Abstract
Given the significance of CD40–CD40 ligand interactions in chronic inflammatory diseases including atherosclerosis, the transcriptional regulation of CD40 expression as a potential therapeutic target was investigated in human umbilical vein cultured endothelial cells. Exposure to interferon-γ (IFN-γ) plus tumor necrosis factor-α resulted in a marked synergistic de novo expression of CD40, which, according to electrophoretic mobility shift analysis, was attributable to activation of the transcription factors nuclear factor-κB (NF-κB), signal transducer and activator of transcription-1 (STAT-1), and interferon regulatory factor-1 (IRF-1). Subsequent time-course studies revealed that de novo synthesis of IRF-1 preceded that of CD40. Decoy oligodeoxynucleotide (ODN) neutralization of STAT-1 or IRF-1, but not of NF-κB, inhibited cytokine-stimulated CD40 expression by 60% at both the mRNA and protein levels, and this effect was mimicked by antisense ODN blockade of IRF-1 synthesis. In contrast, CD40 expression in response to IFN-γ stimulation was sensitive to neutralization of STAT-1 only. These findings suggest that depending on the cytokine composition, CD40 expression in human endothelial cells under proinflammatory conditions is governed by STAT-1 either directly or indirectly through de novo synthesis of IRF-1. Moreover, decoy ODN neutralization of these transcription factors may provide a novel therapeutic option for interfering with CD40–CD40 ligand-mediated inflammatory responses in vivo.
Introduction
CD40 (or TNFR5) is a cell-surface receptor belonging to the tumor necrosis factor (TNF) receptor superfamily that is principally expressed by B cells, but also by other antigen-presenting cells and, in addition, by a variety of nonimmune cells such as smooth muscle cells, fibroblasts, and endothelial cells.1,2 The corresponding ligand (CD40L or CD154) has been cloned and identified as a CD4+ T-cell activation antigen.3 CD40–CD154 interactions play a critical role in the regulation of both humoral and cellular immunity.4 In endothelial cells, CD40 stimulation causes a TNF-α–like increase in expression of adhesion molecules and chemokines that promote the homing and extravasation of leukocytes at sites of inflammation.5,6 Moreover, in addition to monocytes, endothelial cells produce bioactive interleukin-12 (IL-12) in response to CD40 stimulation.7 IL-12 is a potent differentiation factor for naive T-helper cells, promoting their clonal expansion into Th1 cells.8
These aforementioned events also seem to be important for the development of atherosclerosis, and all of the principal cells present in human atherosclerotic lesions, such as endothelial cells, macrophages, smooth muscle cells, and T-helper cells, express CD40, CD154, or both.9,10 Moreover, anti-CD154 antibodies are capable of reducing the size of atherosclerotic lesions in hyperlipidemic mice11 and limiting heart-transplant atherosclerosis in the same species.12 However, because of adverse side effects, the use of such antibodies may be limited in patients with chronic inflammatory diseases.13 Moreover, as yet no low-molecular-weight antagonist for CD40 has been developed, and anti-CD40 antibodies stimulate rather than inhibit CD40 signaling in cells expressing the receptor.14 Suppression of CD40 expression in CD154 target cells may thus provide a feasible therapeutic alternative.
Cytokine-inducible expression of CD40 in rat vascular smooth muscle cells is mediated by the transcription factors nuclear factor-κB (NF-κB) and signal transducer and activator of transcription-1 (STAT-1).15 In the mouse macrophage cell line RAW 264.7, STAT-1 and 2 Ets family members (PU.1 and Spi-B) are involved in interferon-γ (IFN-γ) induction of CD40 gene expression.16 Although sequenced in part, the promoter of the human CD40 gene has not been functionally characterized,17 so the transcription factors governing CD40 expression in human cells are not yet known. Therefore, we investigated the transcriptional regulation of cytokine-induced CD40 expression in human umbilical vein cultured endothelial cells (HUVECs) and in the premonocytic cell line THP-1.
Materials and methods
Cell culture
HUVECs were isolated from freshly collected umbilical cords, as described previously,7 and cultured in medium M199 containing 20% fetal bovine serum (FBS; Life Technologies, Karlsruhe, Germany), 50 U/mL penicillin, 50 μg/mL streptomycin, 10 U/mL nystatin, 5 mM HEPES and 5 mM N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid (TES), 1 μg/mL heparin (Sigma-Aldrich, Deisenhofen, Germany), and 40 μg/mL endothelial cell growth factor (c.c. pro, Neustadt/W., Germany). They were grown to 90% to 100% confluence within 4 to 5 days either in 6-well plates (1.5 × 106 cells/well) for reverse transcription–polymerase chain reaction (RT-PCR) analyses or in 60-mm-diameter Petri dishes (5 × 106 cells/dish) for electrophoretic mobility shift analyses (EMSAs), Western blot, or fluorescence-activated cell sorting (FACS) analyses. They were identified by positive immunofluorescence for von Willebrand factor, positive RT-PCR and FACS analysis for platelet endothelial cell adhesion molecule-1 (PECAM-1), and negative immunofluorescence for smooth muscle α-actin.
The human monocytic cell line THP-1 (American Type Culture Collection, Rockville, MD) and the mouse myeloma cell line P3xTB.A7 (stably transfected with human CD154) were cultured in RPMI 1640 medium (Life Technologies) containing 10% FBS and antibiotics, as described before.7
RT-PCR analysis
Total RNA was isolated from cultured cells by solid-phase extraction with the RNeasy kit from Qiagen (Hilden, Germany). RT-PCR analyses for human CD40, E-selectin, vascular cell adhesion molecule-1, monocyte chemoattractant protein-1, and peptide elongation factor (EF-1) were performed essentially as described previously.7 Amplification of EF-1 cDNA served as an internal standard (housekeeping gene). For human interferon regulatory factor-1 (IRF-1), the following primers (with the respective GenBank accession number, position of the PCR product in the coding sequence, and predicted size) were used for amplification (X14454, position 92-470, 378-bp fragment): 5′-TTCCCTCTTCCACTCGGAGT-3′ (sense) and 5′-GATATCTGGCAGGGAGTTCA-3′ (antisense). The identity of the amplification product for CD40 and IRF-1 was verified by direct sequencing with a model 373 stretch DNA sequencer (Applied Biosystems, Weiterstadt, Germany).
Western blot analysis
Analysis of CD40 receptor and IRF-1 protein expression in HUVECs or THP-1 cells was performed as described.18 Protein extracts (30 μg protein per lane) were separated by denaturing 10% polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate, according to standard protocols, and then transferred to a BioTrace polyvinylidene fluoride transfer membrane (Pall, Dreieich, Germany). Transferred proteins were probed by a polyclonal rabbit antihuman CD40 antibody (1:2000 dilution; Research Diagnostics, Flanders, NJ) or a polyclonal anti–IRF-1 antibody (1:2000 dilution; Santa Cruz Biotechnology, Heidelberg, Germany). Visualization of the protein bands was achieved by using a secondary antirabbit antibody conjugated to horseradish peroxidase (1:3000 dilution; Sigma-Aldrich) and the SuperSignal chemiluminescent substrate (Pierce Chemical, Rockford, IL) followed by exposure to autoradiography film (Hyperfilm MP; Amersham Pharmacia Biotech, Freiburg, Germany). Loading and transfer of equal amounts of protein in each lane were verified by reprobing the membrane with a monoclonal anti–β-actin antibody from mouse ascites fluid and an anti–mouse IgG (whole molecule) peroxidase conjugate (both antibodies obtained from Sigma-Aldrich; 1:3000 dilution), followed by densitometry.18
Electrophoretic mobility shift analysis
Preparation of nuclear extracts from the cultured cells and subsequent nondenaturing 4% polyacrylamide gel electrophoresis were performed as described previously.15 The double-stranded gel shift oligodeoxynucleotides (ODNs; Santa Cruz Biotechnology) for STAT-1 (5′-CATGTTATGATTCCTGTAAGTG-3′), Sis-inducible element (5′-GTGCATTTCCCGTAAATCTTGTCTACA-3′), NF-κB (5′-AGTTGAGGGGACTTTCCCAGGC-3′), and IRF-1 (5′-GGAAGCGAAAATGAAATTGAC-3′) were end-labeled with [γ-32P]ATP by using the 5′-end labeling kit from Amersham Pharmacia Biotech. Typically, the binding mixture contained 5 μg nuclear extract, 20 000 cpm of the32P-labeled oligonucleotide probe (0.5 ng), 1 μg poly[d(I-C)], and 1.33 mM DL-dithiothreitol in a total volume of 15 μL binding buffer. For supershift analyses, 2 μL of the appropriate gel supershift antibody (2 mg/mL; Santa Cruz Biotechnology) per 6 μL of nuclear extract was preincubated at room temperature for 60 minutes before the EMSA was performed.
Decoy ODN technique
Double-stranded ODNs were prepared from complementary single-stranded phosphorothioate-bonded ODNs obtained from Eurogentec (Köln, Germany) by melting at 95°C for 5 minutes, followed by a cool-down phase of 3 to 4 hours at ambient temperature. The efficiency of the hybridization reaction was verified with 2.5% agarose gel electrophoresis and usually found to exceed 95%. The sequences of the single-stranded ODNs were as follows (underlined letters denote phosphorothioate-bonded bases): STAT-1,CATGTTATGCATATTCCTGTAAGTG; STAT-1m,CATGTTATGCAGACCGTAGTAAGTG; NF-κB,AGTTGAGGGGACTTTCCCAGGC; IRF-1,GGAAGCGAAAATGAAATTGAC; IRF-1c,CAGAAAAGTGAAACCCTG; and IRF-1m,CAGATGAGTGTAACCCTG. On the basis of previous EMSA and RT-PCR analyses, the maximally effective concentration and optimal preincubation time for all decoy ODNs in the cultured cells were determined to be 10 μM and 4 hours, respectively.18Decoy ODN uptake was achieved without using any cationic lipid or liposomal complex.
Antisense ODNs
HUVECs were treated with the single-stranded ODNs at approximately 80% confluence. Briefly, the antisense ODNs were premixed with 200 μg/mL Lipofectin reagent (Life Technologies) in normal growth medium without heparin and endothelial cell growth factor at room temperature at the desired concentration (4 μg/well). Medium supplements were added to the premix and incubated with the HUVECs for 4 hours at 37°C. Thereafter, the ODN-containing medium was replaced by fresh medium, and the cells were stimulated with the cytokines for the indicated times. The IRF-1 antisense ODN had the sequence 5′-CGAGTGATGGGCATGTTGGC-3′, thus targeting the translation initiation site in the IRF-1 mRNA.19 The sequence of the missense control was 5′-CGAGTGGTAGACGTA-TTGGC-3′, and that of the scrambled control was 5′-CGAGTGGTAGACGTATTGGC-3′ (underlined letters denote phosphorothioate-bonded bases).
Flow cytometry
HUVECs harvested by scraping were stained at 0°C to 4°C for 30 minutes with either a phycoerythrin-conjugated mouse anti–human CD40 monoclonal antibody or the corresponding isotype control (BD Biosciences, Heidelberg, Germany). Endothelial cells were identified by staining for PECAM-1 using a fluorescein isothiocyanate–conjugated mouse anti–human CD31 monoclonal antibody (BD Biosciences). Flow cytometry was performed with a Coulter Epics XL flow cytometer (Beckman Coulter, Krefeld, Germany). Data are expressed as mean fluorescence intensity.
Nuclear run-on analysis
PCR-based run-on analysis of the de novo expression of CD40 and EF-1 mRNA in isolated nuclei of HUVECs was performed as described previously.15 HUVECs in 100-mm Petri dishes were exposed to either a cytokine mixture or vehicle for 6 hours. Thereafter, the cells were harvested with a cell scraper and the nuclei were isolated. Half of the nuclei were immediately lysed, and the other half were incubated for 30 minutes at 30°C and then lysed. Total RNA was isolated and RT-PCR was performed as described before, except for the use of random primers instead of oligo-dT primers in the reverse transcription step.
Data analysis
Unless indicated otherwise, results are expressed as means ± SEM of n observations. One-way analysis of variance followed by a Dunnett multiple-comparisons test was used to determine differences between the means and the corresponding control value, withP < .05 considered statistically significant.
Results
Cytokine-induced CD40 expression and transcription factor activation
In the cultured HUVECs, exposure to IFN-γ (1000 U/mL) plus TNF-α (100 U/mL) resulted in a marked increase in CD40 expression, which at the mRNA level reached a maximum between 9 and 14 hours (Figure 1A). IFN-γ (1000 U/mL) and TNF-α (1000 U/mL) alone were less effective (203% ± 23% and 242% ± 42%, respectively, of basal CD40 mRNA after 9 hours;P < .05, n = 5-8), whereas IL-1β (60 U/mL) or bacterial lipopolysaccharide (1 μg/mL) had no effect (not shown). Stimulation of the receptor itself by using the mouse myeloma cell line P3xTB.A7 stably transfected with human CD154 (2 × 106cells/well)7 also had no effect on CD40 mRNA expression (135% ± 24% of basal mRNA abundance after12 hours; n = 6).
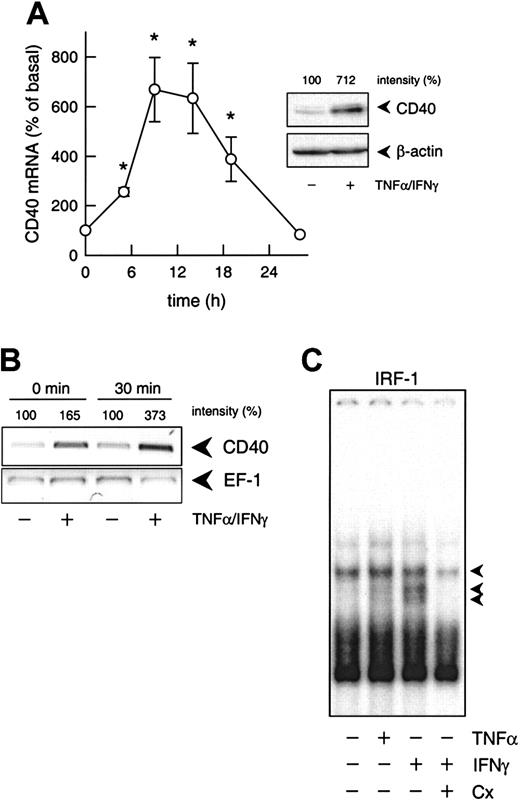
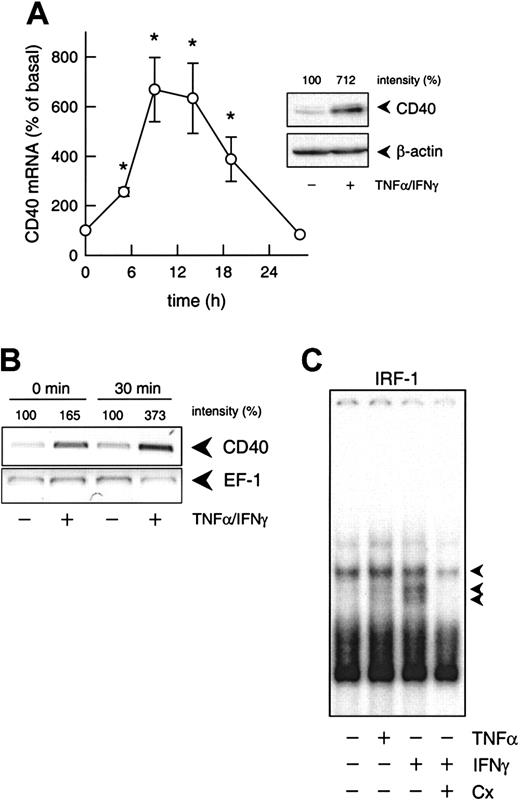
Transcriptional regulation of cytokine-induced CD40 expression.
(A) Time-dependent effect of TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) on CD40 mRNA expression in the cultured HUVECs (calculated as percentage of basal CD40 expression, n = 3-7; *P < .05 versus basal). The inset shows the effects of 100 U/mL TNF-α and 1000 U/mL IFN-γ on CD40 protein expression after 14 hours. Typical Western blot analysis is shown with the relative intensities, as judged by densitometry, indicated at the top. Qualitatively identical results were obtained with at least 2 further batches of HUVECs. Loading and transfer of equal amounts of protein in each lane were verified by reprobing the membrane with an anti–β-actin antibody. (B) PCR-based run-on analysis of the de novo expression of CD40 and EF-1 mRNA in isolated nuclei of cultured HUVECs that had been exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 6 hours. The isolated nuclei were either lysed immediately (0 min) or incubated for 30 minutes at 30°C. The figure depicts the result of one experiment; a qualitatively identical result was obtained in another experiment with a different batch of cells. (C) Effects of IFN-γ (1000 U/mL) or TNF-α (1000 U/mL) on nuclear translocation of IRF-1 (1 constitutive and 2 inducible complexes, as indicated by the arrows) in the cultured HUVECs, and effect of cycloheximide (Cx; 10 μM) on IFN-γ (100 U/mL)–stimulated nuclear translocation of IRF-1. Typical EMSA is shown with nuclear extracts obtained after 3 hours of exposure of the HUVECs to the cytokines. Qualitatively identical results were obtained with at least 4 further batches of HUVECs.
Transcriptional regulation of cytokine-induced CD40 expression.
(A) Time-dependent effect of TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) on CD40 mRNA expression in the cultured HUVECs (calculated as percentage of basal CD40 expression, n = 3-7; *P < .05 versus basal). The inset shows the effects of 100 U/mL TNF-α and 1000 U/mL IFN-γ on CD40 protein expression after 14 hours. Typical Western blot analysis is shown with the relative intensities, as judged by densitometry, indicated at the top. Qualitatively identical results were obtained with at least 2 further batches of HUVECs. Loading and transfer of equal amounts of protein in each lane were verified by reprobing the membrane with an anti–β-actin antibody. (B) PCR-based run-on analysis of the de novo expression of CD40 and EF-1 mRNA in isolated nuclei of cultured HUVECs that had been exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 6 hours. The isolated nuclei were either lysed immediately (0 min) or incubated for 30 minutes at 30°C. The figure depicts the result of one experiment; a qualitatively identical result was obtained in another experiment with a different batch of cells. (C) Effects of IFN-γ (1000 U/mL) or TNF-α (1000 U/mL) on nuclear translocation of IRF-1 (1 constitutive and 2 inducible complexes, as indicated by the arrows) in the cultured HUVECs, and effect of cycloheximide (Cx; 10 μM) on IFN-γ (100 U/mL)–stimulated nuclear translocation of IRF-1. Typical EMSA is shown with nuclear extracts obtained after 3 hours of exposure of the HUVECs to the cytokines. Qualitatively identical results were obtained with at least 4 further batches of HUVECs.
Both blockade by actinomycin D (1 μM; not shown) and nuclear run-on analyses (Figure 1B) confirmed that up-regulation of CD40 expression induced by IFN-γ plus TNF-α was regulated at the transcriptional level. In contrast to the expected20 rapid translocation of NF-κB (p65/p50 heterodimer) and STAT-1 (p91 homodimer) to the nucleus of the cultured HUVECs under these conditions (maximum 30 minutes, not shown), nuclear translocation of IRF-1 was slow (maximum 3 hours, not shown) and sensitive to cycloheximide blockade of protein synthesis (Figure 1C).
The degree of IRF-1 expression correlates with the level of CD40 expression
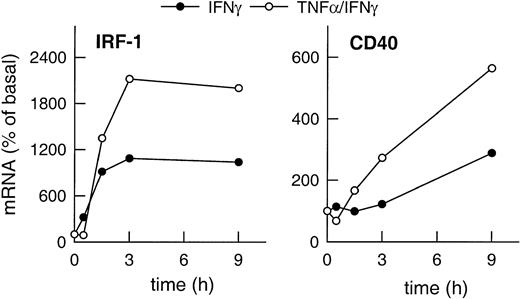
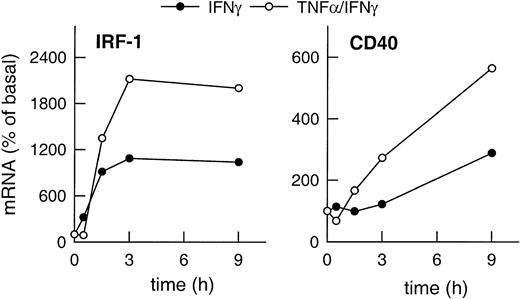
Expression of IRF-1 and CD40 mRNA in HUVECs stimulated with IFN-γ plus TNF-α clearly exceeded the level of expression of both gene products induced by IFN-γ alone (Figure2). Moreover, CD40 expression was up-regulated faster in the presence of IFN-γ plus TNF-α as compared with cells stimulated with IFN-γ alone (Figure 2), whereas IRF-1 expression clearly preceded that of CD40 regardless of the stimulus.
Correlation of cytokine-induced expression of IRF-1 and CD40.
Time-dependent effects of IFN-γ (1000 U/mL) or of TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) on IRF-1 and CD40 mRNA expression in the cultured HUVECs, as judged by RT-PCR analysis (calculated as percentage of basal gene expression). Qualitatively identical results were obtained with at least 2 further batches of cells.
Correlation of cytokine-induced expression of IRF-1 and CD40.
Time-dependent effects of IFN-γ (1000 U/mL) or of TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) on IRF-1 and CD40 mRNA expression in the cultured HUVECs, as judged by RT-PCR analysis (calculated as percentage of basal gene expression). Qualitatively identical results were obtained with at least 2 further batches of cells.
Effects of different decoy ODNs on CD40 expression
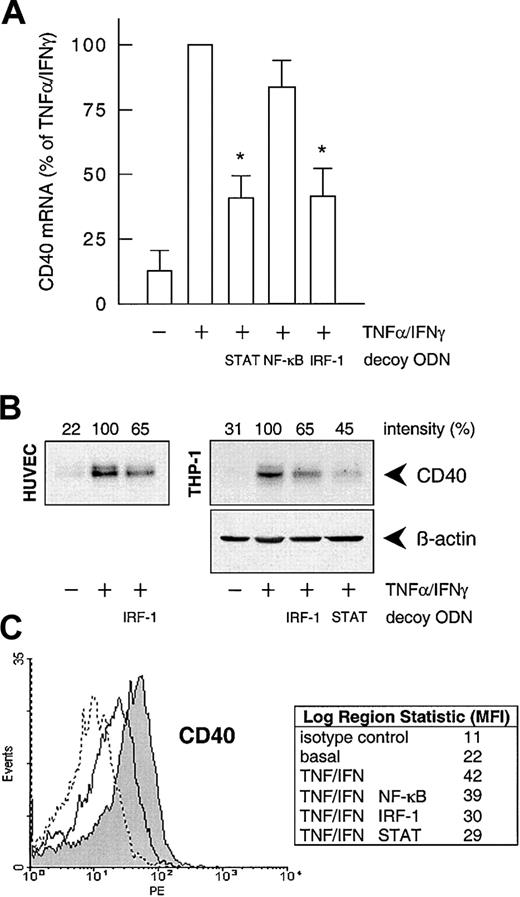
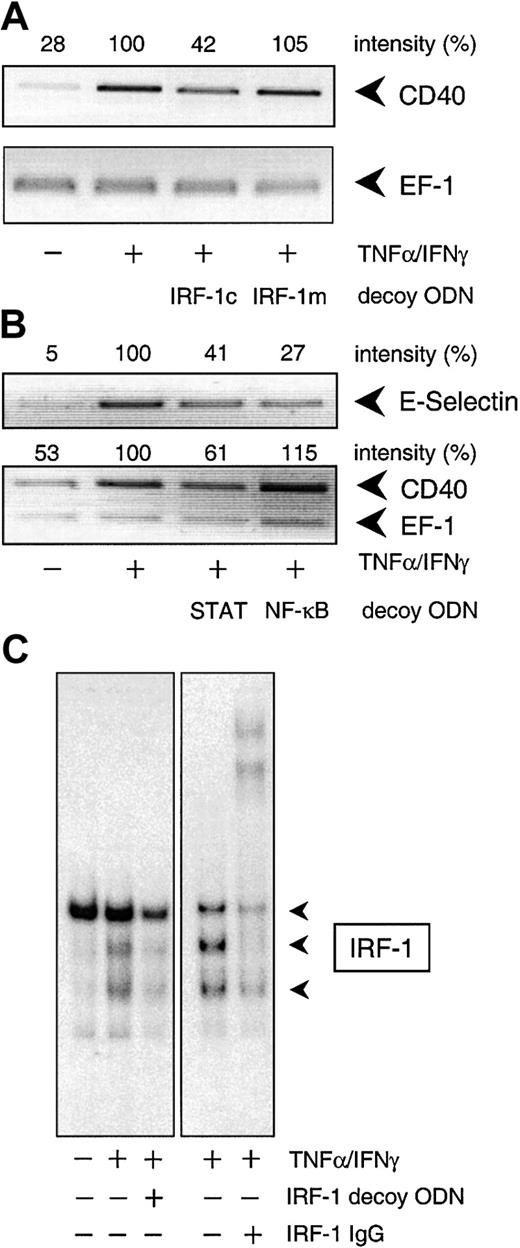
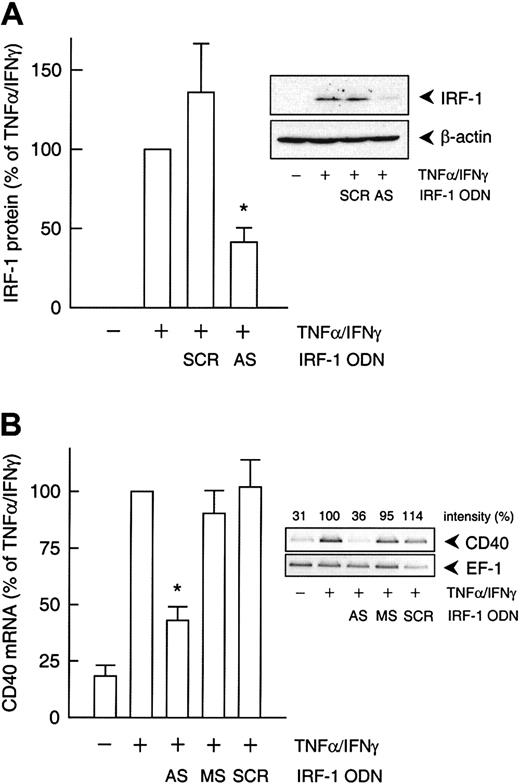
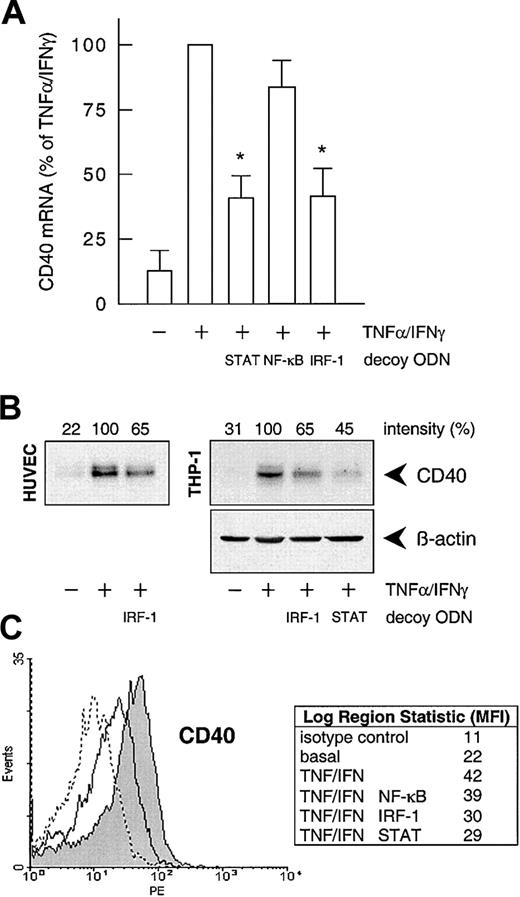
To confirm the involvement of NF-κB, STAT-1, or IRF-1 in cytokine-stimulated CD40 expression, we used the decoy ODN technique. Both CD40 mRNA and protein expression after exposure to IFN-γ plus TNF-α were significantly reduced to approximately 40% of the control value following pretreatment of the HUVECs with a STAT-1 or IRF-1 consensus decoy ODN (Figure 3). These effects of the decoy ODN were specific; for example, CD40 mRNA expression was significantly reduced in cells treated with the IRF-1 consensus but not with a corresponding mutant ODN (Figure4A; cf. Figure 6B for the STAT-1 decoy ODN). In contrast, the NF-κB–specific decoy ODNs had no significant effect on either CD40 mRNA (Figure 3A) or protein expression (Figure3C). Cytokine-stimulated E-selectin mRNA expression, on the other hand, was markedly inhibited by preincubation with the NF-κB decoy ODN, thus confirming its efficacy (Figure 4B). Moreover, as judged by EMSA, the IRF-1 decoy ODN clearly affected the nuclear translocation of IRF-1 in IFN-γ/TNF-α–stimulated THP-1 cells (Figure 4C), and cytokine-stimulated CD40 protein expression in these cells was significantly reduced after pretreatment with the STAT-1 or IRF-1 decoy ODN (Figure 3B).
Effects of the different decoy ODNs on cytokine-stimulated CD40 expression.
(A) Statistical summary of the effects of the STAT-1, NF-κB, or IRF-1 consensus decoy ODNs (10 μM, 4 hours preincubation) on CD40 mRNA expression (calculated as percentage of the stimulated control) induced by TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) in the cultured HUVECs (n = 5; *P < .05 versus TNF-α/IFN-γ). (B) Effect of the STAT-1 or IRF-1 consensus decoy ODN (10 μM, 4 hours preincubation) on TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated CD40 protein expression in HUVECs and THP-1 cells after 14 hours. Typical Western blot analyses are shown; qualitatively identical results were obtained with at least 2 further batches of endothelial or THP-1 cells. (C) Effects of the STAT-1, NF-κB, or IRF-1 consensus decoy ODNs on cell-surface expression of CD40 protein in HUVECs exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 20 hours, as judged by flow cytometry. Data are expressed as mean fluorescence intensity (MFI). Representative histograms depict isotype control (dashed line), basal expression (solid line), and stimulated CD40 expression (solid line, area under the curve highlighted in gray). Qualitatively identical results were obtained with at least 3 further batches of cells.
Effects of the different decoy ODNs on cytokine-stimulated CD40 expression.
(A) Statistical summary of the effects of the STAT-1, NF-κB, or IRF-1 consensus decoy ODNs (10 μM, 4 hours preincubation) on CD40 mRNA expression (calculated as percentage of the stimulated control) induced by TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) in the cultured HUVECs (n = 5; *P < .05 versus TNF-α/IFN-γ). (B) Effect of the STAT-1 or IRF-1 consensus decoy ODN (10 μM, 4 hours preincubation) on TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated CD40 protein expression in HUVECs and THP-1 cells after 14 hours. Typical Western blot analyses are shown; qualitatively identical results were obtained with at least 2 further batches of endothelial or THP-1 cells. (C) Effects of the STAT-1, NF-κB, or IRF-1 consensus decoy ODNs on cell-surface expression of CD40 protein in HUVECs exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 20 hours, as judged by flow cytometry. Data are expressed as mean fluorescence intensity (MFI). Representative histograms depict isotype control (dashed line), basal expression (solid line), and stimulated CD40 expression (solid line, area under the curve highlighted in gray). Qualitatively identical results were obtained with at least 3 further batches of cells.
Specificity of the decoy ODN technique in cultured HUVECs and THP-1 cells.
(A) Example of the specificity of the consensus decoy ODN for IRF-1. Cytokine-stimulated (100 U/mL TNF-α, 1000 U/mL IFN-γ) CD40 expression is significantly reduced in HUVECs pretreated with the consensus (IRF-1c) but not with the mutant (IRF-1m) decoy ODN. (B) Effects of the STAT-1 or NF-κB consensus decoy ODN on CD40 and E-selectin expression in HUVECs exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 9 hours. Typical RT-PCR analyses are shown; comparable results were obtained in 2 further experiments each with different batches of cells. (C) Effect of a 4-hour preincubation with the IRF-1 consensus decoy ODN (10 μM) on nuclear translocation of IRF-1 in THP-1 cells exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 3 hours. Typical EMSA is shown, with the appropriate supershift analysis (IRF-1 IgG) performed with a single batch of THP-1 cells. Identical results were obtained with another batch of cells.
Specificity of the decoy ODN technique in cultured HUVECs and THP-1 cells.
(A) Example of the specificity of the consensus decoy ODN for IRF-1. Cytokine-stimulated (100 U/mL TNF-α, 1000 U/mL IFN-γ) CD40 expression is significantly reduced in HUVECs pretreated with the consensus (IRF-1c) but not with the mutant (IRF-1m) decoy ODN. (B) Effects of the STAT-1 or NF-κB consensus decoy ODN on CD40 and E-selectin expression in HUVECs exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 9 hours. Typical RT-PCR analyses are shown; comparable results were obtained in 2 further experiments each with different batches of cells. (C) Effect of a 4-hour preincubation with the IRF-1 consensus decoy ODN (10 μM) on nuclear translocation of IRF-1 in THP-1 cells exposed to TNF-α (100 U/mL) plus IFN-γ (1000 U/mL) for 3 hours. Typical EMSA is shown, with the appropriate supershift analysis (IRF-1 IgG) performed with a single batch of THP-1 cells. Identical results were obtained with another batch of cells.
Effects of an IRF-1 antisense ODN
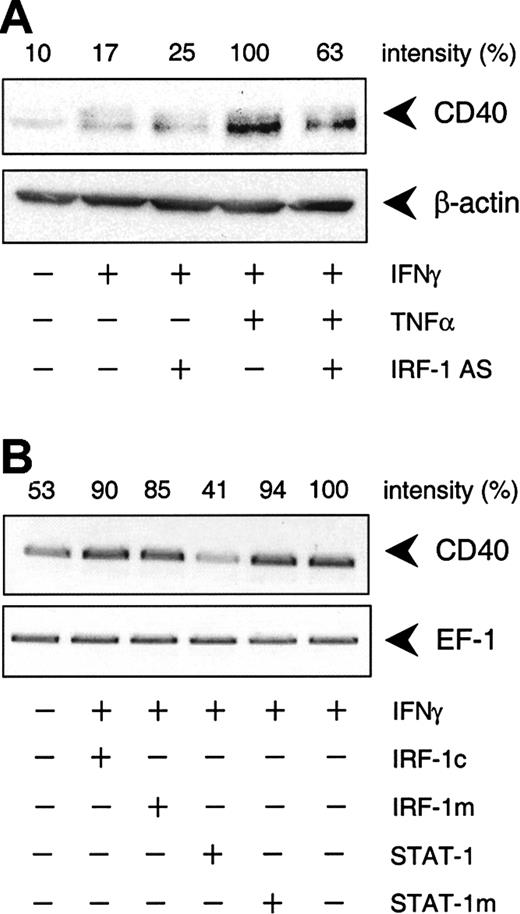
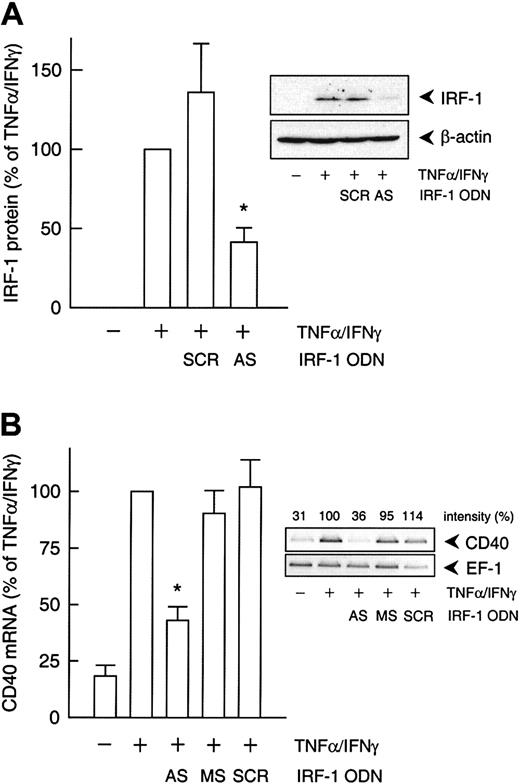
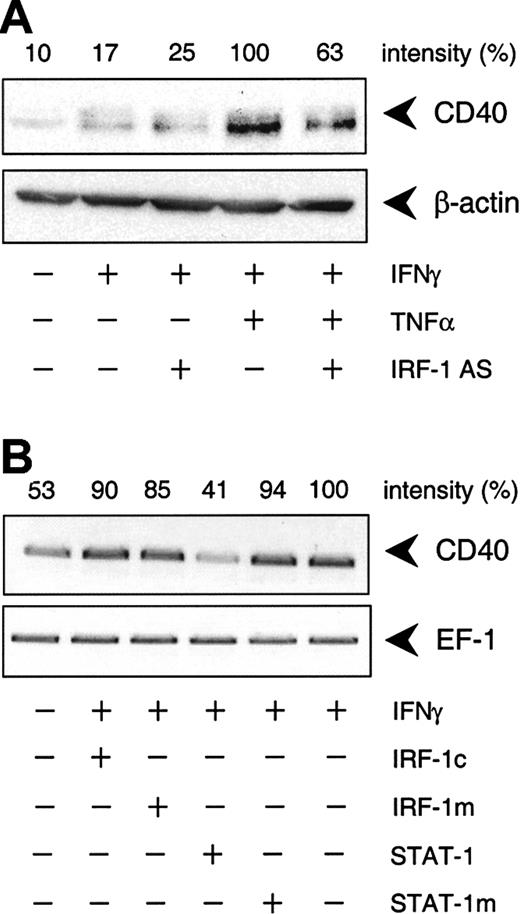
To delineate which of the 2 IFN-γ–stimulated transcription factors, STAT-1 or IRF-1, was responsible for the cytokine-induced increase in CD40 expression, we used an antisense ODN to suppress IRF-1 protein synthesis. Incubation of the cultured HUVECs with the IRF-1 antisense ODN before IFN-γ plus TNF-α stimulation resulted in a 55% reduction of cytokine-induced IRF-1 protein expression, whereas the scrambled control ODN had no such effect (Figure5A). Moreover, the IRF-1 antisense ODN inhibited IFN-γ plus TNF-α–induced CD40 expression to a similar extent at both the mRNA (Figure 5B) and protein levels (Figure6A). However, the antisense ODN did not seem to exert an effect on CD40 protein expression induced by IFN-γ alone (Figure 6A). In line with this finding, the IRF-1 decoy ODN had no effect on the increase in CD40 mRNA abundance in HUVECs stimulated with IFN-γ alone, whereas the STAT-1 decoy ODN was clearly effective (Figure 6B).
IRF-1 antisense ODN effect on cytokine-induced CD40 expression.
(A) Statistical summary of IRF-1 protein expression after 3 hours (calculated as percentage of cytokine-stimulated IRF-1 expression) in TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated HUVECs pretreated with an IRF-1 antisense (AS) or scrambled control (SCR) ODN, as described in “Materials and methods” (n = 3; *P < .05 versus TNF-α/IFN-γ). (B) Statistical summary of the effects of the IRF-1 antisense (AS) ODN or the corresponding missense (MS) or scrambled (SCR) control ODN on TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated CD40 mRNA expression after 9 hours (calculated as percentage of the stimulated control) in the cultured HUVECs (n = 3; *P < .05 versus TNF-α/IFN-γ). The insets show a representative Western blot and RT-PCR analysis, respectively.
IRF-1 antisense ODN effect on cytokine-induced CD40 expression.
(A) Statistical summary of IRF-1 protein expression after 3 hours (calculated as percentage of cytokine-stimulated IRF-1 expression) in TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated HUVECs pretreated with an IRF-1 antisense (AS) or scrambled control (SCR) ODN, as described in “Materials and methods” (n = 3; *P < .05 versus TNF-α/IFN-γ). (B) Statistical summary of the effects of the IRF-1 antisense (AS) ODN or the corresponding missense (MS) or scrambled (SCR) control ODN on TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated CD40 mRNA expression after 9 hours (calculated as percentage of the stimulated control) in the cultured HUVECs (n = 3; *P < .05 versus TNF-α/IFN-γ). The insets show a representative Western blot and RT-PCR analysis, respectively.
IFN-γ–stimulated CD40 protein expression is predominantly STAT-1 dependent.
(A) Effect of the IRF-1 antisense (AS) ODN on IFN-γ (1000 U/mL) or TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated CD40 protein expression in the cultured HUVECs after 14 hours. Representative Western blot analysis is shown. (B) Effects of the STAT-1 or IRF-1 consensus decoy ODN (STAT-1, IRF-1c) or mutant control ODN (STAT-1m, IRF-1m) on CD40 expression in cells exposed to IFN-γ (1000 U/mL) alone for 9 hours. Representative RT-PCR analysis is shown; comparable results were obtained with 2 further batches of cells.
IFN-γ–stimulated CD40 protein expression is predominantly STAT-1 dependent.
(A) Effect of the IRF-1 antisense (AS) ODN on IFN-γ (1000 U/mL) or TNF-α (100 U/mL) plus IFN-γ (1000 U/mL)–stimulated CD40 protein expression in the cultured HUVECs after 14 hours. Representative Western blot analysis is shown. (B) Effects of the STAT-1 or IRF-1 consensus decoy ODN (STAT-1, IRF-1c) or mutant control ODN (STAT-1m, IRF-1m) on CD40 expression in cells exposed to IFN-γ (1000 U/mL) alone for 9 hours. Representative RT-PCR analysis is shown; comparable results were obtained with 2 further batches of cells.
Discussion
The involvement of CD40–CD154 interactions in both Th1- and Th2-cell–mediated chronic inflammatory diseases, such as rheumatoid arthritis or asthma (for review see Van Kooten and Bancheraeu1), as well as in atherosclerosis or transplant vasculopathy,21 has been well documented. Apart from CD154-neutralizing antibodies, low-molecular-weight antagonists for CD40 are not yet available, and antibodies against CD40 activate rather than inhibit CD154 signaling.22 Targeting CD40 gene expression in inflammatory conditions by neutralizing the principal transcription factors involved therein may thus represent an alternative therapeutic approach. This decoy ODN strategy has already been successfully applied to block the expression of different target genes both in vitro and in vivo (for reviews see Morishita et al23 and Mann and Dzau24).
STAT-1 is a key transcription factor in cytokine-induced CD40 expression in rodents,15 16 and preliminary evidence from this laboratory suggests that treatment with the corresponding decoy ODN exerts profound therapeutic effects in mouse or rat models of allergen-induced asthma, antigen-induced arthritis, or transplant rejection. However, species differences with respect to the transcriptional regulation of a given gene are rather frequent. To appropriately use the decoy ODN approach in humans, therefore, the aim of the present study was to identify the key transcription factor(s) involved in cytokine-stimulated CD40 expression in human cells.
Human endothelial cells express CD40 constitutively, and this basal expression can be markedly enhanced after exposure to certain proinflammatory cytokines, namely the combination of TNF-α with IFN-γ,25 26 a typical Th1 cytokine. This effect was abolished by the RNA synthesis inhibitor actinomycin D, and nuclear run-on analyses confirmed de novo transcription of the CD40 gene. Interestingly, stimulation of the receptor itself did not result in an increase in CD40 expression.
The combination of IFN-γ and TNF-α is the most potent stimulus for CD40 expression also in rat vascular smooth muscle cells15,27 and mouse microglia cells/macrophages.16 This synergistic effect of the 2 cytokines might be due to a cooperation between STAT-1 and NF-κB; for example, by binding of STAT-1 to an IFN-γ activation site (GAS) in the 5′-flanking region of the CD40 gene that is overlapped by a nonconsensus binding site for NF-κB, as demonstrated for other proinflammatory gene products such as intercellular adhesion molecule-1, Mig (monokine induced by γ-interferon), or RANTES (regulated on activation normal T cells expressed and secreted).28 According to MatInspector29analysis, there are indeed 2 GASs overlapping with an NF-κB site at −528 and −124 in the promoter of the human CD40 gene (coding sequence 53 997-65 558 of the human clone RP4-599F21 of chromosome 20q12-12.12; GenBank accession number AL035662). Unexpectedly, however, the IRF-1 decoy ODN displayed virtually the same inhibitory effect as the STAT-1 decoy ODN on CD40 expression stimulated by IFN-γ plus TNF-α.
In contrast to NF-κB and STAT-1, IRF-1 is not normally expressed constitutively but must be induced de novo, as demonstrated by the inhibitory effect of cycloheximide. Although de novo synthesis of IRF-1 is primarily induced by IFN-γ through activation of STAT-1, it can also be induced, albeit less effectively, by TNF-α. When combined, the 2 cytokines synergistically induce IRF-1 expression,20,28 an effect that depends on a cooperation between STAT-1 and NF-κB similar to the one described before. However, whereas IFN-γ directly induces binding of STAT-1 to the GAS element, NF-κB binding to the nonconsensus site is activated by TNF-α only in the presence of IFN-γ.30
The notion that de novo synthesis of IRF-1 is a prerequisite for IFN-γ plus TNF-α up-regulation of CD40 expression in the cultured HUVECs is supported by the following findings: (1) The magnitude of IRF-1 synthesis under these conditions correlated with the level of expression of CD40; (2) both IRF-1 synthesis and its translocation to the nucleus preceded CD40 expression; (3) decoy ODN neutralization of IRF-1 effectively inhibited CD40 expression; and, most important, (4) this effect was mimicked by antisense ODN suppression of IRF-1 synthesis. Moreover, according to MatInspector analysis, the promoter of the human CD40 gene indeed contains a binding site for IRF-1 at −1424.
Whereas decoy ODN neutralization of STAT-1 was as effective as that of IRF-1 under the chosen experimental conditions (IFN-γ plus TNF-α stimulation), neutralization of NF-κB was not. The most likely reason for this somewhat unexpected finding is that the NF-κB decoy ODN cannot completely neutralize the transcription factor, so that the remaining activity of the transcription factor is sufficient for its cooperative effect on IRF-1 and hence on CD40 expression. On the other hand, there is a strict requirement for STAT-1 to induce IRF-1, hence CD40 expression, under these conditions, which, as a consequence, is more sensitive to a decrease in STAT-1 activity. Indeed, IFN-γ plus TNF-α stimulation has been shown to induce IRF-1 mRNA expression in a synergistic manner only in normal and not in STAT-1–deficient mouse fibroblasts.27 Nonetheless, reporter gene analysis will have to verify the precise roles of these transcription factors and their binding sites in cytokine-stimulated CD40 expression in human cells.
Collectively, the aforementioned findings suggest that in cultured HUVECs, combined treatment with IFN-γ and TNF-α induces an IRF-1–dependent de novo expression of CD40. Although this effect is indirectly mediated by STAT-1 through induction of IRF-1 synthesis, STAT-1 directly induces CD40 expression in HUVECs exposed to IFN-γ alone. It is thus the cytokine composition under the given proinflammatory conditions that determines which transcription factor is primarily responsible for the increase in endothelial CD40 expression. This transcriptional control of CD40 expression appears to be similar in the premonocytic cell line THP-1. Decoy ODN neutralization of either transcription factor may thus provide an interesting therapeutic option for interfering with CD40–CD154-mediated acute or chronic inflammatory responses in vivo, even though or simply because the expression of other IFN-γ–dependent genes with a comparable disease potential may be affected as well.
We are indebted to Nicole Gottlieb for expert technical assistance and Bianca Lienenlüke for performing the experiments with the mouse myeloma cell line P3xTB.A7.
Supported by the Deutsche Forschungsgemeinschaft (SFB 402/C9).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Markus Hecker, Dept of Cardiovascular Physiology, University of Goettingen, Humboldtallee 23, D-37073 Goettingen, Germany; e-mail: hecker@veg-physiol.med.uni-goettingen.de.