Granulocyte-macrophage colony-stimulating factor (GM-CSF) is critical for promoting the long-term survival of lung- or airway-based eosinophils. Previously, we have shown that fibronectin and tumor necrosis factor α induced autocrine production of GM-CSF that markedly enhanced eosinophil survival. Cytokine release was preceded by and dependent on messenger RNA (mRNA) stabilization. Here, we show that mitogen-activated protein kinase (MAPK) activation is responsible for GM-CSF mRNA stabilization in peripheral blood eosinophils (pbeos). Activation of extracellular signal-regulated kinase (ERK) but not p38 correlated with GM-CSF mRNA stability. Although ERK inhibition completely prevented GM-CSF mRNA stabilization, p38 inhibition had a partial effect. To establish which MAPK was crucial, we transduced pbeos with dominant-active TatMEK1(E) or TatMKK3b(E) proteins that selectively phosphorylate ERK or p38, respectively. These studies showed that ERK but not p38 was sufficient for GM-CSF mRNA stabilization. These data are in contradistinction to the c-Jun NH2-termainal kinase–mediated regulation of interleukin 2 and 3 mRNAs and suggest unique regulatory features for GM-CSF mRNA in eosinophils.
Introduction
Eosinophils are a major source of proinflammmatory mediators in the airway and lung parenchyma of atopic and asthmatic patients.1 In the asthmatic lung, normally short-lived eosinophils become resistant to apoptosis through the autocrine production and release of granulocyte-macrophage colony-stimulating factor (GM-CSF).2 This process can be initiated by engagement of many cell-surface receptors (CD9, CD32, β1, and β7) or exposure to cytokines (tumor necrosis factor α [TNF-α]), suggesting a multiplicity of signaling cascades that result in GM-CSF production and secretion.3-6
Specifically, TNF-α in association with extracellular matrix protein fibronectin altered T-cell function.7,8 Recently, we have shown that eosinophils treated with the combination of TNF-α plus fibronectin (Fn) increased GM-CSF messenger RNA (mRNA) stability, leading to GM-CSF secretion and enhanced in vitro survival. Of note, GM-CSF mRNA stability in long-lived eosinophils derived from bronchoalveolar lavage after in vivo allergen challenge was essentially identical to peripheral eosinophils treated with TNF-α plus Fn.2 We have also shown that Y Box binding protein (YB-1) interacted with the 3′UTR AU-rich elements (AREs) preventing GM-CSF mRNA decay.9 Presumably the YB-1–GM-CSF mRNA interaction blocked RNAse recognition or cleavage.
To better understand the signaling cascades leading to mRNA stabilization, several studies have focused on the role of mitogen-activated protein kinases (MAPKs). Indeed, c-jun-NH2-terminal kinase (JNK) activation was required for interleukin 2 (IL-2) and IL-3 mRNA stabilization in T-cell or mast cell lines.10,11 Here, we investigated if MAPKs (ERK, p38, or JNK) were responsible for GM-CSF mRNA stabilization in TNF-α plus Fn-activated peripheral blood eosinophils (pbeos). We show that ERK and p38 were rapidly activated after pbeos were treated with TNF-α plus Fn, whereas JNK was largely unaffected. TNF-α alone activated p38 but not ERK and was unable to induce or to stabilize GM-CSF mRNA.2 ERK inhibitors (PD98059) blocked GM-CSF mRNA stabilization, and anti-p38 drugs (SB203580) had an intermediate effect, suggesting ERK was the dominant MAPK involved. Finally, by using the constitutively active MAPK kinases (MAPKKs), MEK1(E) and MKK3b(E) that activate, respectively, ERK and p38, we demonstrate that ERK is sufficient to stabilize GM-CSF mRNA, whereas p38 alone has no effects.
Materials and methods
Subjects and eosinophil preparation
Peripheral blood was obtained by venipuncture from patients with allergic rhinitis or asthma. All informed consent was acquired according to a protocol approved by the University of Wisconsin Human Subjects Committee.
Eosinophils (typically > 98% pure) were purified by using a negative immunomagnetic procedure as previously described.12 After isolation, eosinophils were maintained in RPMI 1640 medium, 10% fetal calf serum, and 50 μg/mL gentamicin (all from Gibco Life Technologies, Grand Island, NY), at 37°C in a 5% CO2environment.
Reagents and eosinophil activation
Antiactive ERK, antiactive JNK, and antitotal ERK antibodies were purchased from Promega (Madison, WI) and were used for Western blotting as recommended by the company. Antiactive p38 antibody was purchased from Calbiochem (La Jolla, CA). The ERK inhibitor (PD98059) and p38 inhibitor (SB203580) were supplied, respectively, by New England Biolabs (Beverly, MA) and A.G. Scientific (San Diego, CA). The inhibitors were dissolved in dimethyl sulfoxide and added to cell cultures 30 minutes before activation with TNF-α plus Fn.
Recombinant human TNF-α was purchased from R&D Systems (Minneapolis, MN). Human cellular Fn was purchased from Sigma (St Louis, MO). Human plasma Fn-coated 96-well tissue culture plates were purchased from Becton Dickinson (Bedford, MA). Eosinophils were activated as previously described2 with TNF-α (10 ng/mL) in 96-well tissue culture plates. For Fn experiments, eosinophils were cultured in Fn-coated plates with media supplemented with 20 μg/mL soluble cellular Fn. Eosinophils were activated for 30 minutes or 5 hours at 1 × 106 cells/mL before cell lysis for Western blotting or GM-CSF mRNA transfection, respectively.
Cell lysates and Western blotting
Following incubation, the cells were pelleted and resuspended in the lysis buffer (20 mM Tris, 137 mM NaCl, 1 mM EDTA, 0.1 mM sodium orthovanate, 10 mM sodium fluoride, 10 mM β-glycerol phosphate and protease inhibitors, pH 7.4, 1% Triton X-100, 0.25% deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]), as previously described.13 The cells were passed 10 times through a 29-gauge needle and incubated on ice for 10 minutes. The eosinophil lysates were centrifuged at 12 000g for 2 minutes to remove the insoluble material, and the supernatants were stored at −80°C before immunoblotting. The primary antibodies were detected with an antirabbit horseradish peroxidase–conjugated secondary antibody and developed by using the enhanced chemiluminescence system (Amersham, Piscataway, NJ). The signals obtained by autoradiography were quantified on an AlphaImager 2200 analysis system (Innotech, San Leandro, CA).
Plasmid constructions, mRNA transfection, and Northern blotting
Complementary DNA (cDNA) coding for human GM-CSF was obtained from the American Type Culture Collection (Rockville, MD). The plasmid for in vitro, wild-type GM-CSF mRNA synthesis has been described previously.14
Particle-mediated gene transfer of in vitro–transcribed mRNAs into cultured cells was performed by using the Accell Gene-Gun (Powerject, Madison, WI), as previously described.12
At indicated times, cells were pelleted and lysed in TRIreagent (Molecular Research Center, Cincinnati, OH), and total RNA was quantitatively isolated and analyzed by Northern blotting with a radioactively labeled GM-CSF or actin cDNA probes as described previously.12 GM-CSF mRNA signals were normalized to those for actin mRNA to accommodate any differences in the extraction, gel loading, and transfer of total RNA. After stringent washing at 50°C for 5 minutes with 0.1 × sodium chloride/sodium citrate and 0.1% SDS, the blots were quantitated by phosphorimaging (Model 445SI; Molecular Dynamics, Sunnyvale, CA).
MEK1(E) or MKK3b(E) transduction
The coding region of constitutively active human MAPKK (MEK1(E) or MKK3b(E))15,16 were generously provided by Jiahui Han, La Jolla, CA. The full-length coding region was cloned into pTatHA9,17,18 coding for a fusion protein containing from N to C terminus: 6-histidine, Tat translocation sequence, hemagglutinin epitope, and MEK1(E) or MKK3b(E) that was expressed in BL21 cells. As previously described,9 expressing bacterial cultures were dissolved in 8 M urea, 50 mM Tris pH 8.0, 150 mM NaCl, sonicated, and centrifuged at 15 000g. Cleared lysate was mixed with NiNTA resin (Qiagen, Valencia, CA) in the presence of 10 mM imidazole, and His-tagged protein was allowed to bind. The resin was washed 3 times with 8 M urea, 50 mM Tris pH 8.0, 150 mM NaCl, 10 mM imidazole. Protein was eluted with 1 M imidazole, 50 mM Tris pH 8.0, 150 mM NaCl and dialyzed against phosphate-buffered saline. Protein was more than 90% pure by Coomassie-stained SDS-polyacrylamide gel electrophoresis analysis. Tat fusion protein (10 nM; TatMEK1(E) or TatMKK3b(E)) was added to 8 × 106 cells for 2 hours at 37°C that were then transfected with the Gene-Gun as described above. After washing, GM-CSF mRNA stability (half-life time) was determined by Northern blot as described above, and for survival experiments the cells were cultured for 1 hour after transduction at 1 × 107cells/mL, diluted 1:10, and cultured for 4 days. Viability was determined as previously described.19
Statistical analysis
Results were expressed as mean ± SD or SEM. Statistical analysis was performed by using paired and unpaired Studentt tests. Correlation data were analyzed for statistical significance by using the Pearson test. P values < .05 were considered as statistically significant.
Results
Eosinophils treated with TNF-α plus Fn-activated ERK and p38 but not JNK
We have recently demonstrated that eosinophils treated with TNF-α plus Fn showed increased GM-CSF mRNA accumulation, stability, and GM-CSF protein production.2 In an effort to understand the signaling cascades underlying these effects, we evaluated MAPK activation. These kinases were essential for IL-2 and IL-3 mRNA stabilization in T and mast cells.10,11 As shown in Figure1A,B, the treatment of pbeos with TNF-α plus Fn activated ERK and p38 but not JNK, which was slightly phosphorylated in resting eosinophils. Of note, kinetic experiments showed that ERK and p38 reached their activation maxima between 5 and 60 minutes after stimulation, whereas JNK phosphorylation was unchanged (data not shown), consistent with another report.20PD98059 completely abrogated ERK phosphorylation but had no significant effect on the quantity of phosphorylated p38 (Figure 1A,B, lane 5). In contrast, U0126, another specific ERK inhibitor, prevented ERK and p38 phosphorylation equally well (not shown). Treatment of pbeos with TNF-α alone predominantly activated p38, although some phosphorylated ERK was detected, whereas Fn alone had the reverse effect, mostly activating ERK and only slightly p38 (Figure 1A-B). These data are consistent with prior studies in eosinophils.21 22 Thus, it is likely that Fn or TNF-α activated the appropriate pbeos pathways in our hands and suggested that selective MAPK activation might control GM-CSF mRNA stability.
ERK and p38 are activated by Fn or TNF-α in peripheral blood eosinophils.
Eosinophils were preincubated for 30 minutes with dimethyl sulfoxide alone or 50 μM PD98059 (PD), followed by 30 minutes with medium alone (Resting), TNF-α, fibronectin (Fn) (as described in “Materials and methods”), or TNF-α plus Fn (T + Fn). The cells were lysed, analyzed on a 10% SDS-polyacrylamide gel electrophoresis, and immunoblotted with antiactive ERK, antiactive p38, antiactive JNK, or anti-ERK1/ERK2 (Total-ERK). The signals obtained by autoradiography are shown (A) and are representative of 3 different donors. (B) The signals were quantified and normalized to total-ERK. Each value represents the mean (SEM) of 3 different donors.
ERK and p38 are activated by Fn or TNF-α in peripheral blood eosinophils.
Eosinophils were preincubated for 30 minutes with dimethyl sulfoxide alone or 50 μM PD98059 (PD), followed by 30 minutes with medium alone (Resting), TNF-α, fibronectin (Fn) (as described in “Materials and methods”), or TNF-α plus Fn (T + Fn). The cells were lysed, analyzed on a 10% SDS-polyacrylamide gel electrophoresis, and immunoblotted with antiactive ERK, antiactive p38, antiactive JNK, or anti-ERK1/ERK2 (Total-ERK). The signals obtained by autoradiography are shown (A) and are representative of 3 different donors. (B) The signals were quantified and normalized to total-ERK. Each value represents the mean (SEM) of 3 different donors.
ERK activation was correlated with GM-CSF mRNA stabilization
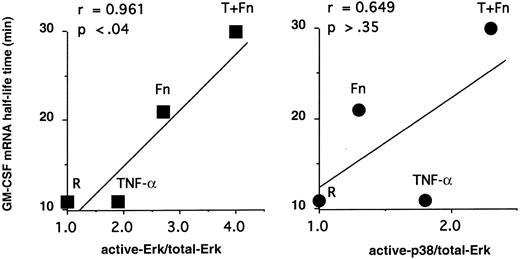
To connect MAPK with mRNA decay, we correlated GM-CSF mRNA stability in pbeos after exposure to TNF-α, Fn, or both agents with the level of MAPK activation (Figure 2). Clearly, GM-CSF mRNA stability was highly correlated with ERK activation (r = 0.961; P < .04), whereas p38 phosphorylation was not (Figure 2; r = 0.649;P > .35). Consistent with these data, GM-CSF mRNA never accumulated nor was stabilized in eosinophils treated with TNF-α2 that activates p38 (Figure 1).
GM-CSF mRNA stability is correlated with ERK activation but not p38.
The ratios of act-ERK/total-ERK and p38/total-ERK determined in Figure1 were plotted versus GM-CSF mRNA half-life times (minutes) determined previously.2 The Pearson test was performed, confirming a statistical correlation between GM-CSF mRNA stabilization and ERK activation but not p38 activation.
GM-CSF mRNA stability is correlated with ERK activation but not p38.
The ratios of act-ERK/total-ERK and p38/total-ERK determined in Figure1 were plotted versus GM-CSF mRNA half-life times (minutes) determined previously.2 The Pearson test was performed, confirming a statistical correlation between GM-CSF mRNA stabilization and ERK activation but not p38 activation.
ERK inhibitors block GM-CSF mRNA stabilization
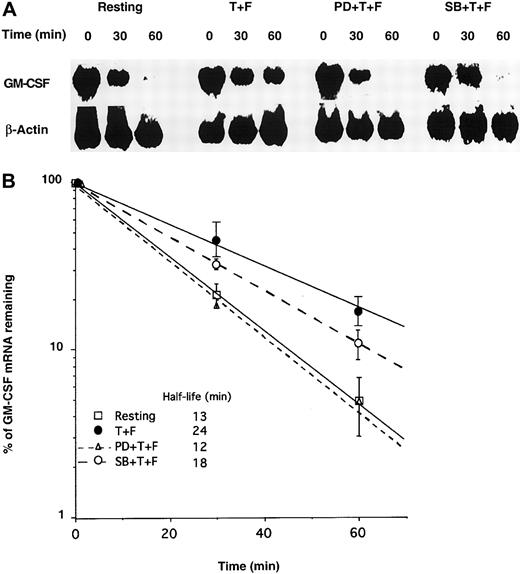
To confirm the respective roles of ERK and p38, we transfected GM-CSF mRNA into pbeos following treatments with MAPK inhibitors and TNF-α plus Fn. The decay rate of exogenous GM-CSF mRNA was measured by Northern blotting. As previously reported by our laboratory,2 TNF-α plus Fn stabilized GM-CSF mRNA in pbeos (GM-CSF mRNA half-life = 24 minutes) (Figure3A-B), which was completely prevented by the ERK inhibitor (PD98059) (GM-CSF mRNA half-life = 12 minutes) (Figure 3A-B), suggesting that ERK activation was necessary for GM-CSF mRNA stabilization. The p38 inhibitor (SB203580) had an intermediate effect, attenuating GM-CSF mRNA stabilization by approximately 50%, suggesting that p38 contributed to GM-CSF turnover in cooperation with ERK in the context of TNF-α plus Fn-activated pbeos. However, it remained possible that SB203580 nonspecifically affected the Erk pathway.
PD98059 inhibits GM-CSF mRNA stabilization in eosinophils activated by TNF-α plus Fn.
Eosinophils were incubated for 30 minutes with medium alone, 50 μM PD98059 (PD), or 10 μM SB203580 (SB). Then cells were activated for 5 hours with TNF-α + Fn (T + F) or medium alone (Resting) prior to transfection with GM-CSF mRNA by particle-mediated gene transfer. (A) At the indicated time points, equal numbers of cells were harvested, and total RNA was quantitatively isolated and Northern blotted with 32P-labeled GM-CSF or β-actin cDNA probes. Signals were visualized by using a PhosphorImager. (B) Radioactive signals were quantified and normalized to β-actin mRNA and plotted versus time. Each point is the mean (SD) of 3 experiments with 3 different donors.
PD98059 inhibits GM-CSF mRNA stabilization in eosinophils activated by TNF-α plus Fn.
Eosinophils were incubated for 30 minutes with medium alone, 50 μM PD98059 (PD), or 10 μM SB203580 (SB). Then cells were activated for 5 hours with TNF-α + Fn (T + F) or medium alone (Resting) prior to transfection with GM-CSF mRNA by particle-mediated gene transfer. (A) At the indicated time points, equal numbers of cells were harvested, and total RNA was quantitatively isolated and Northern blotted with 32P-labeled GM-CSF or β-actin cDNA probes. Signals were visualized by using a PhosphorImager. (B) Radioactive signals were quantified and normalized to β-actin mRNA and plotted versus time. Each point is the mean (SD) of 3 experiments with 3 different donors.
ERK activation was sufficient for GM-CSF mRNA stabilization and increased eosinophil survival
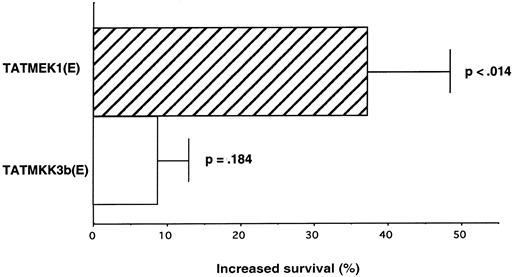
To demonstrate whether ERK or p38 activation was sufficient for GM-CSF mRNA stabilization, we transduced pbeos with either the constitutively active TatMEK1(E) or TatMKK3b(E) that respectively and selectively activate ERK or p38.15,16 As previously described with TatYB-1 protein,9 pbeos were incubated for 2 hours with TatMEK1(E) or TatMKK3b(E) prior to analysis of their effects on GM-CSF mRNA stability. Table 1shows that MEK1(E) stabilized GM-CSF mRNA by more than 2-fold (t1/2 from 12 to 27 minutes; P < .048, n = 5), whereas MKK3b(E) had no effect (t1/2 from 13 to 12 minutes).
As a consequence of increased GM-CSF mRNA stability, we anticipated eosinophil survival would be prolonged compared with untreated controls. Therefore, viability was determined 4 days after TatMEK1(E) or TatMKK3B(E) transduction and GM-CSF mRNA transfection. As previously described12 GM-CSF mRNA transfection increased eosinophil in vitro survival at 4 days from about 10% for control eosinophils to approximately 30%. The survival of TatMEK1(E)-transduced cells was increased by an additional 35% over cells solely transfected with GM-CSF mRNA that was statistically significant as calculated by the paired Student t test (P < .014). Consistent with its failure to alter GM-CSF mRNA stability, TatMKK3b(E) had no significant effect on eosinophil survival (Figure 4;P = .184) compared with GM-CSF mRNA-transfected controls. Finally, cells transfected but then transduced with Tatβgal also failed to show enhanced survival,9 demonstrating the Tat tag did not have nonspecific effects.
MEK1(E) increases eosinophil survival after GM-CSF mRNA transfection.
Eosinophils were transduced for 2 hours with 10 nM MEK1(E) or MKK3b(E) and transfected with GM-CSF mRNA by using the Gene-Gun. One hour after transfection, the cell cultures were diluted by 10-fold (1 × 106 cells/mL), and the survival was determined 4 days later by trypan blue. The survival for transduced pbeos was compared with the nontransduced control and calculated as follows: (% survival with TatMKKinase/% survival with transfected but nontransduced cells) − 1 × 100. Data were analyzed by paired Student t test.
MEK1(E) increases eosinophil survival after GM-CSF mRNA transfection.
Eosinophils were transduced for 2 hours with 10 nM MEK1(E) or MKK3b(E) and transfected with GM-CSF mRNA by using the Gene-Gun. One hour after transfection, the cell cultures were diluted by 10-fold (1 × 106 cells/mL), and the survival was determined 4 days later by trypan blue. The survival for transduced pbeos was compared with the nontransduced control and calculated as follows: (% survival with TatMKKinase/% survival with transfected but nontransduced cells) − 1 × 100. Data were analyzed by paired Student t test.
Discussion
In this present study, we have investigated signaling pathways that mediate GM-CSF posttranscriptional regulation in eosinophils after TNF-α plus Fn activation. Our data demonstrate that the combination of TNF-α plus Fn led to a rapid and significant phosphorylation of ERK and p38 (Figure 1). Correlation and inhibitor studies (Figures 2 and 3) showed that ERK activation was highly associated with and likely essential for increased GM-CSF mRNA stability. Finally, transduction of the constitutively active TatMEK1 demonstrated that ERK regulates GM-CSF mRNA stability (Table 1) despite our observation that TNF-α plus Fn induced the phosphorylation of both ERK and p38. In addition, p38 activation was not correlated with GM-CSF mRNA stabilization (Figure 2) nor did constitutively active TatMKK3b(E) alter GM-CSF mRNA half-life time (Table 1). However, as the p38 inhibitor (SB203580) prevented 50% of the increase in GM-CSF mRNA stability after TNF-α plus Fn activation (Figure 3), it seems likely that p38 acts as a cofactor with ERK. It remains possible that SB203580 nonspecifically affects ERK or MAPK-activated protein kinases (MAPKAPKs) downstream of ERK. Several studies have described p38 and ERK signaling converge at multiple MAPKAPKs, including MAPKAPK5, 3pK, or MnK1/2.23-26 These kinases could be downstream of MAPK in the pathways leading to GM-CSF mRNA stability. Although we were unable to observe SB203580 modulation of ERK phosphorylation (data not shown), Ludwig et al27 noted such an effect in human embryonic kidney cells. Because of the small number of available cells, we did not determine if ERK activity was partially inhibited by SB203580.
Most of the studies analyzing the role of MAPK on mRNA stability have used transformed cell lines (Jurkat, Hela, THP-1, fibroblastlike, or mast cell–like) as models. These studies have generally implicated p38 or JNK as critical kinases.10,11,28-32 ERK has, however, been linked to macrophage inflammatory protein-2 or nucleolin mRNA stability in fresh rat peritoneal neutrophils or human peripheral blood mononuclear cells, respectively.33-35 Whether mRNA regulation in primary cells, including eosinophils specifically, depends on ERK phosphorylation remains unclear. Many previous studies have shown ERK activation as a consistent event after in vitro activation of eosinophils. For example, eotaxin induced both ERK and p38 activation in eosinophils, whereas JNK was constitutively phosphorylated.20 Similarly, IL-5 activated both MAPK (ERK and p38),36,37 whereas guinea pig eosinophil activation with leukotriene B4 or human eosinophil adhesion to Fn was rapidly followed by only ERK phosphorylation.22,38 Macrophage inflammatory protein-3α–induced human eosinophil migration also involved ERK activation.39
The additive effects of TNF-α plus Fn on ERK phosphorylation were somewhat unexpected. TNF-α has been shown to facilitate the association of Fn with integrins.7,8 Given the rapidity of ERK activation, it is unlikely that TNF-α induces an up-regulation of cell surface β1 integrin expression that in turn promotes Fn signaling.40 For similar reasons, ERK phosphorylation could not be a result of GM-CSF release that occurs several hours later. Thus, we conclude that the combination of TNF-α plus Fn redirects and amplifies intracellular signaling toward ERK.
Previously, we demonstrated that TNF-α plus Fn-induced eosinophil survival was completely dependent on GM-CSF release and its subsequent action on eosinophils.2 Here, we show that direct ERK activation either by TNF-α plus Fn or TatMEK1 leads to the same result. Thus, despite prior data suggesting that ERK directly antagonizes Bcl-XL/Bcl-2–associated death promoter (BAD)–mediated cell death through Rsk activation,41 our aggregate data suggest GM-CSF does not prolong survival by ERK but rather by the Janus kinase/STAT pathway42 or phosphatidylinositol 3′-kinase.43
Our data also represent an important link in understanding how GM-CSF mRNA and other cytokine mRNAs are stabilized on cell activation. This process likely involves several ARE-specific binding proteins, including HuR, hnRNP A/C, nucleolin, and YB-1.9,35,44 45 These activities likely target the ARE and modify the recognition of labile cytokine mRNAs by the decay machinery. However, despite a common 3′ untranslated ARE, GM-CSF and IL-2 mRNA stability are dependent on ERK/p38 or JNK, respectively. This dependency suggests that novel effectors must also be involved that are differentially regulated by these distinct MAPKs.
We thank the other members of the SCOR-asthma group, particularly Julie Sedgwick for providing pbeos and Mary Ellen Bates for her assistance with the MAPK analyses. We also thank Beth Capowski and Syrus Soltaninassab for their assistance for Tat protein production.
Supported by Project 5 of SCOR-asthma-P50HL56396 from the National Institutes of Health (J.S.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James S. Malter, Department of Pathology and Laboratory Medicine K4/812-CSC, University of Wisconsin Hospital and Clinic, 600 Highland Ave, Madison, WI 53792; e-mail:jsmalter@facstaff.wisc.edu.