Adducins are a family of cytoskeletal proteins encoded by 3 genes (alpha, beta, and gamma). Platelets express alpha and gamma adducins, in contrast to red blood cells that express alpha and beta adducins. During platelet activation with thrombin, calcium ionophore A23187, or phorbol 12-myristate 13-acetate, alpha and gamma adducins were phosphorylated by protein kinase C (PKC) as detected by an antibody specific for a phosphopeptide sequence in the highly conserved carboxy terminus. Platelet activation also led to adducin proteolysis; inhibition by calpeptin suggests that the protease was calpain. The kinase inhibitor staurosporine inhibited PKC phosphorylation of adducin and also inhibited proteolysis of adducin. Experiments with recombinant alpha adducin demonstrated that the PKC-phosphorylated form was proteolyzed at a significantly faster rate than the unphosphorylated form. The concentration of adducin in platelets was estimated at 6 μM, similar to the concentration of capping protein. Fractionation of platelets into high-speed supernatant (cytosol) and pellet (membrane and cytoskeleton) revealed a shift of PKC-phosphorylated adducin to the cytosol during platelet activation. Platelet aggregation detected turbidometrically was decreased in the presence of staurosporine and was completely inhibited by calpeptin. Thrombin-induced changes in morphology were assessed by confocal microscopy with fluorescein phalloidin and were not prevented by staurosporine or calpeptin. Our results suggest that regulation of adducin function by PKC and calpain may play a role in platelet aggregation.
Introduction
After stimulation with agonists, platelets undergo a complex set of events described as activation, adhesion (to endothelial cells), aggregation (to each other), and clot retraction. Activation includes secretion of granule contents by fusion with the open canalicular system,1 shape changes such as extension of filopodia and lamellipodia,2 and changes in the fibrinogen receptor, integrin αIIbβ3 or GPIIbIIIa,3that enable it to bind fibrinogen and to create cell-cell bridges. Numerous signaling pathways are involved in the coordination of these cellular events, and regulation of cytoskeletal protein interactions is essential to normal platelet function.4 5
One of the major cytoskeletal components of platelets is actin, which exists in the platelet as monomeric, ie, G-actin, and filamentous, ie, F-actin. In the resting platelet, most of the actin is in the monomeric state, and some exists as short actin filaments with capping proteins preventing polymerization into longer filaments. When platelets are activated, most of the G-actin polymerizes into F-actin, causing extensions of the platelet membrane into filopodia and lamellipodia.6 In addition to the cytoplasmic G- and F-actin in platelets, there is a membrane skeleton, a specialized structure composed of crosslinked short actin filaments with linkages to integral membrane proteins.7,8 The membrane skeleton has been most studied in red blood cells in which short actin filaments are crosslinked by spectrin tetramers.9 Platelets also contain spectrin in their membrane skeleton,10 but a more abundant component of the platelet membrane skeleton is actin-binding-protein 280 (ABP-280) or filamin.11
Although many components of the red cell membrane skeleton are expressed in platelets, platelets also have cytoplasmic actin filaments and microtubules contributing to their shape. The complex cytoskeleton of platelets requires careful coordination to regulate physiologic changes in these structures. We have identified another cytoskeletal component of platelets, adducin, that may have a role in the cytoskeletal changes occurring during platelet activation.
Adducin was first described in red blood cells12,13 but is found in all human cells examined14 and may function in the cytoplasm as well as on the membrane skeleton. Adducins (alpha, beta, and gamma) are a family of cytoskeleton proteins encoded by 3 distinct genes.15 The functions of adducin purified from red blood cells have been well studied by in vitro assays. Purified red cell adducin consists of alpha and beta polypeptides tightly associated into dimers or tetramers.16 The in vitro functions of red cell adducin (alpha/beta) include cross-linking spectrin and actin filaments,17 capping actin filaments,18 and bundling actin filaments.19 Adducin's interaction with spectrin and actin is regulated via phosphorylation by several protein kinases20-23 and by calcium/calmodulin.17Gamma adducin was discovered as a protein kinase C (PKC)–binding protein by screening a kidney complementary DNA expression library with PKC as a probe.24 25
In contrast to the alpha/beta adducin complex found in red cells, the adducin complex found in platelets and most nonerythroid cells consists of alpha and gamma subunits.14 Platelet adducin (alpha/gamma) may have different properties from the red cell alpha/beta complex, and these properties may be crucial to adducin's function in platelets and other nonerythroid cells. In experiments reported here, we found that alpha and gamma adducins were phosphorylated at a PKC phosphorylation site upon activation of platelets by thrombin, calcium ionophore A23187, or phorbol 12-myristate 13-acetate (PMA). PKC phosphorylation of adducin was associated with a shift of adducin from the high-speed pellet (membrane and cytoskeleton) to the high-speed supernatant (cytosol). Platelet activation also was associated with rapid proteolysis of adducin that was inhibited by the calpain-specific inhibitor calpeptin. In vitro experiments demonstrated that PKC-phosphorylated adducin was proteolyzed more rapidly than unphosphorylated adducin.
These results provide groundwork toward understanding a possible role of adducin in platelet function. Assays of platelet function demonstrated that platelet aggregation was decreased in the presence of staurosporine and completely inhibited by calpeptin. Thrombin-induced platelet shape change (extrusion of filopodia and lamellipodia) was not prevented by staurosporine or calpeptin. Our results suggest that regulation of adducin function by PKC and calpain may play a role in platelet aggregation.
Materials and methods
Platelet preparation
After informed consent, blood was collected from healthy adult donors by a 19-gauge butterfly needle. The first 5 mL was discarded, and a second syringe was attached. Blood (30 mL) was then collected into a syringe prefilled with 5 mL acid citrate dextrose anticoagulant (85 mM sodium citrate, 111 mM dextrose, and 71 mM citric acid) and equilibrated to 37°C. The blood was centrifuged in 4 polypropylene tubes (15 mL) at 350g at room temperature. The platelet-rich plasma was then transferred gently into 2 polypropylene tubes (15 mL) and prostaglandin I2 (Sigma Chemical, St Louis, MO) was added at a concentration of 0.05 μg/mL to prevent platelet activation during processing. Unstimulated platelets were isolated by gel filtration as described by Fox et al.26 Buffers were prewarmed to 37°C and contained prostaglandin I2 at a concentration of 0.05 μg/mL. Sepharose 2B (packed bed volume, 20 mL; Pharmacia, Peapack, NJ) was washed successively in a plastic chromatography column (Bio-Rad, Hercules, CA) with 20 mL of 0.15 M NaCl and 20 mL column buffer (136 mM NaCl, 2 mM KCl, 10 mM NaH2CO3, 0.5 mM NaH2PO4, 0.4 mM MgCl2, 4.4 mM glucose, 10 mM HEPES, pH 7.4). Platelet-poor plasma was used to pre-equilibrate the column, followed by 50 mL column buffer. The platelet-rich plasma was then loaded onto the column, followed by 10 mL column buffer. Elutions were collected in 2-mL fractions, and those with the highest platelet concentrations as determined by a resistive-particle counter were combined into one tube (average platelet count for pooled fractions was 110 000-340 000 platelets/μL). Column-derived samples were incubated at 37°C for 3 hours to allow prostacyclin activity to wear off.
Platelet activation
Platelet activation involved the addition of human thrombin (Sigma), calcium ionophore A23187 (Sigma), or PMA (Sigma). Platelet activators and inhibitors, except for thrombin, were dissolved in dimethyl sulfoxide (DMSO). Thrombin was dissolved in a solution containing 150 mM NaCl and 10 mM Tris-HCl, pH 7.4. Calcium chloride was added to all platelet samples just before activation for a final concentration of 2.5 mM. Platelets were incubated with the kinase inhibitor staurosporine (Calbiochem, La Jolla, CA) or the calpain inhibitor calpeptin (Novabiochem, La Jolla, CA) for 10 to 30 minutes before activation with thrombin. For each experiment, platelets were divided into 1 mL microfuge tubes and kept at 37°C in a water bath. After platelet activation by thrombin, calcium ionophore, or PMA, the samples were inverted 6 times and incubated at 37°C for different lengths of time depending on the experiment. To end the experiment, a volume of Laemmli sample buffer equal to 25% of the platelet volume was added, and the sample was boiled for 10 minutes. Laemmli sample buffer (Biorad) contained 62.5 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate (SDS), 25% glycerol, 5% beta-mercaptoethanol, and 0.01% Bromphenol Blue.
Western blotting and Coomassie blue-stained gels
Samples were separated by SDS polyacrylamide gel electrophoresis on 4% to 15% gradient gels (Biorad) at 100 V. The same sample volume was loaded for the corresponding Coomassie blue-stained gels and the gels that were used for the Western blots. Gels were stained for 15 minutes in Coomassie Brilliant Blue R-250 (0.1% Coomassie blue, 50% methanol, 10% acetic acid), followed by destaining (10% methanol, 7% acetic acid). Samples run on duplicate gels were transferred to nitrocellulose electrophoretically and hybridized with antibodies. Nitrocellulose blots were incubated overnight at 4°C in buffer containing phosphate-buffered saline (PBS), 4% bovine serum albumin (BSA; Fraction V; Sigma), 0.1% Triton X-100, and primary antibody. Blots were washed 3 times at room temperature in the same buffer without BSA, then incubated with protein A conjugated to horseradish peroxidase (Biorad) in buffer with BSA for 60 minutes at room temperature. Blots were washed again 4 times, then incubated with chemiluminescence substrate (Amersham, Piscataway, NJ) and exposed to film. The antibodies used were either antiphosphoadducin (rabbit immunoaffinity-purified immunoglobulin G; Upstate Biotechnology, Lake Placid, NY), or a general antiadducin antibody made in our laboratory. The antiadducin antibody was generated in rabbits by using several synthetic adducin peptides conjugated with glutaraldehyde to rabbit serum albumin (Sigma). The rabbit serum was affinity purified on a column of recombinant human alpha adducin, yet it reacts with alpha, beta, and gamma adducins.
Recombinant alpha adducin
Full-length human alpha adducin was expressed in BL21(DE3) pLysSEscherichia coli by using the pCRT7/NT-TOPO expression vector (Invitrogen, Carlsbad, CA). The vector was constructed so that the amino terminus of alpha adducin is synthesized in frame with a 6-histidine tag. The 6-histidine tag permitted purification of the expressed polypeptide by using immobilized metal affinity chromatography. For purification, the cobalt-containing Talon Superflow Resin (Clontech, Palo Alto, CA) was used. Extraction and elution buffers were prepared according to the user manual for isolation under native conditions. Recombinant adducin was further concentrated by using centriplus (YM-50) centrifuge filters (Millipore, Bedford, MA).
Quantitation of adducin in platelets
Adducin levels in platelet samples were determined by using optical density measurements from Western blots and reference to known adducin levels in samples of recombinant alpha adducin. The concentration of recombinant alpha adducin was determined by using the Biorad protein assay kit with BSA as standard. Several dilutions of recombinant alpha adducin and platelet samples from 2 individuals were run on a 4% to 15% polyacrylamide gel and transferred to nitrocellulose. Western blotting with the antiadducin antibody was performed as described earlier. Adducin levels in the standards were calibrated to an optical density scale by using light intensity measurements of scanned gel images analyzed by using the public domain National Institutes of Health (NIH) Image program. The amount of alpha adducin in each platelet sample was calculated from the standard curve. The number of platelets was determined by a resistive-particle counter after column purification, and the concentration of alpha adducin per platelet was calculated by dividing the amount of adducin from Western blot by the number of platelets loaded on the gel.
Calpain proteolysis of recombinant adducin
Recombinant alpha adducin (240 ng/μL) was phosphorylated by incubation in a reaction-containing adenosine triphosphate 1.6 mM, CaCl2 4.2 mM, PMA 100 nmol/L, recombinant PKCα 0.83 ng/μL (Upstate Biotechnology), NaCl 0.1 mol/L, EGTA 0.8 mM, and NaPO4 0.02 M pH 7.4. Unphosphorylated alpha adducin was prepared by incubating in the same reaction mix with water replacing PKC. Mixes were incubated for 30 minutes in a 30°C water bath. After the incubation at 30°C, samples were divided into 1.5-mL microcentrifuge tubes for the time course of calpain proteolysis. Twenty microliters of human erythrocyte Calpain I (diluted to 0.003 U/μL; Calbiochem) was added to each tube containing 9.6 μg alpha adducin (PKC phosphorylated or unphosphorylated), and samples were incubated for different lengths of time, ranging from 15 to 60 seconds. The reactions were stopped with 20 μL Laemmli sample buffer and boiled for 10 minutes. Samples were analyzed by Western blotting, and relative intensity of bands was calculated from the optical density of bands determined from scanned gel images by using NIH Image software. For each time point, the amount of adducin remaining was calculated from relative intensity and plotted as a curve. P for the difference between the slopes of the curves was determined by Tukey studentized range test.
Platelet fractionation
Column-purified platelets were divided into 1.5-mL Beckman microfuge tubes at 37°C (900 μL per tube). Thrombin (0.1 U/mL or 1 U/mL) and CaCl2 (2.5 mM) were added, and samples were inverted 6 times and kept at 37°C. Some samples were preincubated with calpeptin (200 μg/mL) for 30 minutes. At indicated time points after activation, 100 μL 10× concentrated lysis buffer was added (final concentration 1% Triton X-100, 2.5 mM EGTA, 4 mM Pefabloc-[Roche, Indianapolis, IN], 0.01 mg/mL Aprotinin [Sigma], 10 mM sodium orthovanadate [Calbiochem], 0.01 mg/mL Leupeptin [Sigma], 1 mM phenylmethylsulfonyl fluoride [Sigma]). Samples were vortexed and put on ice for 20 minutes. Samples were then centrifuged in a Beckman TLA 100.4 ultracentrifuge at 55 000 rpm (126 203g) for 3 hours. Gel samples were made by adding 400 μL Laemmli sample buffer to 800 μL supernatant and resuspending the pellet with 750 μL Laemmli sample buffer. Samples were boiled for 10 minutes and loaded proportionately (45 μL supernatant to 22.5 μL pellet) onto 4% to 15% gradient polyacrylamide gels as described earlier. Western blotting was performed as described above, and relative optical density was determined for adducin and PKC-phosphorylated adducin by using NIH Image software.
Platelet aggregation assay
Column-purified platelets were assayed by using a 4-channel platelet aggregation chromogenic kinetic system (PACKS-4; Helena Laboratories, Beaumont, TX) with constant stirring (1000 rpm) at 37°C. Platelets were incubated with 0.3 μM staurosporine, 200 μg/mL calpeptin, or 0.8% DMSO (vehicle control) for 30 minutes before activation. After 30 minutes, samples were added to siliconized glass tubes with a stir bar and activated with 1 U/mL thrombin in the presence of 2.5 mM CaCl2. Maximum aggregation within 2 minutes was determined by the aggregometer and averaged for 3 experiments. P was determined by Tukey studentized range test.
Fluorescence microscopy
Platelets were prepared by column purification as described above. Duplicate samples were made for Western blotting and for microscopy. After incubation with the inhibitor staurosporine (0.3 μM) or calpeptin (200 μg/mL) for 30 minutes or none, platelets were activated with thrombin for 60 seconds. Activation was stopped by addition of glutaraldehyde (final concentration 2%). AlexaFluor 488 phalloidin (Molecular Probes, Eugene, OR) was added to each sample (final concentration 0.002 U/μL phalloidin), and samples were kept at 4°C overnight or longer. On the day of microscopy, samples were pelleted in a microfuge at 10 000 rpm (6720g) for 5 minutes. After the supernatant was removed, the pellet was resuspended with 1 mL PBS, vortexed, and spun again. Pellets were resuspended in 200 μL PBS, and aliquots were placed on glass slides with a coverslip for examination with a Leica confocal microscope.
Results
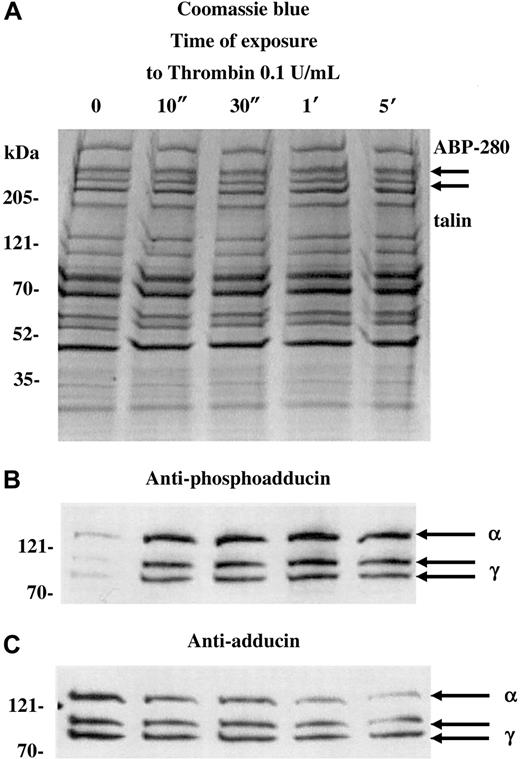
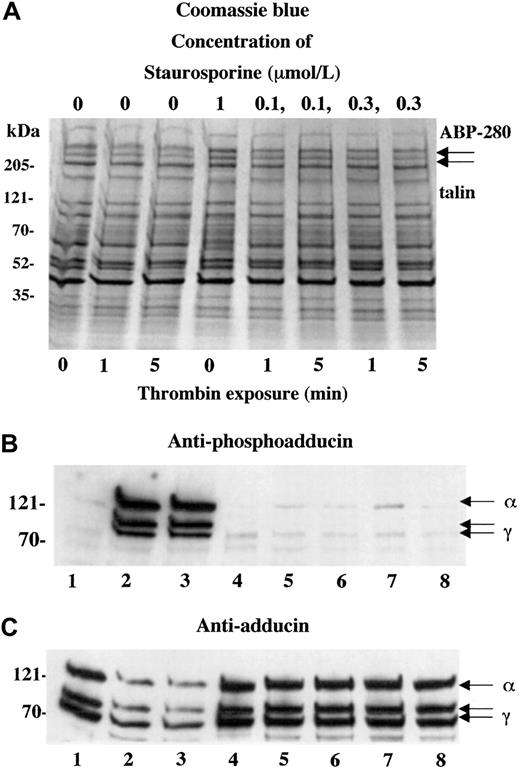
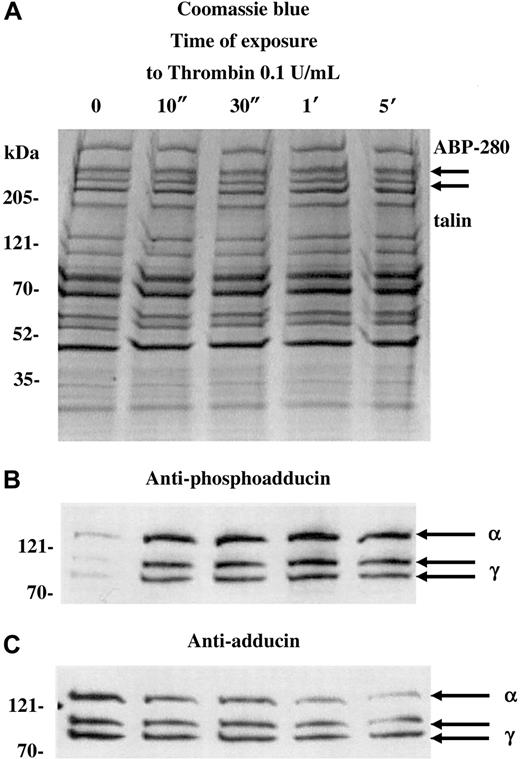
Figure 1 shows a Coomassie blue–stained gel (A) and Western blots (B,C) of platelets activated with 0.1 U/mL thrombin for increasing amounts of time (10 seconds to 5 minutes). For this experiment, a dilute concentration of thrombin was used to capture the state of phosphorylated adducin with minimal proteolysis. Standard doses of thrombin (1 U/mL) caused rapid proteolysis of adducin, making it difficult to assess the level of phosphorylation (see below). As a control for the degree of platelet activation, arrows in panel A indicate the positions of ABP-280 and talin, cytoskeletal components that also undergo proteolysis during platelet activation.27 Although immunoblotting would be necessary to assess minor proteolysis of ABP-280 or talin, panel A shows that dilute thrombin did not cause major proteolysis of ABP-280 or talin (compare with figures below).
PKC phosphorylation and proteolysis of adducin in thrombin-activated platelets.
Column-purified human platelets were exposed to dilute thrombin (0.1 U/mL) in the presence of 2.5 mM CaCl2 for 10 seconds to 5 minutes. (A) Coomassie blue–stained gel. (B) Western blotting with an antibody specific for the carboxy terminal PKC phosphorylation site of adducin demonstrated rapid phosphorylation of alpha and gamma adducins. Western blotting with a general anti-adducin antibody (C) demonstrated increasing proteolysis of adducin with time of exposure to thrombin.
PKC phosphorylation and proteolysis of adducin in thrombin-activated platelets.
Column-purified human platelets were exposed to dilute thrombin (0.1 U/mL) in the presence of 2.5 mM CaCl2 for 10 seconds to 5 minutes. (A) Coomassie blue–stained gel. (B) Western blotting with an antibody specific for the carboxy terminal PKC phosphorylation site of adducin demonstrated rapid phosphorylation of alpha and gamma adducins. Western blotting with a general anti-adducin antibody (C) demonstrated increasing proteolysis of adducin with time of exposure to thrombin.
The antibody used in panel B is specific for the PKC-phosphorylated form of adducin. This antibody was raised against the phosphopeptide CKKFRTP[pS]FLKKNK, corresponding to amino acids 656-668 of human gamma adducin (Upstate Biotechnologies). The serine residue in this site is phosphorylated by PKC and is conserved among alpha, beta, and gamma adducins.15 In resting platelets (lane 0), there was a low level of PKC phosphorylation of adducin, but, within 10 seconds of addition of thrombin, there was a large increase in the level of PKC phosphorylation of adducin. The upper band, migrating at approximately 125 kDa is alpha adducin, and the 2 lower bands are alternatively spliced isoforms of gamma adducin (80 and 90 kDa) (for review see Suriyapperuma et al15). Beta adducin is not expressed in platelets.14 The antibody used in panel C recognizes alpha and gamma adducins independent of their state of phosphorylation. This antibody was used as a control to demonstrate that adducin was present (but relatively unphosphorylated) in the resting platelets, lane 0. In addition, the general adducin antibody demonstrated that the amount of adducin decreased with time of exposure to thrombin and started as quickly as 10 seconds. The intensity of the thrombin-induced PKC-phosphorylated bands appeared constant from 10 seconds to 5 minutes (panel B), yet the decreased amount of total adducin from 10 seconds to 5 minutes (panel C) would suggest that there is an increased proportion of phosphorylated adducin with time of exposure to thrombin. From this experiment it is not possible to determine whether the adducin being proteolyzed is phosphorylated or not; the PKC-phosphorylated adducin could be preferentially proteolyzed while PKC phosphorylation of unphosphorylated adducin would continue, giving a net result that the level of phosphorylated adducin would appear unchanged. It should be noted that if platelets were prepared without prostacyclin in the column buffers, then the adducin was already PKC phosphorylated before addition of thrombin, and a lower yield of platelets was obtained (data not shown).
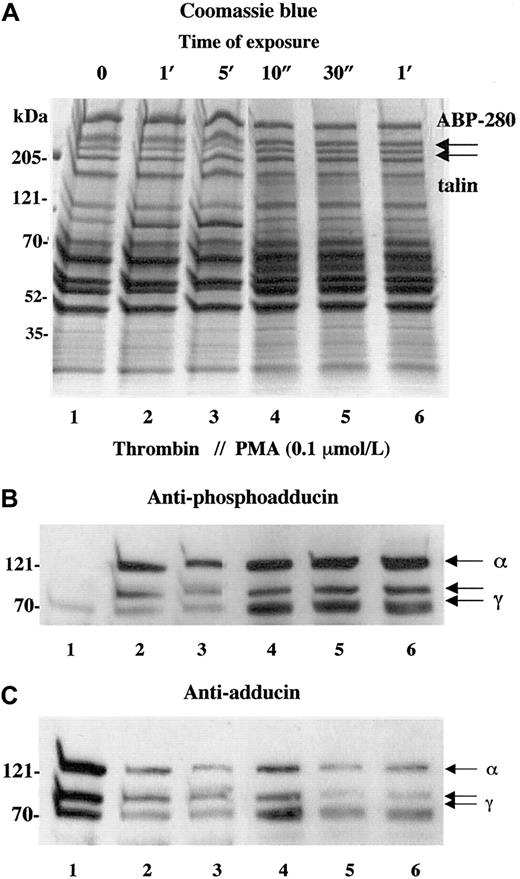
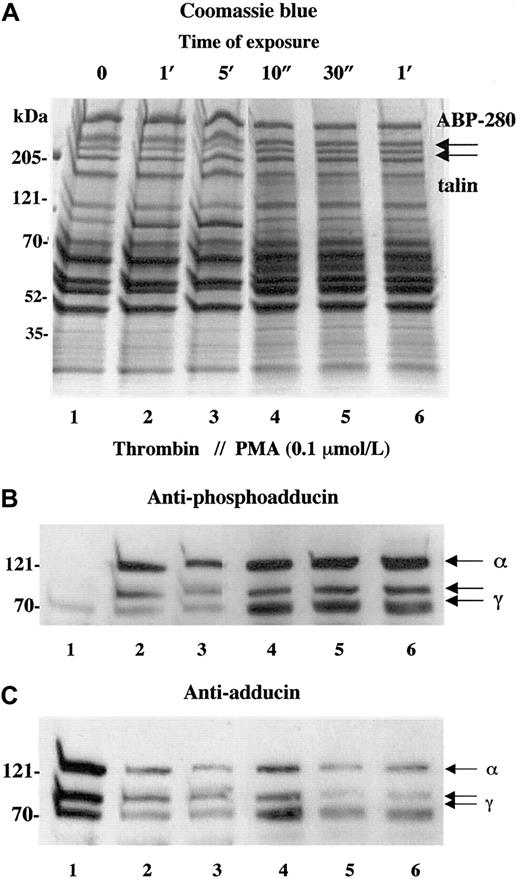
Activation of PKC is a well-studied component of thrombin-induced platelet activation.28-30 To verify that the phosphorylation of adducin detected with the antiphosphoadducin antibody was due to the action of PKC, platelets were stimulated with PMA that directly activates PKC. Figure 2shows a comparison of thrombin activation (1 U/mL) versus activation with PMA (100 nmol/L). In panel B, the phosphospecific antibody demonstrated very low levels of adducin phosphorylation in resting platelets (lane 1) and increased adducin phosphorylation with exposure to thrombin (lanes 2 and 3) or PMA (lanes 4-6). In panel C, the general adducin antibody again demonstrated a large amount of adducin in resting platelets (lane 1) and decreased adducin in both thrombin and PMA-activated platelets (lanes 2-6). These results confirm that the thrombin-induced phosphorylation of adducin detected with this antibody was due to the action of PKC.
Activation of platelets with PMA mimics the effect of thrombin on adducin phosphorylation and proteolysis.
Platelets were exposed to thrombin (1 U/mL) or PMA (0.1 μM) for increasing times. Direct activation of PKC with PMA caused rapid phosphorylation of adducin (B) as well as rapid proteolysis of adducin (C).
Activation of platelets with PMA mimics the effect of thrombin on adducin phosphorylation and proteolysis.
Platelets were exposed to thrombin (1 U/mL) or PMA (0.1 μM) for increasing times. Direct activation of PKC with PMA caused rapid phosphorylation of adducin (B) as well as rapid proteolysis of adducin (C).
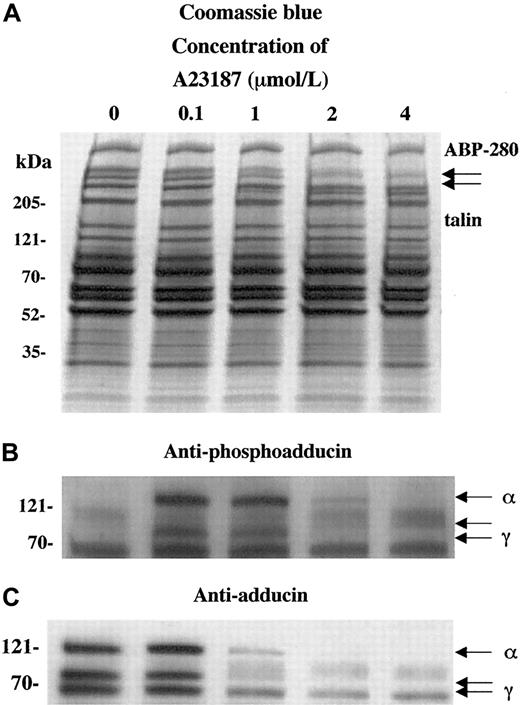
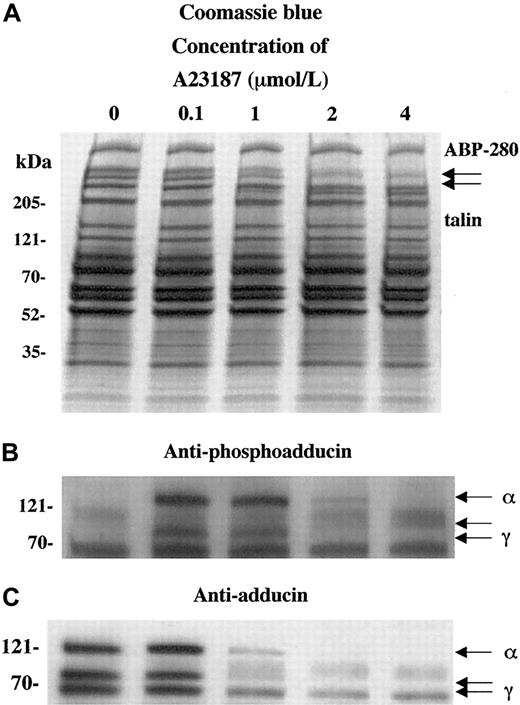
To characterize the calcium dependence of the adducin phosphorylation and proteolysis, platelets were activated with the calcium ionophore A23187. Figure 3 shows platelet activation by using increasing concentrations of calcium ionophore A23187 (0.1 to 4 μM for 2 minutes). Panel A shows that extensive proteolysis of ABP-280 and talin occurred at higher concentrations of A23187 (1-4 μM). This proteolysis was blocked by preincubation for 5 minutes with 5 mM EGTA to chelate extracellular calcium before the exposure to A23187 and calcium (data not shown).
Activation of platelets with calcium ionophore A23187 (and 2.5 mM CaCl2) mimics the effect of thrombin on adducin phosphorylation and proteolysis.
Direct activation of PKC by influx of calcium ions caused PKC phosphorylation of adducin (B) as well as proteolysis (C). At low concentrations of ionophore, phosphorylation occurred without proteolysis (lane 2).
Activation of platelets with calcium ionophore A23187 (and 2.5 mM CaCl2) mimics the effect of thrombin on adducin phosphorylation and proteolysis.
Direct activation of PKC by influx of calcium ions caused PKC phosphorylation of adducin (B) as well as proteolysis (C). At low concentrations of ionophore, phosphorylation occurred without proteolysis (lane 2).
In panel B, phosphorylation of adducin was seen at lower concentrations of A23187 (0.1 and 1 μM), but at higher concentrations (2 and 4 μM) less phosphorylation was seen because the adducin was already proteolyzed (panel C). At 0.1 μM A23187, adducin was phosphorylated by PKC but not yet proteolyzed, suggesting that PKC phosphorylation precedes proteolysis of adducin in platelets. At lower concentrations of A23187 (1-50 nmol/L), PKC phosphorylation of adducin was not detected (data not shown). EGTA blocked both the phosphorylation and the proteolysis of adducin (data not shown). These results suggest that the phosphorylation of adducin is calcium dependent, which confirms the results above, suggesting that PKC phosphorylated adducin during platelet activation.
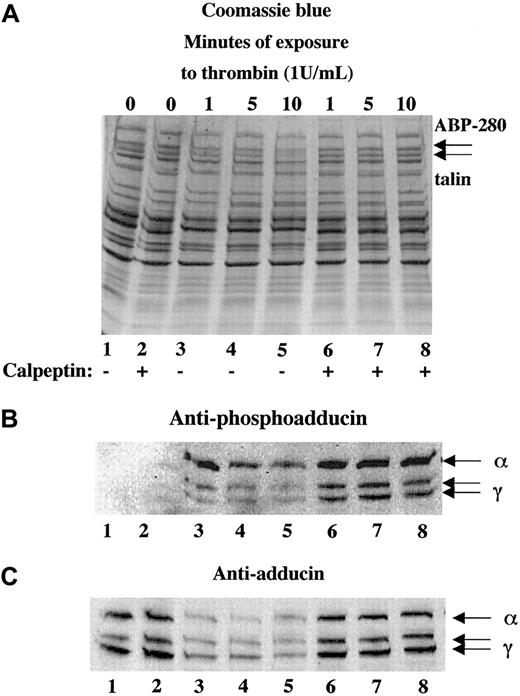
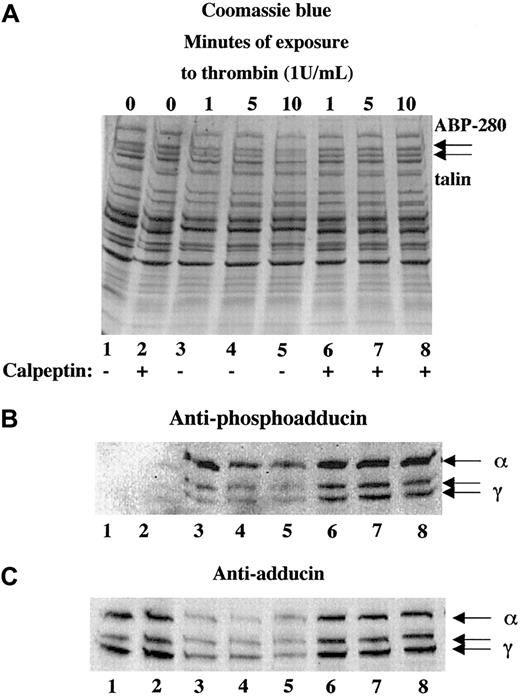
Platelets contain a calcium-dependent protease, calpain, that is known to proteolyze the cytoskeletal proteins ABP-280, talin, spectrin, and cortactin10,27,31 as well as the integrin β332,33 during platelet activation. To test whether calpain was responsible for the proteolysis of adducin during platelet activation, platelets were activated with thrombin in the presence or absence of calpeptin, a membrane-permeable calpain inhibitor.34 Panel A of Figure4 shows thrombin-induced proteolysis of ABP-280 and talin in lanes 3 to 5. This proteolysis was blocked by preincubation of platelets with 100 μg/mL calpeptin before the addition of thrombin, shown in lanes 6 to 8. Panel B (lanes 1 and 2) shows lack of PKC phosphorylation of adducin in resting platelets and platelets incubated with calpeptin alone (200 μg/mL). Activation of platelets with thrombin caused PKC phosphorylation of adducin (lanes 3-8). The intensity of the PKC phosphorylated bands was much stronger in the samples pretreated with calpeptin (lanes 6-8) because proteolysis of adducin was blocked. Panel C demonstrates this inhibition of proteolysis using the general antiadducin antibody. Lanes 3 to 5 show a decreased amount of adducin in thrombin-activated platelets compared with the controls (lanes 1 and 2). The amount of adducin in lanes 6 to 8 was the same as the control, demonstrating that calpeptin blocked proteolysis of adducin.
The calpain-specific inhibitor, calpeptin, inhibited proteolysis of adducin.
Platelets were activated with thrombin (and 2.5 mM CaCl2) in the presence (lanes 6-8) or absence (lanes 3-5) of calpeptin (100 μg/mL preincubation for 10 minutes). Phosphorylation of adducin was more prominent in the samples treated with calpeptin (B) because the proteolysis of adducin was inhibited (C).
The calpain-specific inhibitor, calpeptin, inhibited proteolysis of adducin.
Platelets were activated with thrombin (and 2.5 mM CaCl2) in the presence (lanes 6-8) or absence (lanes 3-5) of calpeptin (100 μg/mL preincubation for 10 minutes). Phosphorylation of adducin was more prominent in the samples treated with calpeptin (B) because the proteolysis of adducin was inhibited (C).
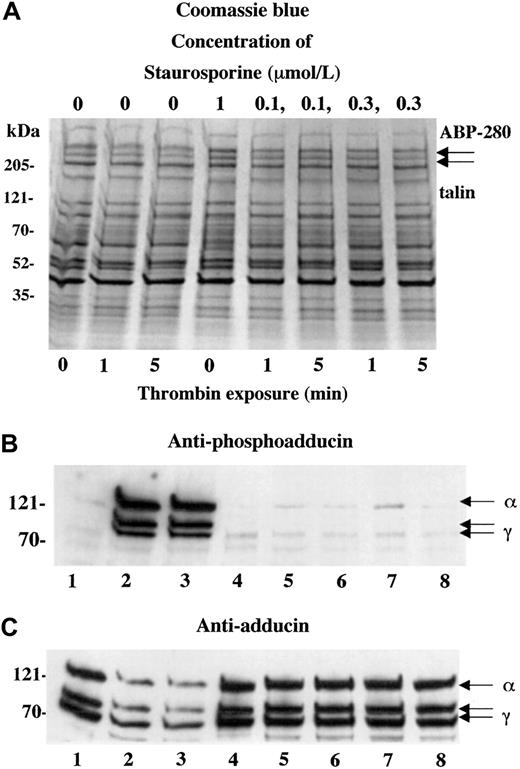
The observation of rapid PKC phosphorylation and calpain proteolysis of adducin during platelet activation suggested that PKC phosphorylation might promote the proteolysis of adducin. To test this possibility, we preincubated platelets with a kinase inhibitor to determine whether blocking phosphorylation of adducin would also block proteolysis of adducin. Figure 5 shows the results of preincubating platelets with staurosporine (0.1 and 0.3 μM)29 before activation with thrombin. Panel B shows the typical PKC phosphorylation of adducin with addition of thrombin (lanes 2 and 3) but no PKC phosphorylation of adducin in platelets preincubated with staurosporine before addition of thrombin (lanes 5-8). Lane 1 shows resting platelets, and lane 4 shows platelets preincubated with staurosporine and not activated with thrombin. Panel C, using the general adducin antibody, shows the typical proteolysis of adducin in thrombin-activated platelets (lanes 2 and 3) but no proteolysis in the platelets pretreated with staurosporine before thrombin activation (lanes 5-8). These results suggest that proteolysis of adducin could be dependent on PKC phosphorylation of adducin. It is also possible that staurosporine inhibited proteolysis of adducin by indirectly inhibiting the activation of calpain.
Staurosporine blocked thrombin-induced PKC phosphorylation and proteolysis of adducin.
Preincubation of platelets with the kinase inhibitor staurosporine (10 minutes) completely blocked thrombin-induced PKC phosphorylation of adducin (B) and proteolysis (C). Thrombin concentration, 1 U/mL; CaCl2, 2.5 mM.
Staurosporine blocked thrombin-induced PKC phosphorylation and proteolysis of adducin.
Preincubation of platelets with the kinase inhibitor staurosporine (10 minutes) completely blocked thrombin-induced PKC phosphorylation of adducin (B) and proteolysis (C). Thrombin concentration, 1 U/mL; CaCl2, 2.5 mM.
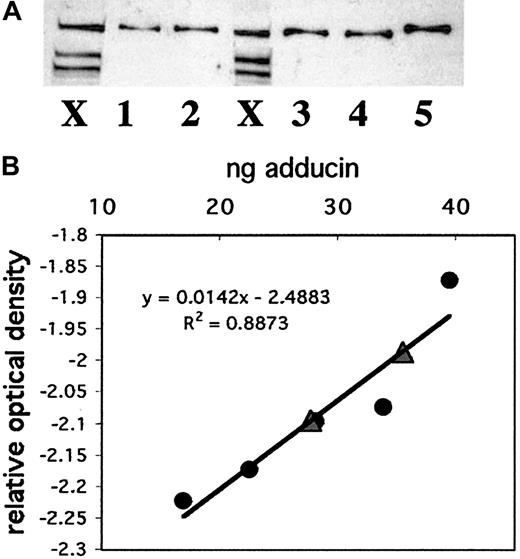
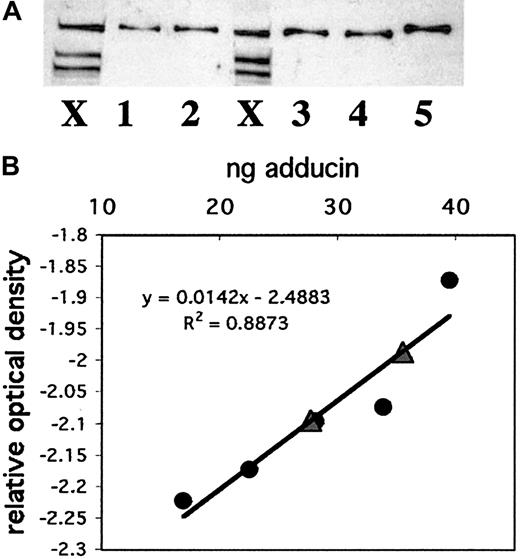
To compare adducin with other actin-binding proteins in platelets and to better understand its possible role in platelet activation, we quantitated the amount of adducin in platelets. Recombinant alpha adducin was used to generate a standard curve for analysis of the concentration of adducin in platelets by Western blot. Figure6A shows a Western blot with whole platelet extracts from 2 individuals (lanes marked X) and 5 dilutions of recombinant alpha adducin (lanes 1-5). Panel B shows the standard curve derived from this Western blot and the positions of the 2 platelet samples (triangles) on the curve. The amount of adducin per platelet based on these 2 samples was determined as 3.5 × 10−6 ng and 4.3 × 10−6 ng (platelet samples contained 7 939 850 and 8 320 000 platelets). Given a mean platelet volume of 8 fL, a mean adducin value of 3.9 × 10−6 ng/platelet, and a molecular weight of 81 kDa (based on the amino acid sequence rather than mobility), the concentration of alpha adducin in a platelet was calculated as 6 μM. This concentration corresponds to approximately 28 800 molecules of alpha adducin per platelet. It is not known whether alpha and gamma adducins exist as a complex in platelets, although evidence from other cells suggests that they are tightly associated.25 If the complex consists of 2 alpha subunits and 2 gamma subunits, then the concentration of the adducin tetramer would be 3 μM, and there would be approximately 14 424 molecules per platelet.
Quantitation of adducin in platelets.
(A) Western blotting was performed with known amounts of recombinant alpha adducin (lanes 1-5) for comparison to platelet samples from 2 individuals (lanes marked X). (B) A standard curve was generated by using NIH Image software to quantitate bands from (A) and convert to relative optical density. Circles correspond to known amounts of adducin; triangles correspond to platelet samples.
Quantitation of adducin in platelets.
(A) Western blotting was performed with known amounts of recombinant alpha adducin (lanes 1-5) for comparison to platelet samples from 2 individuals (lanes marked X). (B) A standard curve was generated by using NIH Image software to quantitate bands from (A) and convert to relative optical density. Circles correspond to known amounts of adducin; triangles correspond to platelet samples.
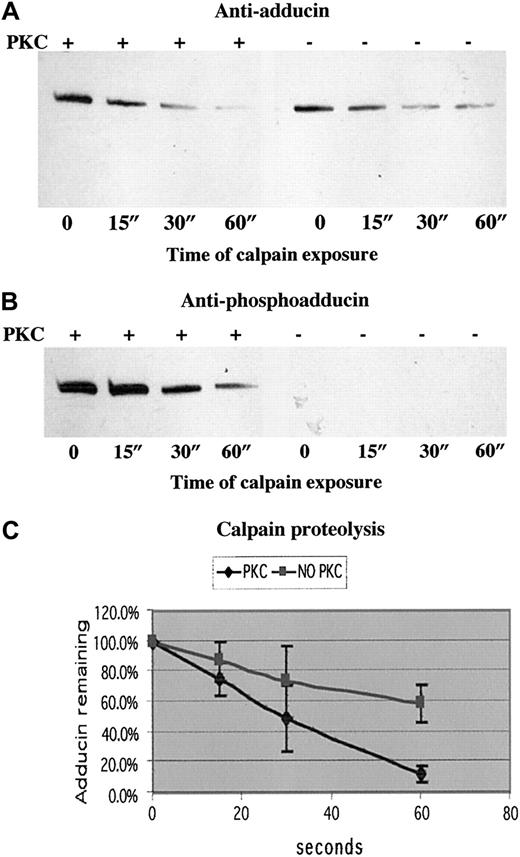
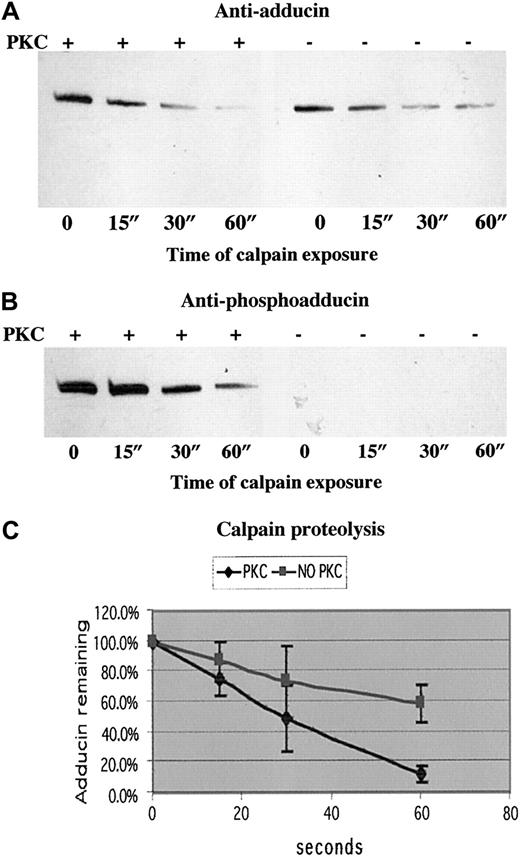
To test our hypothesis that PKC phosphorylation of adducin promotes proteolysis by calpain, we examined calpain proteolysis of recombinant alpha adducin with and without phosphorylation by PKC. Figure7 shows the results of a typical experiment in panels A (anti-adducin Western blot) and B (anti-phosphoadducin Western blot). PKC-phosphorylated adducin decreased to a level of 12% after 60 seconds of incubation with calpain I, whereas unphosphorylated adducin decreased to a level of 58% after 60 seconds of incubation with calpain I (average of 3 experiments quantitated in panel C). On very long exposures, there was a band corresponding to an intermediate product of proteolysis at about 55 kDa, but this band was also proteolyzed over time, confirming our results with platelets that stable intermediate products of proteolysis are not seen after platelet activation. On Coomassie blue–stained gels, the disappearance of the 125 kDa alpha adducin band confirmed that adducin was proteolyzed by calpain and not just undergoing a change in conformation, making it no longer recognizable by antibodies. Quantitation of calpain proteolysis shown in panel C demonstrated that the rate of proteolysis of PKC-phosphorylated adducin was faster than the rate of proteolysis of unphosphorylated adducin. The difference between the slopes of the 2 curves was statistically significant atP < .001.
PKC-phosphorylated adducin was proteolyzed more quickly than unphosphorylated adducin.
Western blotting with (A) anti-adducin antibody and (B) anti-phosphoadducin antibody from a typical time course of μ-calpain proteolysis of recombinant alpha adducin. In lanes 1 to 4, adducin was phosphorylated by PKC before addition of calpain. In lanes 5 to 8, reaction mix included everything as in lanes 1 to 4 except PKC. (C) Quantitation of density of bands from 3 experiments measured with NIH Image and converted to relative optical density. The slope of the line for proteolysis of PKC-phosphorylated adducin is significantly different (P < .001) from the slope of the line for proteolysis of unphosphorylated adducin.
PKC-phosphorylated adducin was proteolyzed more quickly than unphosphorylated adducin.
Western blotting with (A) anti-adducin antibody and (B) anti-phosphoadducin antibody from a typical time course of μ-calpain proteolysis of recombinant alpha adducin. In lanes 1 to 4, adducin was phosphorylated by PKC before addition of calpain. In lanes 5 to 8, reaction mix included everything as in lanes 1 to 4 except PKC. (C) Quantitation of density of bands from 3 experiments measured with NIH Image and converted to relative optical density. The slope of the line for proteolysis of PKC-phosphorylated adducin is significantly different (P < .001) from the slope of the line for proteolysis of unphosphorylated adducin.
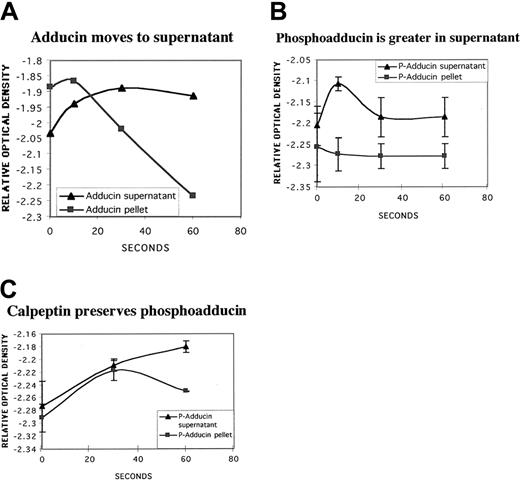
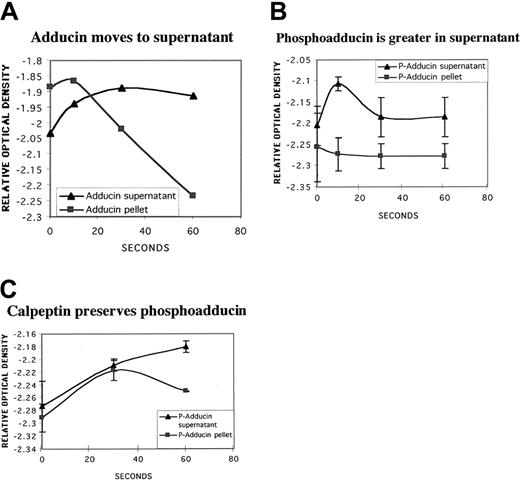
PKC phosphorylation of adducin has been shown to be associated with a shift of adducin from the membrane skeleton to the cytosol in MDCK cells treated with PMA.35 To determine whether the PKC phosphorylation of adducin in platelets was associated with a shift in subcellular location, we fractionated platelets into high-speed (126 000g × 3 hours) supernatant and pellet fractions at time points after addition of dilute thrombin (0.1 U/mL). Western blots were performed on supernatant and pellet fractions, and the optical density of adducin and PKC-phosphoadducin bands were quantitated with NIH Image. Figure 8A shows increased adducin in the supernatant and decreased adducin in the pellet versus time of exposure to thrombin. Although the amount of adducin in the pellet continued to decrease with time, the amount in the supernatant remained constant, so that total adducin decreased over time, consistent with proteolysis. Figure 8B shows the amount of PKC-phosphorylated adducin in supernatant versus pellet fractions (thrombin 0.1 U/mL, n = 2). There was a rapid increase of PKC-phosphorylated adducin in the supernatant fraction (10 seconds), followed by a decrease consistent with proteolysis.
Movement of PKC-phosphorylated adducin to cytosol with thrombin activation.
Column-purified platelets were activated with dilute thrombin (0.1 U/mL) for increasing times and fractionated by high-speed centrifugation (126 000g for 3 hours) into supernatant and pellet fractions. Samples were run on SDS-polyacrylamide gel electrophoresis and Western blotted; relative optical density of bands was quantitated by using NIH Image software. (A) Typical experiment showing increasing adducin in supernatant versus decreasing adducin in pellet. (B) Distribution of PKC-phosphorylated adducin between supernatant and pellet fractions over time (n = 2). (C) Platelets were preincubated with calpeptin (200 μg/mL for 30 minutes), stimulated with thrombin (1 U/mL), then fractionated as above. PKC-phosphorylated adducin continued to increase in the supernatant while decreasing in the pellet fraction.
Movement of PKC-phosphorylated adducin to cytosol with thrombin activation.
Column-purified platelets were activated with dilute thrombin (0.1 U/mL) for increasing times and fractionated by high-speed centrifugation (126 000g for 3 hours) into supernatant and pellet fractions. Samples were run on SDS-polyacrylamide gel electrophoresis and Western blotted; relative optical density of bands was quantitated by using NIH Image software. (A) Typical experiment showing increasing adducin in supernatant versus decreasing adducin in pellet. (B) Distribution of PKC-phosphorylated adducin between supernatant and pellet fractions over time (n = 2). (C) Platelets were preincubated with calpeptin (200 μg/mL for 30 minutes), stimulated with thrombin (1 U/mL), then fractionated as above. PKC-phosphorylated adducin continued to increase in the supernatant while decreasing in the pellet fraction.
To examine the effect of PKC phosphorylation of adducin without proteolysis, platelets were preincubated with calpeptin (200 μg/mL) before thrombin activation (1 U/mL, n = 2). Figure 8C shows that PKC-phosphorylated adducin in the supernatant continued to increase over time in contrast to the decrease seen without calpeptin (Figure8B). PKC-phosphorylated adducin in the pellet also increased up to the 30-second time point and then decreased, consistent with movement of phosphoadducin to the supernatant. Together, these data suggest an association between PKC phosphorylation of adducin and movement to the cytosol during thrombin activation of platelets.
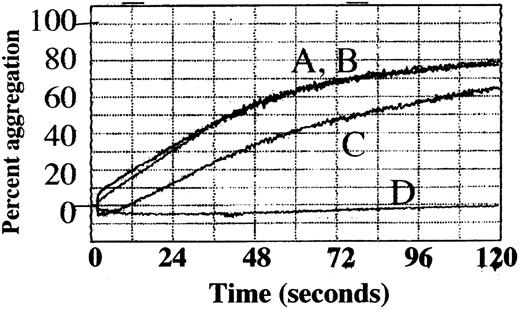
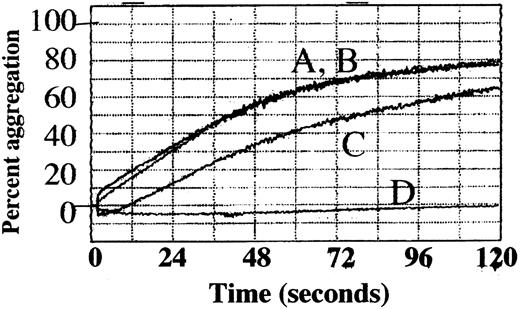
To determine how adducin phosphorylation by PKC and proteolysis by calpain may be involved in platelet function, platelet aggregation was assayed by using a 4-channel aggregometer. Four samples were tested simultaneously: untreated control, staurosporine-treated, calpeptin-treated, or DMSO-treated control. Thrombin (1 U/mL) was used to stimulate aggregation in the presence of 2.5 mM CaCl2. Results of a typical experiment are shown in Figure9. Untreated platelets (A) and DMSO-treated control platelets (B) showed no difference and aggregated within 2 minutes to a maximum of 77% ± 0.8% and 76.8% ± 1.7% (mean of 3 experiments). Aggregation of staurosporine-treated platelets (C) was inhibited to a maximum of 64.1% ± 0.5%; the difference from control was statistically significant (P = .003). Aggregation of calpeptin-treated platelets (D) was completely inhibited with a maximum of 3.8% ± 1.9%; the difference from control was statistically significant (P = .003). These results suggest that PKC phosphorylation is not absolutely required for platelet aggregation but, if inhibited, leads to a delay in platelet aggregation. These results also show that calpain function is essential for platelet aggregation under our conditions of platelet preparation and activation. Inhibition of adducin phosphorylation by PKC and proteolysis by calpain could have a role in the observed inhibition of platelet aggregation, although PKC and calpain have a large number of substrates other than adducin.
Platelet aggregation was delayed by staurosporine and inhibited by calpeptin.
Column-purified platelets were stimulated with thrombin (1 U/mL) in the presence of 2.5 mM CaCl2, and the percentage of aggregation was followed as a measure of change in light transmission over 120 seconds. (A) Control, (B) 0.8% DMSO (vehicle), (C) 0.3 μM staurosporine 30 minutes preincubation, (D) 200 μg/mL calpeptin 30 minutes preincubation.
Platelet aggregation was delayed by staurosporine and inhibited by calpeptin.
Column-purified platelets were stimulated with thrombin (1 U/mL) in the presence of 2.5 mM CaCl2, and the percentage of aggregation was followed as a measure of change in light transmission over 120 seconds. (A) Control, (B) 0.8% DMSO (vehicle), (C) 0.3 μM staurosporine 30 minutes preincubation, (D) 200 μg/mL calpeptin 30 minutes preincubation.
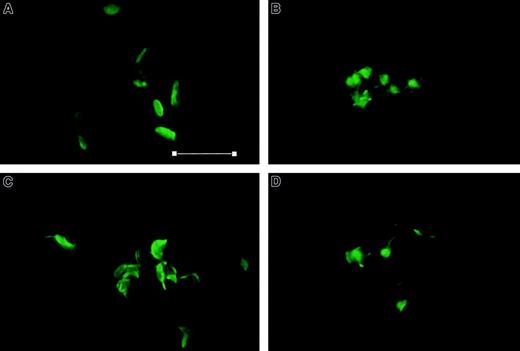
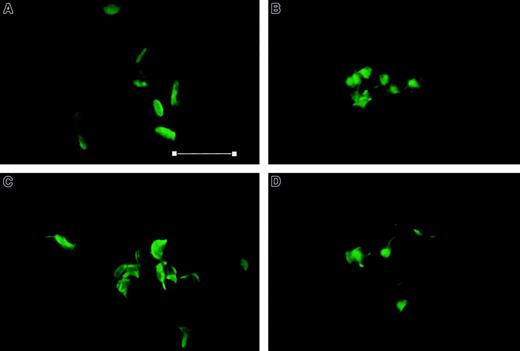
Aggregation of platelets is a complex process comprising several steps: secretion of granules, extension of filopodia and lamellipodia by F-actin polymerization, and modulation of the integrin αIIbβ3 to permit binding of fibrinogen and generation of cell-cell linkages. We have demonstrated that adducin is present in sufficient quantity to cap all barbed ends of actin filaments in resting platelets and that adducin moves from skeleton to cytosol during platelet activation. It is possible that phosphorylation by PKC causes adducin to be released from actin-capping sites, thereby allowing polymerization of short actin filaments into long actin filaments. Thrombin-stimulated platelets were examined by confocal microscopy under conditions shown above that inhibit adducin phosphorylation by PKC and/or proteolysis by calpain. Fixed platelets were labeled with AlexaFluor 488 phalloidin to stain F-actin, and results are shown in Figure10. Panel A shows control platelets that maintained the characteristic disc morphology of unactivated platelets. Panel B shows platelets after thrombin stimulation with extension of filopodia and lamellipodia and F-actin polymerization. Platelets in panels C and D were incubated with staurosporine (C) or calpeptin (D) before addition of thrombin and showed similar extension of filopodia and lamellipodia with F-actin polymerization. The results shown are typical of 3 independent experiments on different days using different platelet donors. These results demonstrated that PKC phosphorylation and calpain proteolysis of adducin were not necessary for the changes in morphology observed by confocal microscopy with phalloidin staining.
Thrombin-induced morphologic changes are not affected by staurosporine and calpeptin.
Column-purified platelets were stimulated with thrombin 1 U/mL for 60 seconds, fixed with glutaraldehyde, and stained with AlexaFluor 488 phalloidin. (A) Control, (B) thrombin, (C) staurosporine (0.3 μM) for 30 minutes, then thrombin, and (D) calpeptin (200 μg/mL) for 30 minutes, then thrombin. All platelets stimulated with thrombin show extension of filopodia and lamellipodia with F-actin polymerization. Lack of aggregation in (D) is apparent. Bar = 7 μm. Magnification × 2400.
Thrombin-induced morphologic changes are not affected by staurosporine and calpeptin.
Column-purified platelets were stimulated with thrombin 1 U/mL for 60 seconds, fixed with glutaraldehyde, and stained with AlexaFluor 488 phalloidin. (A) Control, (B) thrombin, (C) staurosporine (0.3 μM) for 30 minutes, then thrombin, and (D) calpeptin (200 μg/mL) for 30 minutes, then thrombin. All platelets stimulated with thrombin show extension of filopodia and lamellipodia with F-actin polymerization. Lack of aggregation in (D) is apparent. Bar = 7 μm. Magnification × 2400.
Discussion
We have shown that adducin was rapidly phosphorylated by PKC in platelets activated with thrombin, PMA, or calcium ionophore A23187, as defined by the use of a PKC site-specific phosphopeptide antibody. There are several possibilities for how PKC phosphorylation may regulate the function of adducin. Studies on the interaction of gamma adducin with PKC suggested that unphosphorylated gamma adducin can bind PKC, and, when PKC phosphorylates gamma adducin, it is released from its association.25 Further studies are needed to determine whether PKC and unphosphorylated adducin exhibit this interaction in platelets. Phosphorylation of recombinant alpha and beta adducin by PKC has been shown to decrease21 their interactions with spectrin and actin in vitro, and treatment of MDCK cells with PMA caused a redistribution of adducin from membrane to cytosol as seen by light microscopy.35 We have shown that adducin moves from high-speed pellet (membrane and cytoskeleton fraction) to supernatant (cytosolic fraction) during platelet activation and levels of PKC-phosphorylated adducin are much higher in the cytosol versus the pellet. These observations suggest that activation-induced PKC phosphorylation of adducin releases it from binding sites in the membrane skeleton or cytoskeleton.
Erythrocyte adducin (alpha/beta) has been shown to cap actin filaments at the barbed end,18 and PKC phosphorylation reduces the F-actin capping activity of adducin.21 Platelets contain 0.5 μM F-actin barbed ends,36 based on measurements of average filament length and amount of polymerized actin per platelet.2 We have demonstrated that sufficient adducin is present (3-6 μM) to cap all actin filaments in the resting platelet. The concentration of adducin in platelets is similar to the 2 to 5 μM concentration of capping protein.36,37 However, the dissociation constant for binding to barbed ends is only 100 nmol/L for adducin18 versus 1 to 2 nmol/L for capping protein.38 In addition, it has not yet been demonstrated that platelet adducin (alpha/gamma) has actin-capping activity.
Adducin is also a substrate for rho kinase, and phosphorylation by rho kinase, in contrast to phosphorylation by PKC, increases adducin's interaction with F-actin.22 Rho kinase is also activated during platelet activation,39 so it is not yet known how the combination of phosphorylation by PKC and rho kinase will affect adducin function in platelets.
One important observation from our studies is that adducin was proteolyzed during platelet activation. Inhibition of the proteolysis by calpeptin suggests that calpain is likely to be the protease involved. Calpain is an important component of platelet activation, causing highly specific calcium-induced proteolysis of several cytoskeleton proteins, including ABP-280, talin, spectrin, and cortactin.10,27,31 In addition, calpain proteolyzes the β3 subunit of the αIIbβ3 integrin, the fibrinogen receptor, during platelet activation.3,32,33,39 This is the first report of adducin as a substrate for calpain proteolysis and may represent an important pathway for regulation of adducin function in platelets as well as other cells. Results of in vitro experiments with recombinant alpha adducin demonstrated that PKC-phosphorylated adducin was proteolyzed by calpain more quickly than unphosphorylated adducin. Demonstrating the influence of PKC phosphorylation on adducin proteolysis in the absence of binding proteins such as spectrin or actin suggests a mechanism whereby PKC phosphorylation of adducin directly causes a conformational change of adducin, permitting rapid proteolysis by calpain. A similar result has recently been shown in which the PKC-phosphorylated form of adducin was preferentially proteolyzed by caspase-3 in cisplatin-treated renal epithelial cells.40 Phosphorylation of adducin by PKC may be a common mechanism for regulating adducin proteolysis by several proteases.
Others have shown that inhibition of calpain blocks platelet granule secretion, aggregation, and spreading on glass.41 Our results inhibiting proteolysis of adducin and platelet aggregation with calpeptin are consistent with adducin having a role in platelet aggregation. Targeted deletion of the μ-calpain gene in mice demonstrated that μ-calpain is crucial to normal platelet aggregation and clot retraction.42 The proteolysis of ABP-280, talin, and β3 integrin was not inhibited in platelets from the μ-calpain null mice, suggesting that proteolysis of the cytoskeleton is not required for platelet aggregation or clot retraction. However, platelets from the μ-calpain null mice still contain m-calpain, so it is not possible to conclude definitively about the role of the cytoskeleton in platelet aggregation from the μ-calpain null mice.
We have shown that adducin is crucial to the normal architecture of red blood cells.14 Targeted disruption of the beta adducin gene in mice caused a phenotype similar to the human disease hereditary spherocytosis. Platelets were unaffected in these mice because platelets do not express beta adducin. Future studies with targeted disruption of alpha and gamma adducin genes are likely to provide answers about the specific role of adducin in platelets.
We thank Laura Stewart for help with aggregation assays and Doug Bolgiano for help with statistical analyses.
Supported by grant R01-DK55005 from the National Institutes of Health, Bethesda, MD, and by a grant from the March of Dimes Birth Defects Foundation, Mamaroneck, NY.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Diana M. Gilligan, Puget Sound Blood Center, 921 Terry Ave, Seattle, WA 98104; e-mail: dianag@psbc.org.