This study aimed at determining the prevalence of INK4deletions and their impact on outcome in 125 children with acute lymphoblastic leukemia (ALL) at first relapse using real-time quantitative polymerase chain reaction. Patients were enrolled into relapse trials ALL-REZ BFM (ALL-Relapse Berlin–Frankfurt–Münster) 90 and 96. The prevalence of p16INK4a and p15INK4bhomozygous deletions was 35% (44 of 125) and 30% (38 of 125), respectively. A highly significant association of both gene deletions was found with the 2 major adverse prognostic factors known for relapsed childhood ALL: T-cell immunophenotype and first remission duration. There was no correlation between INK4 deletions and probability of event-free survival. These findings argue against an independent prognostic role of INK4 deletions in relapsed childhood ALL.
Introduction
Homozygous deletions of the tumor suppressor genes p16INK4a and p15INK4b are found in approximately 25% of childhood acute lymphoblastic leukemia (ALL) at first presentation (5% to 20% in B-cell precursor and 60% to 80% in T-cell ALL). The prognostic importance of these alterations is controversial. Whereas an adverse prognostic effect of p16INK4a and p15INK4bdeletions has been found in some studies on pediatric1,2and adult ALL,3 others studying a larger number of patients were not able to confirm these findings.4,5However, the coincidence of these deletions with T-cell immunophenotype and high white blood cell (WBC) count is unequivocal.6 7The prevalence of INK4 deletions in relapsed ALL and their prognostic importance have not been determined yet.
We investigated bone marrow (BM) samples from 125 children with first relapse of ALL enrolled into relapse trials ALL-REZ BFM (ALL-Relapse Berlin–Frankfurt–Münster) 90/96 for INK4deletions to compare the prevalence with that in initial disease and to elucidate their possible prognostic importance in ALL relapse.
Study design
Patients and treatment
BM samples (containing more than 75% BM blasts) collected prior to chemotherapy at first relapse of ALL from 125 children enrolled into trials ALL-REZ BFM 90 and 968 were analyzed retrospectively. The studied patients are representative of the overall population enrolled in these trials with regard to sex, time of relapse, blast count, and immunophenotype. Treatment consisted of chemotherapy and central nervous system irradiation; bone marrow transplantation was performed when indicated. BM samples were obtained after informed and written consent. Median follow-up was 894 days (range, 176-2058 days).
DNA extraction and real-time quantitative polymerase chain reaction
Preparation of cell samples and DNA extraction were performed as described previously.9
We developed a gene-dosage assay using TaqMan real-time quantitative (rq-) polymerase chain reaction (PCR), with p16INK4a and p15INK4b as test sequences (TS) and β-globin as a reference gene (RG). The normalized ratio of TS/RG yields a value of approximately 1 in samples without the deletion, whereas ratios with values close to 0 indicate homozygous deletion.10,11 Rq-PCR reactions were carried out in separate tubes with specific primers and dually labeled TaqMan probes for each sequence. Serial dilutions of genomic DNA with known concentrations were amplified simultaneously to generate a calibration curve. Samples were analyzed in duplicate (TS and RG), and diagnosis was made only when concordant results were achieved in 3 independent runs. The cutoff value between homozygous deletion and absence of homozygous deletion was set at 0.3 (contaminating normal cells up to 25% + 5% variation). Because of considerations discussed in detail previously,12 values greater than 0.3 were interpreted as absence of homozygous deletions, and hemizygous deletions were not assessed. The assay was validated using serial dilutions of cell line Jurkat DNA (INK4−/−) in the background of normal DNA, as described by Carter et al.13 The correlation coefficient for the linear function obtained in these experiments was 0.97, demonstrating the capability of the assay to measure normal cell contamination. Oligonucleotides and TaqMan probes used were as follows: p16INK4a: p16se 5′-GGGGCACCAGAGGCAGTA, p16as 5′-GGGCTGAACTTTCTGTGCTG, E2 TaqMan: 5′-FAM-ATGCCCGCATAGATGCCGCGGAAG-TAMRA; p15INK4b: 15F 5′-AGCCGCCCACAACGACTT, 15R 5′-CGTTGGCAGCCTTCATCGAA, 15TM 5′-FAM-TACCCAATTTCCCACCCCCACCCACC-TAMRA;β-globin: PCO3: 5′-CTGACACAACTGTGTTCACTAGC, KM38: 5′-TATTGGTCTCCTTAAACCTGTCTTG, TMglobin03: 5′-FAM-TCCTGAGGAGAAGTCTGCCGTTAC-TAMRA (TIB Molbiol, Berlin, Germany).
Reactions were performed in a final volume of 50 μL with 50 ng genomic DNA, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 3 mM MgCl2, 200 μM each deoxyribonucleotide triphosphate (GIBCO BRL, Life Technologies, Karlsruhe, Germany), 0.5 U Platinum-Taq (GIBCO BRL), 0.2 μM each oligonucleotide, and 0.1 μM each TaqMan probe (both TIB Molbiol). PCR was performed as follows: An initial denaturation step of 10 minutes at 97°C was followed by 5 cycles at 97°C for 60 seconds and 65°C for 60 seconds; then 35 cycles at 95°C for 20 seconds and 65°C for 45 seconds. Fluorescence was detected at the end of the 65°C step.
Results and discussion
A homozygous deletion of p16INK4a and p15INK4b was found in 44 (35%) and 38 (30%) of the 125 samples, respectively (Table 1). Six samples showed an isolated deletion of p16INK4a, and one sample had only a p15INK4b deletion. The homozygous deletion frequencies of p16INK4a and p15INK4b were 30% and 25% in 109 B-cell precursor (BCP) ALL samples, respectively, and 73% for both in 15 T-cell ALL samples (P = .001). The median first-remission duration was significantly shorter (P < .01) in the groups with p16INK4a and p15INK4b deletions (773 and 739 days, respectively) than in those without deletions (both cohorts 1108 days) (Table 1). Furthermore, we observed a significant association between homozygous p16INK4a deletion and higher peripheral blast cell and WBC counts (median 2410 blasts/μL for patients with p16INK4a deletion and 440 blasts/μL for those without this alteration;P = .004). Patients with p15INK4b deletion had median peripheral blast counts of 1700 blasts/μL, compared with 500 blasts/μL in patients without the deletion (not significant; Table 1).
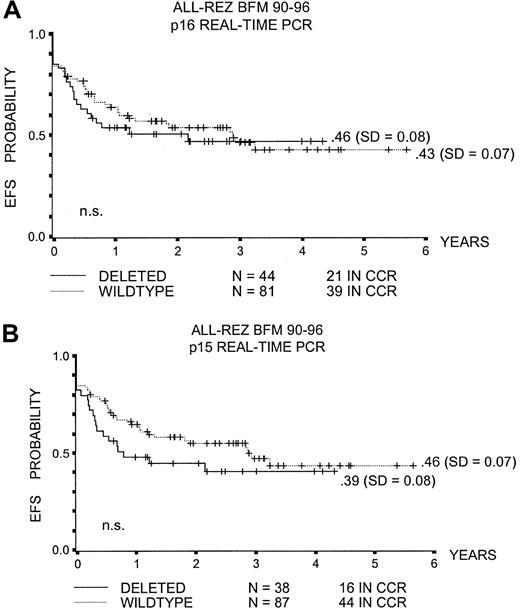
The probability of event-free survival (pEFS) at 5 years for children with p16INK4a or p15INK4bdeleted leukemic blasts was not significantly different from that in children without these alterations (.46 ± .08 and .39 ± .08 versus .43 ± .07 and .46 ± .07, respectively; Figure1A,B).
Event-free survival with p16INK4a and p15INK4b deletion.
(A) pEFS and p16INK4a deletion. (B) pEFS and p15INK4b deletion. CCR indicates complete continuous remission; n.s., not significant.
Event-free survival with p16INK4a and p15INK4b deletion.
(A) pEFS and p16INK4a deletion. (B) pEFS and p15INK4b deletion. CCR indicates complete continuous remission; n.s., not significant.
The deletion prevalences in relapsed childhood ALL were 35% for p16INK4a and 30% for p15INK4b, slightly higher than those reported for initial ALL. In this disease, the reports on deletion prevalences vary considerably, especially with regard to BCP ALL. TheINK4 deletion prevalences in initial BCP ALL reported in 9 different studies ranged from 10% to 20%, and the respective values for T-cell ALL ranged from 60% to 80%.1,4,5,7 14-18 The higher INK4 deletion prevalence in relapsed ALL could be interpreted as an indication of an adverse prognostic role for this alteration in initial ALL; alternatively, some deletions could have been acquired in the interval between initial and relapse diagnosis.
T-cell immunophenotype and high WBC count are known to correlate withINK4 deletion in initial childhood ALL.1,2,4,5We also observed this correlation in our cohort of patients; additionally, we found a significant association of INK4deletion and the major adverse prognostic parameter known for ALL relapse, a short first-remission duration.19Interestingly, despite this correlation, no significant difference in pEFS was found between the group with INK4 deletion and those without, a fact that renders a prognostic role of INK4deletions in relapsed childhood ALL unlikely.
Our data on relapsed patients provide a new possible explanation for the discordant results seen in initial ALL: Patients withINK4 deletion tend to relapse approximately 1 year earlier (median first-remission duration approximately 2.1 years versus approximately 3 years). Accordingly, the follow-up time in some studies could be too short, leading to a bias toward more adverse events in the group of patients with INK4 deletion. The median follow-up time in 2 studies reporting a correlation between INK4deletion and adverse outcome was 3.25 years in one (Heyman et al2) and was not provided in the other.1 In contrast, the study of Rubnitz et al5 examining the largest number of patients to date (n = 155) had a follow-up of more than 5 years in both subgroups and found no prognostic effect.
Carter et al13 reported that hemizygous p16INK4a deletions correlated with adverse outcome in childhood ALL. Because of methodologic and theoretical considerations discussed in detail elsewhere,12 we did not assess hemizygous deletions of the INK4 genes in our study. Theoretically, this could veil an adverse prognostic impact ofINK4 deletions because the potentially present hemizygous deletions would appear in the group with absence of homozygous deletions. However, in contrast to Carter et al,13 Heyman et al2 found no difference in pEFS between patients with hemizygous deletion and those with wild-type INK4 genes. Nevertheless, a validation of our and other's data with an appropriate method (ie, fluorescence in situ hybridization) would be worthwhile.
It remains the task of further studies to examine alternative mechanisms of INK4 tumor suppressor gene inactivation by appropriate methods (ie, immunohistochemistry or methylation-specific PCR) to clarify their prognostic significance.
Supported by Deutsche Kinderkrebsstiftung, Bonn, Germany; Deutsche Krebshilfe, Bonn, Germany; and Deutsche José Carreras Stiftung für Leukämieforschung, Munich, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hagen Graf Einsiedel, Charité Campus Virchow-Klinikum, Otto-Heubner-Centrum für Kinder-und Jugendmedizin, Klinik für Pädiatrie m.S. Onkologie/Hämatologie, Forschungshaus Raum 2.0412, Augustenburger Platz 1, 13353 Berlin, Germany; e-mail: hagenve@charite.de.