
Complementary DNA screening
An oligo dT and random-primed K562 λ gt-11 complementary DNA (cDNA) library (Clontech, Palo Alto, CA) was screened with 2 double-stranded synthetic oligonucleotides. The sequences of the positive strands of these double-stranded oligonucleotides are described as oligonucleotides 11 and 12 in Table1. The specific activity of the radioactive probes was 8 × 107 cpm/μg. The amount of each probe added was 4 × 106 cpm per filter. Two million clones from a λ gt11 cDNA library spread over 40 plates (150 × 12 mm) were screened according to standard protocols.38
Mutagenesis and preparation of plasmid constructs
A polymerase chain reaction (PCR)–based strategy was used to clone the wild-type β-globin promoter and its mutant variants in pGL3 vector. A DNA fragment from −270 to +54 bp served as the template. Forward primers included the mutations of choice and appropriate restriction enzyme sites. After the PCR, the DNA fragments were digested with the restriction enzymes, purified on a 1% agarose gel, ligated to pGL3 plasmid, and transformed into Escherichia coli (DH5α). Unless otherwise indicated all the promoters were cloned at HindIII-XhoI sites and HS2 atSalI-BamHI sites. Wild type and mutant β-promoter constructs were confirmed by DNA sequencing. Large-scale plasmid preparations of 2 clones of each construct were made by the Qiagen maxi-kit protocol.
The initial partial HLTF cDNA clone isolated by screening the λ gt11 cDNA library of K562 cells was cloned into a pTrcHis2TOPO vector and transformed into TOP10 E coli cells (Invitrogen, Carlsbad, CA). A HLTF cDNA coding for aa 2 to aa 332 was generated by PCR, using the HLTF cDNA clone isolated by λ gt11 screen as a template. Oligonucleotides 27 and 28 (Table 1) were used as forward and reverse primers, respectively. The plasmid DNA from recombinant clones was sequenced to check the proper orientation of the clone and exclude internal mutations. HLTF expression in the bacteria was induced with 1 mM isopropyl thiogalactoside (IPTG) in a 50-mL culture for 4 hours to obtain a 38-kd HLTF protein with Myc and His tags at its C-terminal end. At the end of the induction, the cells were harvested and suspended in 1 mL ice-cold HEPES buffer pH 7.9 containing 300 mM NaCl. The protein extract was prepared by sonicating the cells on a Branson cell disruptor fitted with a micro tip with 60% duty cycles and output control of 6 in a 1-second pulsed mode. The cells were subjected to 6 rounds of 30 pulses each with 2 minutes' incubation on ice between each round. The clear sonicated extract was centrifuged at 15 000g for 15 minutes, and supernatant was used for the EMSA and to detect the induced protein on SDS-PAGE.
Total RNA and reverse transcriptase-PCR
RNAwiz reagent from Ambion (Catalog No. 9736; Austin, TX) was used to isolate total RNA from K562 cells. The RNA isolation procedure was as per the manufacturer's protocol. About 200 μg total RNA was digested with 10 U Rnase-free DNase for 30 minutes at room temperature, extracted with phenol chloroform and stored at −80°C until further use for reverse transcriptase (RT)–PCR. The 20-μL RT reaction contained 10 μg total RNA, 200 ng oligo dT, 0.5 mM dNTP, 1 μL RNase inhibitor, 5 mM dithiothreitol, and 200 U superscript II from GibcoBRL. The reaction was started by the addition of the enzyme and incubated at 42°C for 1 hour. One microliter of this RT reaction was used as a template for the PCR reaction. A typical 25-μL PCR reaction in 1× Perkin-Elmer (Boston, MA) buffer with 2.5 mM MgCl2contained 1 μL RT product as template, 200 μM dNTPs, 0.5 μCi (1.85 × 104 Bq) [α-32P]dCTP, 100 ng of each gene-specific primer set, and 0.2 U Platinum DNA polymerase (Gibco-BRL). The conditions for the 18- and 22-cycle PCR were as follows: 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute. We aligned ε-, γ-, δ-, and β-globin cDNA sequences and designed gene-specific primers from the nonhomologous regions. The gene-specific primer sets are as follows: primers 13 and 14 for β-globin (338-bp PCR product), primers 15 and 16 for ε-globin (194-bp PCR product), primers 17 and 18 for γ-globin (397-bp PCR product), primers 19 and 20 for β-actin (299-bp PCR product), primers 21 and 22 for glyceraldehydes-3-phosphate dehydrogenase (GAPDH) (290-bp PCR product), and primers 23 and 24 for EKLF (239-bp PCR product). The sequences of these primers are listed in Table 1.
Results
Conserved sequences in the β-promoter do not entirely coincide with binding sites for known factors
Alignment of β-globin promoter sequences from mouse to human shows a conserved region, the PCS at −115 to −139 bp from the transcription start site (Figure 1A). Two segments of this region, a GTCATCA sequence and a trinucleotide GCT, are distinctly conserved. The third segment comprising the CCAAGGACAGG sequence at the 5′ end is partially conserved. The GCTGTCA sequence that includes the GCT trinucleotide and the first half of the GTCATCA sequence correspond to the binding site for TGIF, a homeodomain-containing transcription factor.39,40 However, the majority of this conserved region is not a binding site for known transcription factors, and none of the EMSA bands formed with the PCS oligonucleotide are affected by an antibody to TGIF (data not shown). Interestingly, the conserved segments of the β-promoter are homologous to sequences overlapping the AP1/NF-E2 site of HS2 (Figure1C) that are also conserved from mouse to human (Figure 1B) and are needed for the core-enhancing activity of the LCR.25,41 42To emphasize its homology with the PCS, we call the LCR sequence HS2-CS for HS2-conserved sequence. Among the 3 segments, an AGCACAGC sequence from the 5′ portion of the HS2-CS shows partial phylogenetic conservation. A comparative look at these partially conserved sequences on PCS and HS2-CS from mouse to human shows a common AGG/CACAGG/C sequence pattern. Such a homology between the PCS and HS2-CS sequences raises the possibility of related roles for these 2 DNA elements in β-promoter transcription.
Phylogenetically conserved sequences in the β-globin promoter and the HS2.
(A) Conservation of the −112 to −140 bp region from the transcription start site of the adult human β-globin promoter. The conserved sequences are underlined. The number at the left-hand side of the human sequence is the nucleotide number from the human globin locus (GenBank locus HUMHBB). (B) Comparison of the HS2 sequences at the NF-E2 region from mouse to human. The tandem NF-E2/AP1 sequences are double underlined. The numbers at the left-hand side of the sequences are the nucleotide numbers assigned to the respective globin locus. (C) Homology between the −112 to −139 bp β-promoter sequence (PCS) and the NF-E2 site (HS2-CS) of the HS2. The homologous sequences are underlined.
Phylogenetically conserved sequences in the β-globin promoter and the HS2.
(A) Conservation of the −112 to −140 bp region from the transcription start site of the adult human β-globin promoter. The conserved sequences are underlined. The number at the left-hand side of the human sequence is the nucleotide number from the human globin locus (GenBank locus HUMHBB). (B) Comparison of the HS2 sequences at the NF-E2 region from mouse to human. The tandem NF-E2/AP1 sequences are double underlined. The numbers at the left-hand side of the sequences are the nucleotide numbers assigned to the respective globin locus. (C) Homology between the −112 to −139 bp β-promoter sequence (PCS) and the NF-E2 site (HS2-CS) of the HS2. The homologous sequences are underlined.
DNA-protein complexes are formed on the β-PCSs
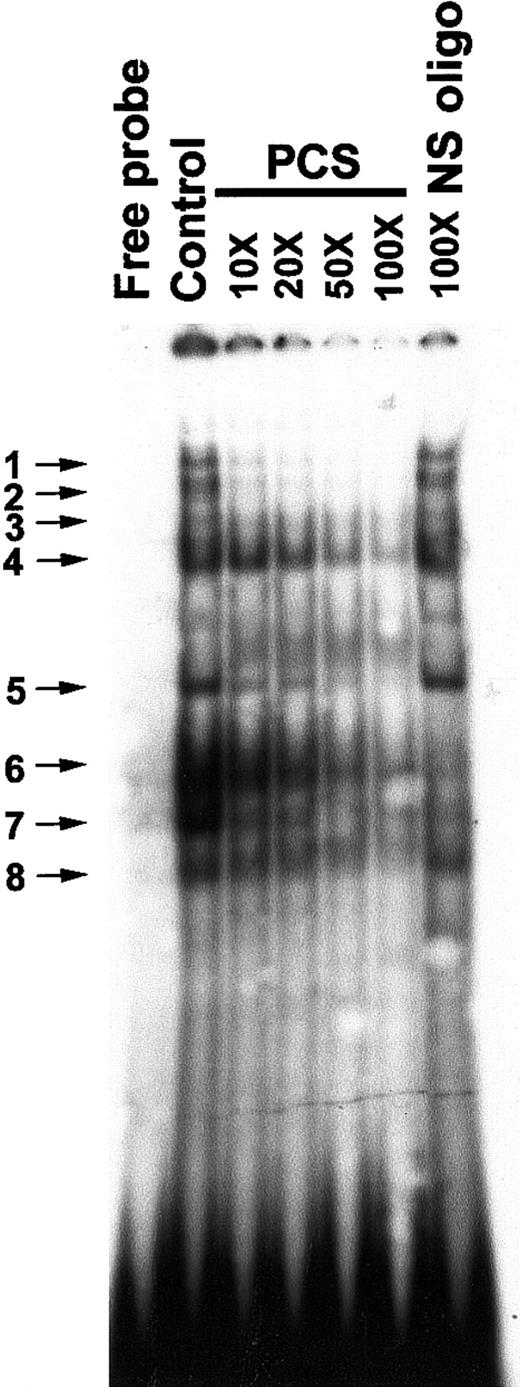
Previously, an in vivo footprint was detected at the PCS region of the β-promoter in human–mouse erythroleukemic hybrid lines, A181γ and A181β.43 To investigate whether DNA-protein complexes are formed on the PCS in K562 cells, we carried out EMSAs with an oligonucleotide representing the −140 to −110 bp region of the β-promoter and K562 nuclear extracts that displayed multiple protein-DNA complexes (Figure2). The EMSA bands could be competed by progressively increasing molecular excess of nonradioactive PCS DNA. The EMSA bands 1 to 5 and 8 were not competed by a 100-fold excess of nonspecific oligonucleotide, suggesting that the DNA-protein complexes in these bands are sequence specific. However, EMSA bands 6 and 7 were competed by the nonspecific oligonucleotide as well, indicating their relaxed sequence specificity for DNA binding (Figure 2).
Electrophoretic mobility shift assay (EMSA) to show in vitro sequence-specific binding of proteins to the PCS.
The control lane displays EMSA bands obtained by incubating 5 μg nuclear extract with 2 ng 32P-labeled double-stranded PCS (oligo 1). In oligonucleotide competition assays, the32P-labeled double-stranded PCS of the β-promoter (oligo 1) was mixed with various amounts (molar excesses) of nonradioactive PCS-DNA as indicated at the top of the figure. A double-stranded oligonucleotide from −142 to −171 bp relative to the transcription start site of the β-promoter was used as a nonspecific (NS) oligonucleotide (oligo 25 and 26). The band between band 4 and 5 inconsistently appears on the gel. The EMSA bands are numbered and indicated by arrows.
Electrophoretic mobility shift assay (EMSA) to show in vitro sequence-specific binding of proteins to the PCS.
The control lane displays EMSA bands obtained by incubating 5 μg nuclear extract with 2 ng 32P-labeled double-stranded PCS (oligo 1). In oligonucleotide competition assays, the32P-labeled double-stranded PCS of the β-promoter (oligo 1) was mixed with various amounts (molar excesses) of nonradioactive PCS-DNA as indicated at the top of the figure. A double-stranded oligonucleotide from −142 to −171 bp relative to the transcription start site of the β-promoter was used as a nonspecific (NS) oligonucleotide (oligo 25 and 26). The band between band 4 and 5 inconsistently appears on the gel. The EMSA bands are numbered and indicated by arrows.
A protein with helicase domains is present in the DNA-protein complexes formed on PCS
We used 2 double-stranded synthetic oligonucleotides of the PCS sequence to screen a K562 cDNA library in the λ gt-11 expression vector (oligonucleotides 11 and 12 described in the “Materials and methods” section). After screening 2 million plaques, we obtained one positive cDNA clone of 2.85 Kb. This clone contained sequences coding for the N-terminal 332 amino acids of a mammalian gene variously known as HLTF, HIP116, RUSH1, and Zbu1.44-48 There is a bias for detecting truncated forms of the cDNA in expression libraries, particularly when the DNA-binding domain is situated at the 5′ end of a relatively large protein like HLTF. The truncated form of HLTF obtained by us could have been derived from incompletely spliced products that retained the DNA-binding domain. The full-length human HLTF gene codes for a 1006 amino acid protein that has been shown to possess DNA-dependent ATPase activity. The predicted amino acid sequence is homologous to the SWI2/SNF2 family of helicases with 7 helicase domains and also has a C3HC4 zinc-binding motif called a ring finger located between the helicase motifs in an arrangement similar to that found in the yeast RAD5 and RAD 16 proteins.43 46 The clone that we have isolated is a short fragment of HLTF spanning the DNA-binding domain and an NTP-binding domain. Structural analysis of the predicted amino acid sequence with the COILS program on the Expasy web site (www.ch.embnet.org/software/coils_form.html) revealed 3 coiled coil regions between aa 104 to aa 125, aa 151 to aa 168, and aa 202 to aa 231. Each of these coiled coil structures harbors 3 leucine heptads.
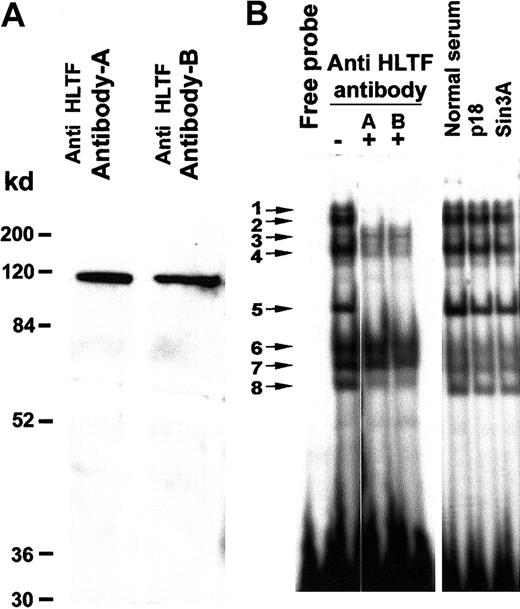
To establish if the bands in the gel mobility shift assay contain the HLTF protein, we used 2 affinity-purified antibodies. Western blotting of K562 nuclear extracts with these antibodies exhibited a single prominent 115-kd band (Figure 3A). HLTF antibody A as well as antibody B neutralized gel shift bands 1, 2, 4, 5, and 8 in an EMSA with K562 nuclear extracts, confirming the presence of HLTF in these DNA-protein complexes (Figure 3B). Normal rabbit serum and purified antibodies against Sin3A and p18 Maf did not disturb any of these EMSA bands. Similarly, we have tested antibodies against YB1, HDAC1, Fos, and NF-E2. None of them had any effect on these EMSA bands (data not shown). In another control experiment, these antibodies did not disturb EMSA bands obtained with an oligo that binds the ATF transcription factor and K562 nuclear extracts (data not shown). These control experiments together with appearance of a single 115-kd band on the Western blot (Figure 3A) demonstrate the specificity of the antibodies for HLTF. The DNA-binding region of the HLTF lies between aa 123 to aa 219.47 Antibody A was raised against amino acids 8 to 38 and antibody B was raised against amino acids 370 to 387. As these antibodies were raised against polypeptides that are near the DNA-binding site of the HLTF, we observe disappearance of the gel shift bands instead of super shifts.
HLTF is present in several major DNA-protein complexes formed on the PCS.
(A) Western blot of the K562 nuclear extract with HLTF antibody A and B shows a prominent band of 115 kd. A minor faint band below may be the degradation product. K562 nuclear extract (50 μg) was resolved on a 12% SDS-PAGE. The HLTF antibodies are indicated on the top, and the molecular weight markers are indicated at the left-hand side. (B) EMSA with 32P-labeled PCS oligonucleotide (oligo 1) and K562 nuclear extracts in the presence (+) and absence (−) of HLTF antibodies A and B. Normal rabbit serum and antibodies against p18 Maf and Sin3A served as control. In a standard 20-μL EMSA reaction mixture, 2 μL each of the antibody mentioned at the top of the panel and 5 μg nuclear extracts were preincubated for 10 minutes before the addition of the 32P-labeled DNA probe. The binding mix was incubated for an additional 10 minutes at room temperature after the addition of the DNA probe and analyzed on a 5% polyacrylamide gel.
HLTF is present in several major DNA-protein complexes formed on the PCS.
(A) Western blot of the K562 nuclear extract with HLTF antibody A and B shows a prominent band of 115 kd. A minor faint band below may be the degradation product. K562 nuclear extract (50 μg) was resolved on a 12% SDS-PAGE. The HLTF antibodies are indicated on the top, and the molecular weight markers are indicated at the left-hand side. (B) EMSA with 32P-labeled PCS oligonucleotide (oligo 1) and K562 nuclear extracts in the presence (+) and absence (−) of HLTF antibodies A and B. Normal rabbit serum and antibodies against p18 Maf and Sin3A served as control. In a standard 20-μL EMSA reaction mixture, 2 μL each of the antibody mentioned at the top of the panel and 5 μg nuclear extracts were preincubated for 10 minutes before the addition of the 32P-labeled DNA probe. The binding mix was incubated for an additional 10 minutes at room temperature after the addition of the DNA probe and analyzed on a 5% polyacrylamide gel.
HLTF is also present in some of the DNA-protein complexes formed on the HS2-CS sequence
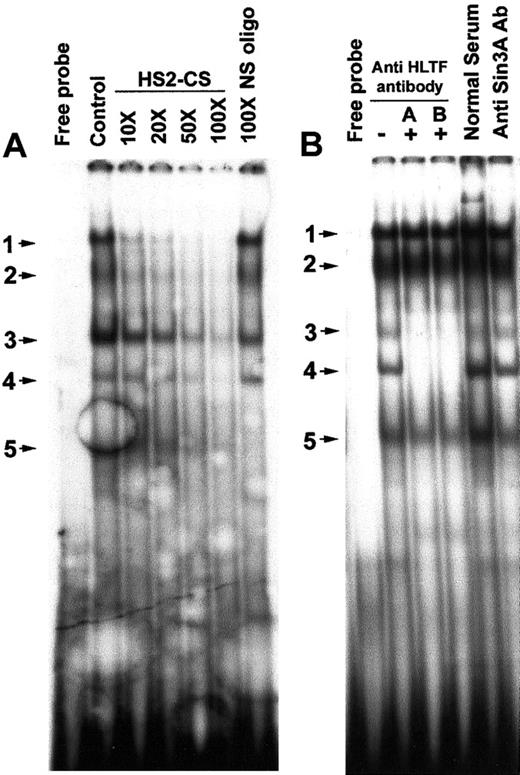
We looked for the presence of HLTF in the DNA-protein complexes formed on the HS2-CS site of the HS2, because this site has significant homology with the PCS. EMSA with an oligonucleotide containing the HS2-CS sequence displayed 5 major bands of which bands 1 to 4 could be specifically competed by nonradioactive HS2-CS oligonucleotide probe (Figure 4A). EMSA band 5 is nonspecific, as it could also be competed by 100-fold excess of a nonspecific oligonucleotide. Bands 3 and 4 in the EMSA were abolished by the addition of the anti-HLTF antibodies A and B, indicating the presence of HLTF in these DNA-protein complexes (Figure 4B).
Presence of HLTF in some of the DNA-protein complexes formed on the HS2-CS.
(A) In vitro sequence-specific binding of K562 nuclear proteins to the HS2-CS. The DNA probe used in the EMSA is 32P-labeled double-stranded HS2-CS sequence (oligo 10). The control lane is EMSA without nonradioactive HS2-CS oligo. The molar excesses of nonradioactive DNA probe used in the competitive EMSA are shown on the top of the lanes. The NS oligo is described in Figure 2. The EMSA bands are numbered and indicated by arrow marks. Free probe lane displays EMSA pattern in the absence of K562 nuclear extracts. (B) HLTF is present in 2 EMSA bands obtained with a double-stranded HS2-CS sequence and K562 nuclear extracts. Gel supershift/neutralization assay with K562 nuclear extract and 32P-labeled HS2-CS oligonucleotide in the presence (+) and absence (−) of HLTF antibody A and B, normal rabbit serum, and anti-Sin3A antibody. The assay conditions were the same as described in Figure 3.
Presence of HLTF in some of the DNA-protein complexes formed on the HS2-CS.
(A) In vitro sequence-specific binding of K562 nuclear proteins to the HS2-CS. The DNA probe used in the EMSA is 32P-labeled double-stranded HS2-CS sequence (oligo 10). The control lane is EMSA without nonradioactive HS2-CS oligo. The molar excesses of nonradioactive DNA probe used in the competitive EMSA are shown on the top of the lanes. The NS oligo is described in Figure 2. The EMSA bands are numbered and indicated by arrow marks. Free probe lane displays EMSA pattern in the absence of K562 nuclear extracts. (B) HLTF is present in 2 EMSA bands obtained with a double-stranded HS2-CS sequence and K562 nuclear extracts. Gel supershift/neutralization assay with K562 nuclear extract and 32P-labeled HS2-CS oligonucleotide in the presence (+) and absence (−) of HLTF antibody A and B, normal rabbit serum, and anti-Sin3A antibody. The assay conditions were the same as described in Figure 3.
Correlation between β-promoter transcription and binding of HLTF to the PCS
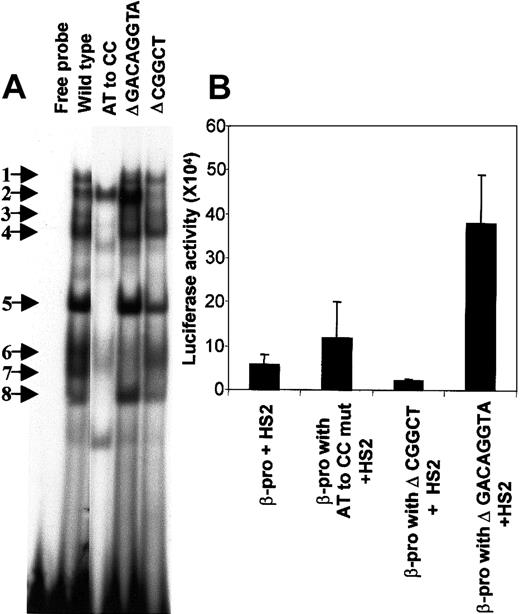
We created mutations in 3 conserved segments of the PCS to determine the bases necessary for the binding of HLTF (Figure5A). The functional consequence of disturbance of various EMSA bands with PCS mutations was investigated by constructing pGL3 vectors with a normal β-promoter and its mutant variants, transiently expressing them in K562 cells, and measuring luciferase activity (Figure 5B). Comparison of the strengths of the β-promoter fragments from −136 to +54 bp that include the PCS and its mutant variants were made in the presence of a 736-bp core HS2 (from nucleotides 8486 to 9222 on the human globin locus sequence) placed at the enhancer site of the pGL3 vector.
Mutations in the PCS affect EMSA bands and β-promoter transcription.
(A) Mutations in the PCS influence the DNA-protein complex formation. EMSA was carried out with the wild-type PCS and its mutant variants by using K562 nuclear extracts. The 20-μL reaction mixture contained 2 ng 32P-labeled double-stranded DNA (∼100 000 cpm) in the binding buffer and 5 μg nuclear extract. The sequences of the sense strand of each of the double-stranded 32P-labeled DNA probes is as follows: wild type, oligo 1; AT to CC substitution, oligo 2; Δ GACAGGTA, oligo 3; and Δ CGGCT, oligo 4 (Table 1). Sequences from the pGL3 plasmid-cloning sites were included in oligo 3 and 4 to maintain their length equivalent to the wild-type PCS oligonucleotide. The free probe lane is the mixture of all the above sequences in the binding reaction mixture without nuclear extracts. The EMSA bands are numbered and indicated by arrows. (B) Functional effects of mutations in the PCS. The wild-type β-promoter from −136 to +54 bp and its mutants were generated by PCR and cloned into the Hind III-XhoI site of the pGL3. The forward primers used in the PCR are as follows: wild-type β-promoter, oligo 5; β-promoter with AT to CC substitution in the PCS, oligo 6; β-promoter with Δ GACAGGTA in the PCS, oligo 7; and β-promoter with Δ CGGCT in the PCS, oligo 8. Oligo 9 served as the reverse primer in all the PCR reactions. A 736-bp HS2 (from nucleotides 8486 to 9222 on the human β-globin locus) was cloned at the BamHI-SalI site of the pGL3. All the values in the bar diagram are the averages of 3 to 8 separate experiments, most of which were done in duplicate. The luciferase activities are normalized as activity per 10 μg total protein. Horizontal lines above each bar indicate 1 SD in the average of the experimental values.
Mutations in the PCS affect EMSA bands and β-promoter transcription.
(A) Mutations in the PCS influence the DNA-protein complex formation. EMSA was carried out with the wild-type PCS and its mutant variants by using K562 nuclear extracts. The 20-μL reaction mixture contained 2 ng 32P-labeled double-stranded DNA (∼100 000 cpm) in the binding buffer and 5 μg nuclear extract. The sequences of the sense strand of each of the double-stranded 32P-labeled DNA probes is as follows: wild type, oligo 1; AT to CC substitution, oligo 2; Δ GACAGGTA, oligo 3; and Δ CGGCT, oligo 4 (Table 1). Sequences from the pGL3 plasmid-cloning sites were included in oligo 3 and 4 to maintain their length equivalent to the wild-type PCS oligonucleotide. The free probe lane is the mixture of all the above sequences in the binding reaction mixture without nuclear extracts. The EMSA bands are numbered and indicated by arrows. (B) Functional effects of mutations in the PCS. The wild-type β-promoter from −136 to +54 bp and its mutants were generated by PCR and cloned into the Hind III-XhoI site of the pGL3. The forward primers used in the PCR are as follows: wild-type β-promoter, oligo 5; β-promoter with AT to CC substitution in the PCS, oligo 6; β-promoter with Δ GACAGGTA in the PCS, oligo 7; and β-promoter with Δ CGGCT in the PCS, oligo 8. Oligo 9 served as the reverse primer in all the PCR reactions. A 736-bp HS2 (from nucleotides 8486 to 9222 on the human β-globin locus) was cloned at the BamHI-SalI site of the pGL3. All the values in the bar diagram are the averages of 3 to 8 separate experiments, most of which were done in duplicate. The luciferase activities are normalized as activity per 10 μg total protein. Horizontal lines above each bar indicate 1 SD in the average of the experimental values.
Deletion of the CGGCT sequence from the PCS selectively abolishes HLTF-containing band 2 and a non-HLTF band 7, and decreases transcription to half (P = .01). However, an AT to CC substitution mutation abolishes the HLTF-containing EMSA bands 1, 4, 5, and 8 along with non-HLTF bands 3, 6, and 7 and stimulates transcription by 2.5-fold (P = .03) (Figure 5B). One possible explanation for the difference in the effects of these 2 mutations would be that HLTF acts as a transcriptional activator and transcription factors present in the non-HLTF EMSA bands down-regulate transcription. This explanation is consistent with the additional observation that deletion of the GACAGGTA sequence from PCS selectively abolishes non-HLTF EMSA bands 6 and 7, and in transient transcription assay this deletion mutation stimulates the β-promoter activity (P = .002). Further characterization of the various proteins in these EMSA bands and their functional significance is needed.
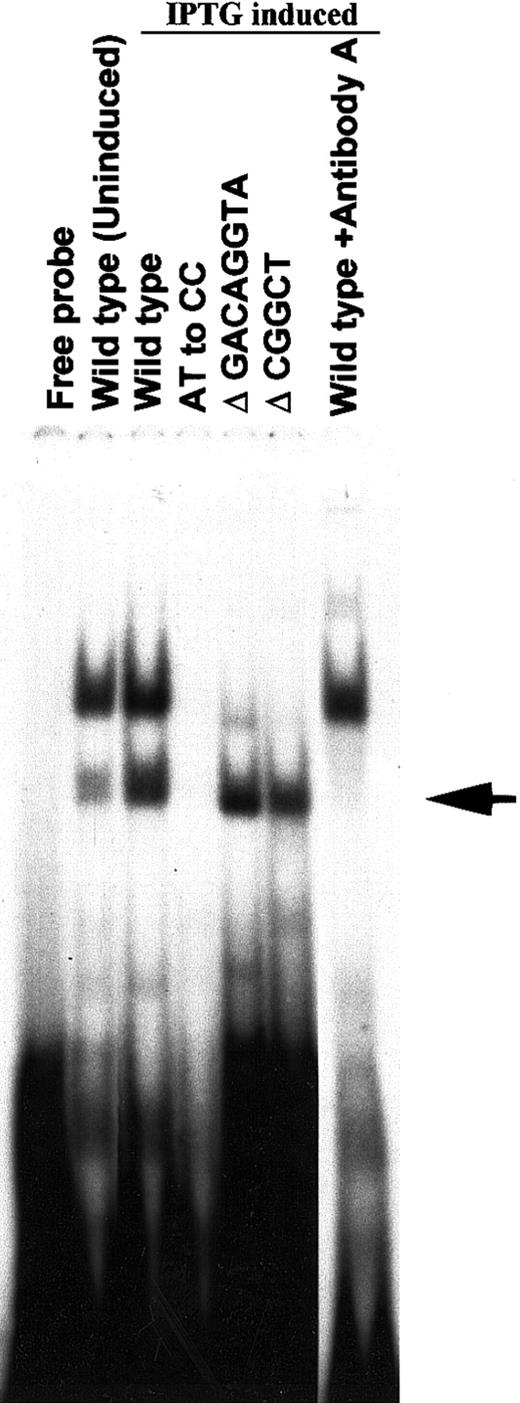
To demonstrate the direct binding of the N-terminal 38-kd initial recombinant HLTF isolated by screening the λ gt11 expression cDNA library of K562 cells, we cloned the cDNA of this protein in pTrcHis2 TOPO vector and expressed it in E coli with Myc and His tags at the C-terminal end. The EMSA with the bacterial extracts expressing the 38-kd HLTF and gel supershift/neutrilization assay with antibody A showed direct binding of the recombinant HLTF with the PCS (Figure6). The binding properties of native HLTF in K562 cells and the recombinant 38-kd HLTF are similarly affected by the AT to CC substitution mutation and Δ GACAGGTA mutation in the PCS (Figures 5 and 6). However, unlike the native HLTF in EMSA band 2 with K562 nuclear extracts, the 38-kd HLTF binds to the PCS with Δ CGGCT mutation. Therefore, there appears to be at least 2 sequence-specific HLTF-containing protein complexes that differentially interact with the PCS in K562 cells.
Binding of the recombinant 38-kd HLTF to wild-type and mutated PCS.
A cDNA clone coding for the N-terminal 38-kd HLTF (aa 2 to aa 332) isolated by screening the λ gt11 cDNA library of K562 cells was cloned into pTrcHis2TOPO vector and expressed in E colicells. The protein extracts of uninduced and IPTG-induced bacterial cells were used in the EMSA. The wild-type and mutant versions of the PCS oligonucleotides are indicated, and their sequences are described in Figure 5. The 20-μL EMSA binding mixture contained 2 ng32P-labeled double-stranded DNA (∼100 000 cpm) in the binding buffer and 1 μL E coli protein extract. The binding reaction was started by the addition of the32P-labeled DNA probe. The reaction mixture was incubated for 10 minutes at room temperature and then analyzed on a 5% polyacrylamide gel. For gel supershift/neutralization assay with antibody A, the bacterial extract, and the antibody were incubated for 5 minutes before the addition of the 32P DNA probe. The EMSA band obtained with 38-kd HLTF is shown by an arrow. The upper EMSA band is due to a bacterial protein in the extract.
Binding of the recombinant 38-kd HLTF to wild-type and mutated PCS.
A cDNA clone coding for the N-terminal 38-kd HLTF (aa 2 to aa 332) isolated by screening the λ gt11 cDNA library of K562 cells was cloned into pTrcHis2TOPO vector and expressed in E colicells. The protein extracts of uninduced and IPTG-induced bacterial cells were used in the EMSA. The wild-type and mutant versions of the PCS oligonucleotides are indicated, and their sequences are described in Figure 5. The 20-μL EMSA binding mixture contained 2 ng32P-labeled double-stranded DNA (∼100 000 cpm) in the binding buffer and 1 μL E coli protein extract. The binding reaction was started by the addition of the32P-labeled DNA probe. The reaction mixture was incubated for 10 minutes at room temperature and then analyzed on a 5% polyacrylamide gel. For gel supershift/neutralization assay with antibody A, the bacterial extract, and the antibody were incubated for 5 minutes before the addition of the 32P DNA probe. The EMSA band obtained with 38-kd HLTF is shown by an arrow. The upper EMSA band is due to a bacterial protein in the extract.
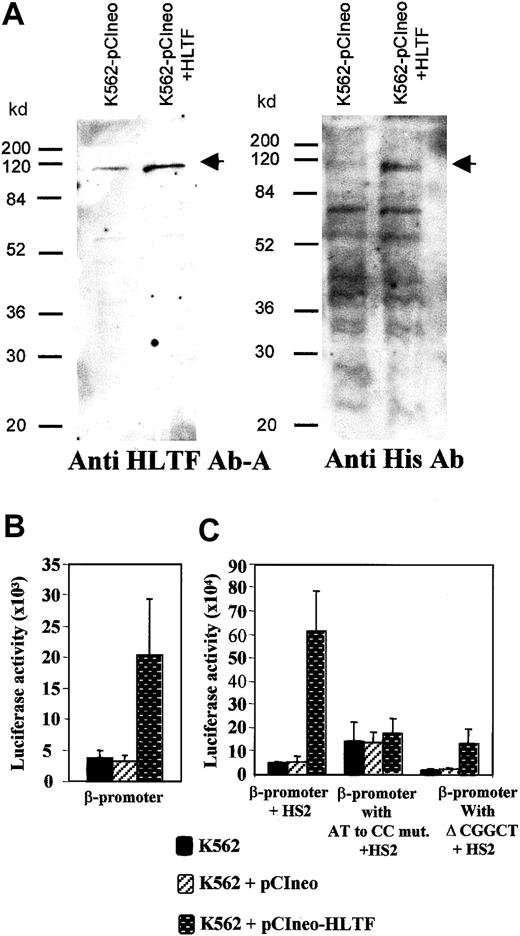
To further investigate the role of HLTF in β-globin transcription, we overexpressed HLTF in K562 cells. For this purpose, we generated K562 cells that were stably transfected with a pCIneo vector containing HLTF tagged with poly histidine at its N-terminal end. Western analysis of the whole cell extracts of these K562 cells with anti-HLTF antibody A and antibodies against the His tag confirmed the overexpression of HLTF (Figure 7A). If HLTF is an activator of β-globin transcription, overexpression of HLTF in K562 cells should enhance transient transcription. We tested the β-promoter activity in these HLTF-overexpressing cells by transient transfection of the pGL3 constructs containing β-promoter in the presence and absence of HS2. There was a 5.5-fold increase in the luciferase activity in HLTF-overexpressing cells that are transfected with pGL3 containing wild-type β-promoter alone (Figure 7B). In the presence of HS2, there was a 13-fold average increase in the transcription in the HLTF-overexpressing cells (Figure 7C). An AT to CC substitution mutation and deletion of CGGCT sequence from the PCS that inhibit HLTF binding (Figure 5A) significantly decreases the β-promoter activity in these HLTF-overexpressing cells (Figure 7C). The P values for the changes in luciferase activities for β-promoter with AT to CC mutation and with Δ CGGCT in comparison to the normal β-promoter activity in the presence of HS2 are .014 and .006, respectively.
Overexpression of HLTF in K562 cells and its effect on the β-globin transcription.
(A) Western blot of total protein extract of K562 cells stably transfected with pCIneo vector alone and pCIneo containing HLTF with poly His tag at its N-terminal end. Protein (70 μg) was loaded in each lane. The cell-free extracts are indicated at the top of the panel, and standard molecular weight marker positions are indicated at the left-hand side. The arrows mark points at the HLTF band. The antibodies against HLTF and poly His used to probe the blot are mentioned at the bottom of the panel. In the blot probed with the anti-His antibody, multiple bands found below the 115-kd HLTF band are common to both the lanes. These bands presumably are due to the binding of anti poly-His antibody to proteins containing short stretches of histidines. (B) Transient transfections of −136 to +54 bp β-promoter (cloned into theHindIII-XhoI site of the pGL3) into normal and HLTF-overexpressing cells. (C) Transient transfection of pGL3 constructs containing β-promoter and its mutant variants and HS2 at the enhancer site. These constructs are described in Figure 5. The values in the bar diagram are the averages of 12 experiments, including 3 separate experiments performed in duplicate with each of the 2 separate pools of K562 cells stably transfected with HLTF. The luciferase activities are normalized as activity per 10 μg total protein. Horizontal lines above each bar indicate 1 SD in the average of the experimental values.
Overexpression of HLTF in K562 cells and its effect on the β-globin transcription.
(A) Western blot of total protein extract of K562 cells stably transfected with pCIneo vector alone and pCIneo containing HLTF with poly His tag at its N-terminal end. Protein (70 μg) was loaded in each lane. The cell-free extracts are indicated at the top of the panel, and standard molecular weight marker positions are indicated at the left-hand side. The arrows mark points at the HLTF band. The antibodies against HLTF and poly His used to probe the blot are mentioned at the bottom of the panel. In the blot probed with the anti-His antibody, multiple bands found below the 115-kd HLTF band are common to both the lanes. These bands presumably are due to the binding of anti poly-His antibody to proteins containing short stretches of histidines. (B) Transient transfections of −136 to +54 bp β-promoter (cloned into theHindIII-XhoI site of the pGL3) into normal and HLTF-overexpressing cells. (C) Transient transfection of pGL3 constructs containing β-promoter and its mutant variants and HS2 at the enhancer site. These constructs are described in Figure 5. The values in the bar diagram are the averages of 12 experiments, including 3 separate experiments performed in duplicate with each of the 2 separate pools of K562 cells stably transfected with HLTF. The luciferase activities are normalized as activity per 10 μg total protein. Horizontal lines above each bar indicate 1 SD in the average of the experimental values.
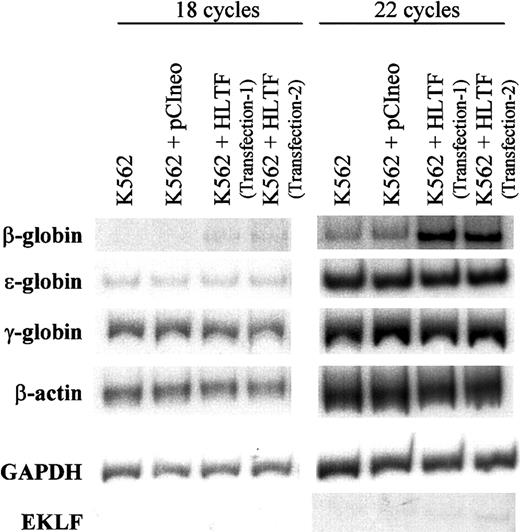
To understand the effect of HLTF on β-promoter transcription in the context of normal chromatin structure, we estimated the levels of endogenous β-globin transcript by semiquantitative RT-PCR experiments using specific primers for ε-, γ-, and β-globins (Figure8). The levels of β-actin and GAPDH messages were similar in the normal K562 cells and also in those stably transfected with pCIneo alone and pCIneo with His-tagged HLTF. In all these cell lines, the levels of ε- as well as γ-globin messages remain unaltered, whereas in HLTF-overexpressing K562 cells, we observed substantial increase in the β-globin message. The RT-PCR band was confirmed to be derived from spliced β-globin mRNA by sequence analysis. Thus, HLTF can selectively enhance adult β-globin transcription in K562 cells.
RT-PCR of total RNA from normal and HLTF-overexpressing K562 cells.
RT-PCR conditions are described in the “Materials and methods” section. After 18 (left) or 22 (right) cycles, the PCR products were run on a 6% polyacrylamide gel for 2.5 hours, dried, and exposed to x-ray film. The data are representative of 3 independent experiments. K562 + HLTF transfection-1 and transfection-2 are the 2 pools of K562 cells stably transfected with pCIneo containing HLTF in 2 independent transfections. The exposure times were adjusted for each set of PCR to obtain bands of optimum intensity. The autoradiogram exposure times are as follows: 16 hours at room temperature (RT) for β-globin, 12 hours at −80°C for EKLF, 5 hours at RT for ε-globin, 15 minutes at RT for γ-globin, 1 hour at RT for β-actin, and 0.5 hours at RT for GAPDH.
RT-PCR of total RNA from normal and HLTF-overexpressing K562 cells.
RT-PCR conditions are described in the “Materials and methods” section. After 18 (left) or 22 (right) cycles, the PCR products were run on a 6% polyacrylamide gel for 2.5 hours, dried, and exposed to x-ray film. The data are representative of 3 independent experiments. K562 + HLTF transfection-1 and transfection-2 are the 2 pools of K562 cells stably transfected with pCIneo containing HLTF in 2 independent transfections. The exposure times were adjusted for each set of PCR to obtain bands of optimum intensity. The autoradiogram exposure times are as follows: 16 hours at room temperature (RT) for β-globin, 12 hours at −80°C for EKLF, 5 hours at RT for ε-globin, 15 minutes at RT for γ-globin, 1 hour at RT for β-actin, and 0.5 hours at RT for GAPDH.
Discussion
In the present study, we have focused on a phylogenetically conserved β-promoter sequence, the PCS, which lies at −115 to −136 bp relative to the transcription start site of the human β-promoter (Figure 1). EMSA with K562 nuclear extracts reveal multiple bands that bind to the PCS in a sequence-specific manner (Figure 2). We inquired whether in K562 cells the protein(s) binding to this sequence acts as either an inhibitor or an activator of transcription of the β-globin gene whose activity is attenuated, perhaps due to the absence of other globin transcription activators such as EKLF.49
By screening a K562 cDNA expression library in λ gt11, we identified a previously described HLTF belonging to the SWI2/SNF2 family that binds to an oligonucleotide derived from the PCS sequence and is part of several of the complexes formed on the PCS in vitro (Figure 3). The proteins belonging to SWI2/SNF2 family are found in cells as part of multiprotein complexes.50-52 HLTF is known to form complexes with SP1 and SP3 in HeLa cells,53 which speaks to its ability to participate in multiprotein complexes. Moreover, HLTF contains a C-terminal ring finger domain that potentially participates in protein-protein interactions. The AT to CC substitution and Δ CGGCT mutations in the PCS selectively abolish one or another HLTF-containing band (Figure 5A), suggesting that there are at least 2 different HLTF complexes that may form on the PCS.
In the β-globin locus, SWI/SNF complexes are recruited to the CACCC site of the β-promoter by the transcription factor EKLF and on a 250-bp AT-rich sequence situated between the γ and δ genes by the IKAROS transcription factor.50,51 These complexes contain a protein called BRG1 that forms their core ATPase subunit. Structurally and functionally, BRG1 and HLTF have some similarities and some distinct differences. Both contain DNA-dependent ATPase activity44 and 7 helicase domains, but neither has been demonstrated to have helicase activity. Although the C-terminal regions of both these proteins contain protein-protein interaction motifs, their structural domains are different. The BRG1 contains a bromo domain, whereas HLTF possesses a ring finger domain. Significantly, BRG1 does not possess a DNA-binding domain, whereas HLTF contains a unique DNA-binding motif at the N-terminal region between aa 123 to aa 219.47
We constructed K562 cells that stably overexpressed HLTF, so as to study the effect of HLTF on endogenous globin genes as well as on a transiently transfected reporter plasmid. Overexpression of HLTF in K562 cells results in an increase in the transcription from transiently transfected β-promoter luciferase constructs, both in the absence and in the presence of HS2 (Figure 7). An AT to CC substitution mutation that removes 4 of the HLTF bands in EMSA significantly diminishes the stimulatory effect of HLTF overproduction (Figure 7). Another Δ CGGCT mutation in the PCS that removes one of the HLTF gel shift bands also inhibits the transcription in transiently transfected normal as well as HLTF-overexpressing K562 cells (Figures 5 and 7). These results argue that HLTF is an activator of β-globin transcription and acts through the PCS in transient transfections.
Overexpression of HLTF also significantly increases the endogenous levels of β-globin message (Figure 8) but does not alter the levels of either ε- or γ-globin messenger RNA (mRNA), suggesting that HLTF has access to and can specifically act on the endogenous β-globin gene. Identification of HLTF as an adult β-globin–specific transcriptional activator strengthens the notion that preferential activation of transcription is crucial for the developmental regulation of β-like genes. Apart from K562 cells, we have detected HLTF in adult β-globin–producing mouse erythroleukemic cells and human MB-02 cells by RT-PCR analysis and gel supershift assays with antibody A (data not shown). Among the transcription factors acting on globin genes, only EKLF has been suggested to preferentially activate β-globin transcription.54 EKLF knock-out mice die of anemia during fetal erythropoiesis because of severe deficiency of adult β-globin.55,56 In transgenic mice carrying the complete human β-like globin locus, expression of the human β-globin gene is severely hampered in an EKLF−/−background.57 58 K562 cells do not synthesize EKLF and lack detectable levels of β-globin mRNA. Because of the absence of EKLF, chromatin-remodeling activity may not be recruited and may severely limit the activity of the β-promoter. In this situation, the exogenous expression of HLTF might have activated EKLF transcription, which, in turn, elevates the β-globin mRNA levels. However, the RT-PCR data suggest that the overexpression of HLTF does not activate EKLF transcription (Figure 8), so that the induction of β-globin transcription in HLTF-overexpressing K562 cells is independent of this factor.
It remains to be seen whether HLTF action is even partly analogous to that of the more extensively studied SWI/SNF complexes. Within the SWI2/SNF2 family of proteins, HLTF is closest in sequence to yeast DNA repair proteins RAD5 and RAD16.44 In a recent report, a DNA repair transcription-coupling factor, CSB, has been shown to possess ATP-dependent chromatin remodeling activity,59 but a similar activity has not yet been demonstrated with HLTF. As the PCS and CACC sites of the β-promoter are adjacent to each other, there may be synergism between the HLTF and EKLF-bound SWI/SNF complex. However, the mechanism of HLTF action in β-globin gene transcription appears to be different than that of EKLF, because overexpression of EKLF in K562 cells does not activate the endogenous β-globin gene.54 Further studies with K562 cells simultaneously overexpressing EKLF and HLTF may shed light on these aspects of the β-globin transcription.
EMSA and transient transfection assays with normal and mutant PCS indicate that, in addition to activator complexes containing HLTF, a transcription inhibitor(s) binds to the PCS (Figure 5). There are at least 2 other instances in the β-globin locus in which transcriptional activators as well as repressors bind to the same site. Bach1, a transcriptional activator, and Bach2, a transcriptional repressor, bind to a common DNA sequence in the HS2.60 In another case, SWI/SNF and NuRD complexes that positively and negatively influence transcription, respectively, bind to the same AT-rich sequence between the γ- and δ-globin genes.52 Similarly, HLTF and an unidentified inhibitor(s) of transcription may both bind to PCS. We are in the process of purification of additional PCS binding proteins to expedite analysis of their role in the overall regulation of β-globin transcription.
We thank Drs Beverly Chilton for providing us the antibodies against HLTF, Alexandra Belayew for the HLTF-pCIneo construct (Université de Mons-Hainaut, Mons, Belgium), and A. Raghunathan and Shigeru Yamaga (Weissman lab) for their help in computer-related work.
Supported by grant CA42556 from the National Cancer Institute, National Institutes of Health, Bethesda, MD. M.C.M. acknowledges a research fellowship from Cooley's Anemia Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sherman M. Weissman, Department of Genetics, Boyer Center for Molecular Medicine, Yale University School of Medicine, 295 Congress Ave, New Haven, CT 06536; e-mail: sherman.weissman@yale.edu.