Inherited factor XI deficiency is an injury-related bleeding disorder that is rare in most populations except for Jews, in whom 2 mutations, a stop mutation in exon 5 (type II) and a missense mutation in exon 9 (type III), predominate. Recently, a cluster of 39 factor XI–deficient patients was described in the Basque population of Southwestern France. In this study, we determined the molecular basis of factor XI deficiency in 16 patients belonging to 12 unrelated families of French Basque origin. In 8 families, a nucleotide 209T>C transition in exon 3 was detected that predicts a Cys38Arg substitution. Four additional novel mutations in the factor XI gene, Cys237Tyr, Tyr493His, codon 285delG, and IVS6 + 3A>G, were identified in 4 families. Expression studies showed that Cys38Arg and Cys237Tyr factor XI were produced in transfected baby hamster kidney cells, but their secretion was impaired. Cells transfected with Tyr493His contained reduced amounts of factor XI and displayed decreased secretion. A survey of 206 French Basque controls for Cys38Arg revealed that the prevalence of the mutant allele was 0.005. Haplotype analysis based on the study of 10 intragenic polymorphisms was consistent with a common ancestry (a founder effect) for the Cys38Arg mutation.
Introduction
Factor XI is a zymogen that, upon activation to factor XIa, promotes coagulation by activating factor IX.1It is currently postulated that factor XI does not play an important role in the initiation of blood coagulation but is essential for its propagation after small amounts of thrombin are generated.2 Factor XI is activated by thrombin preferentially on the surface of platelets, which leads to further thrombin generation even after the clot has formed.3-5 The additional amount of thrombin then activates thrombin-activatable fibrinolysis inhibitor, which results in clot stabilization owing to resistance to fibrinolysis.5
Factor XI circulates as a homodimer of identical 80-kd subunits linked by a disulfide bond. Upon activation by thrombin, factor XI is cleaved at Arg369–Ile370, yielding a heavy chain and a light chain, with the latter harboring the active catalytic site. The heavy chain is composed of 4 “apple domains,” each composed of 90 to 91 amino acids and stabilized by 3 disulfide bridges.6,7 Apple domain 1 contains the binding site for high–molecular weight kininogen, thrombin, and prothrombin8-10; apple domain 3 contains the binding site for factor IX and platelets11,12; and apple domain 4 harbors the binding sites for factor XIIa and for dimerization of the 80-kd subunits.13 14
The human factor XI gene consists of 15 exons and is localized on the long arm of chromosome 4 (4q35).15,16 The gene is approximately 50 kilobases (kb) in length,17 and not 23 kb as initially reported.15 So far, 26 mutations causing factor XI deficiency have been reported.18,19 Of these, 4 are nonsense mutations, 5 are splice-site mutations, 3 are small insertion/deletion mutations, and the remaining 14 are missense mutations. Six of the missense mutations were localized to the apple 4 domain, and expression of 5 of them revealed impaired secretion of factor XI from transfected cells.20,21 For one of the mutations, Phe283Leu (also designated type III mutation), the impaired secretion was attributed to a defect in dimerization of the factor XI monomers.21
Inherited factor XI deficiency is a rare autosomal recessive injury-related bleeding disorder.22 The disorder is common in Jews of mainly Ashkenazi (European) origin, among whom 2 mutations, a stop mutation in exon 5 (type II) and a missense mutation in exon 9 (type III), predominate.23,24 A survey of 125 unrelated Ashkenazi Jewish patients with severe factor XI deficiency disclosed that the type II and type III mutations each accounted for 49.2% of the mutant alleles.25 Another cluster of 39 cases with factor XI deficiency was recently identified among Basques residing in Southern France.26 In this study, we determined the molecular basis of factor XI deficiency in 16 French Basque patients and addressed the question of whether one or more mutations account for the relatively high frequency of the disorder in this population.
Patients, materials, and methods
Patients
The diagnosis of factor XI deficiency (factor XI activity below 50 U/dL) was established in 12 probands and 4 immediate relatives who reside in the Basque region of Southwestern France following a finding of prolonged activated partial thromboplastin time (aPTT). Only 1 of the 16 patients was tested because of hematoma following herniotomy; the remaining patients were routinely tested for aPTT. All patients were questioned about previous spontaneous and injury-related bleeding manifestations. The ethnic origin of the patients was defined according to the place of birth of the individual's grandparents or parents. Blood samples were obtained from factor XI–deficient patients who gave their consent to participate in this study, and from control French Basque subjects referred for routine complete blood count.
DNA purification
DNA was extracted from whole blood of patient's leukocytes by a standard procedure.27 Drops of blood remaining after completion of the complete blood count in controls were mounted on Blood Collection Cards (Gentra Systems, Minneapolis, MN) and shipped to the laboratory in Israel. Small disks were punched out from the cards and treated with DNA purification solution (Gentra Systems) according to the manufacturer's instructions. By this procedure, the DNA remains on the disks and can be used directly for polymerase chain reaction (PCR).
Mutation identification
PCR amplifications of exons 3 through 15 of the factor XI gene were performed with the use of genomic DNA as template, with primers and conditions as described by Imanaka et al.28 Direct sequencing of amplified fragments was carried out by an automatic sequencer (ABI Prism 377, Perkin Elmer, Foster City, CA).
Analysis of polymorphisms
The CA repeat polymorphism in intron B and the AT repeat polymorphism in intron M were determined as previously reported.25,29 The polymorphisms 231C>T and 138C>A in intron A19 were analyzed by amplification of intron A and digestion with BseDI (Fermentas, Vilnius, Lithuania) and MslI (New England Biolabs, Beverly, MA), respectively. The 3 previously described polymorphisms 472C>T in exon 5, 844A>G in exon 8, and 1234T>C in exon 11, which are in strong linkage disequilibrium, were analyzed by PCR amplification of the corresponding exons and digestion with the restriction enzymes AatlI (New England Biolabs),Bsh1236I (Fermentas), and HphI (New England Biolabs), respectively.19,30 Intron E dimorphism, comprising a 150–base pair (bp) fragment (allele 1) or a 140-bp fragment (allele 2), was detected as described.31 The recently reported polymorphisms 1855G>T and 1882G>A in exon 15 were analyzed by amplification of exon 15 and simultaneous digestion withBsrBI and RsaI (New England Biolabs).32 All digestions were carried out according to the manufacturer's instructions and analyzed on either 3% agarose gels (Amresco, Solon, OH) or 4% agarose-SFR (super-fine resolution) gels (Amresco) containing ethidium bromide for UV visualization.
Construction of expression vectors
Human factor XI complementary DNA (cDNA) in pBR322 vector was kindly provided by Dr Dominic Chung from the University of Washington, Seattle, and was subcloned by BamHI (New England Biolabs) digestion into pGEM7 (Promega, Madison, WI) for mutagenesis. The sequence alterations 209T>C, 807G>A, 1059G>T, and 1574T>C were introduced into factor XI cDNA by PCR site-directed mutagenesis by means of, in each instance, 2 overlapping oligonucleotide primers that incorporated the corresponding single base pair substitutions.33 The created mutants were confirmed by sequencing. PCR products containing each of the 4 alterations were cut by specific restriction enzymes for replacement of the corresponding normal cDNA in pGEM7-factor XI. The corresponding 5′ and 3′ restriction enzymes for 209T>C, 807G>A, 1059G>T, and 1574T>C PCR fragments were as follows: BbsI and BsaMI; Tth111I and PstI; PstI and BstEII; andBclI and PpuMI, respectively (New England Biolabs).
Wild-type (WT) and mutated cDNAs were subcloned into theBamHI site of pcDNA3 (Invitrogen, Carlsbad, CA) mammalian expression vector carrying the geneticine resistance gene as a selection marker. Plasmids were transformed inEscherichia coli DH5α and grown in Luria-Bertani medium (DIFCO, Detroit, MI) supplemented with 100 μg/mL ampicillin (Sigma, St Louis, MO) for large-scale preparation of DNA by means of Concert nucleic acid purification system (Gibco BRL, Paisley, United Kingdom).
Cell culture and transfection
Baby hamster kidney cells (BHKs) were grown in Dulbecco modified Eagle medium (DMEM) supplemented by 2 mg/mL L-glutamine and 5% fetal calf serum (FCS). Medium and FCS were purchased from Biological Industries, Beit-Haemek, Israel. The cells were transfected by means of LipofectAMINE reagent (Gibco BRL) with 1 μg either WT factor XI or mutant factor XI pcDNA3s. Mock cells were produced by transfecting BHK cells with 1 μg native pcDNA3 vector. Selection for stably transfected cells was carried out by growing the cells in medium containing 0.7 μg/mL G418 (Gibco BRL) for 2 weeks. Medium was collected for factor XI assays from confluent plates containing about 106 cells grown for 24 hours with 7 mL FCS-free medium (Optimem 1) (Gibco BRL).
Factor XI activity and antigen measurements
Factor XI activity was measured by an aPTT-based assay with severe factor XI–deficient plasma used as substrate (factor XI activity below 1 U/dL) and 1/10 to 1/640 dilutions of normal reference plasma used for construction of a standard curve. The medium of cultured BHK cells was concentrated × 20 by filtration on a centriconPM-30 (Millipore, Bedford, MA) and assayed directly without the usual 1/5 to 1/10 dilution prior to testing. The sensitivity of factor XI activity measurements was thus as low as 0.01 U/dL.
Factor XI antigen was determined by an enzyme-linked immunosorbent assay (ELISA) using 4 μg/mL anti–factor XI monoclonal antibodies for capturing (Enzyme Research, South Bend, IN), followed by incubation with samples containing factor XI antigen and binding of 1:1000 dilution of polyclonal goat anti–factor XI immunoglobulin (Ig)–G (Enzyme Research). Amplification and detection were achieved with biotinylated antigoat IgG and avidin peroxidase by means of the Vectastain ABC kit (Vector Laboratories, Burlingame, CA). Standard curves were constructed with reference plasma diluted 1/25 to 1/800 in Tris-buffered saline (TBS) (50 mM Tris, 150 mM NaCl), pH 7.5. When no factor XI antigen was detected in the medium of transfected cells, ELISA measurements were repeated with 2 additional capture monoclonal antibodies (mAbs), kindly provided by Dr Joost Meijers (University of Amsterdam, The Netherlands). One antibody, mAb-3, was directed against an epitope on apple 2, and the other, mAb-5, was directed against an epitope on apple 1. This precaution was taken to confirm that the absence of factor XI antigen was not due to a loss of a recognition site. Measurement of factor XI antigen was performed either in the medium or in × 10 concentration of the medium. The lower limit of factor XI antigen detection was 0.5 ng/mL.
Western blot analysis of factor XI antigen
Proteins were analyzed by sodium dodecyl sulfate–polyacrylamide gels (SDS-PAGE) and immunoblotting. Factor XI antigen was detected on the gel by means of 1:1000 dilution of polyclonal goat anti–factor XI IgG (Enzyme Research) followed by 1:5000 dilution of peroxidase-labeled antigoat IgG (Sigma) and chemiluminescence (ECL Western blotting detection reagent) (Amersham Pharmacia Biotech, Buckinghamshire, England).
Immunoprecipitation
The medium of BHK cells was filtered through a 0.22-μ membrane (Millipore) and centrifuged for 15 minutes at 14 000g to remove cell debris. One milliliter of medium was incubated for 1 hour at 4°C with 3 μg nonimmune mouse IgG (Sigma) followed by 60 μL 10% Pansorbin cells for 1 hour at 4°C (Calbiochem-Novabiochem, La Jolla, CA). After centrifugation, the supernatant was transferred to a tube containing 3 μg monoclonal antibody against factor XI (Enzyme Research) and incubated on ice for 1 hour. Immune complexes were removed by addition of 60 μL goat antimouse IgG coupled to agarose (Sigma), incubation for 2 hours with mixing at 4°C, and centrifugation. The agarose beads were washed twice with 50 mM Tris (pH 7.5), 0.5 M NaCl, and 0.1% NP-40, followed by a wash with TBS, pH 7.5. Proteins were eluted in sample buffer (0.125 M Tris-HCl, pH 6.8; 5% SDS; 25% glycerol; and bromophenolblue as dye) and incubated for 15 minutes at 60°C before SDS-PAGE.
Metabolic labeling of factor XI in transfected cells
Transfected cells in 6-well plates, each containing 105 cells, were exposed to methionine-free DMEM for 1 hour; then, 150 μCi (5.55 MBq) [35S] protein mix (New England Biolabs) was added for a 30-minute pulse. The cells were chased with cold methionine (2 mM) for 1, 3, 6, and 20 hours and then washed twice with phosphate-buffered saline (PBS) and lysed in 1 mL cold lysis buffer (10 mM Hepes, pH 7.5; 50 mM NaCl; 5 mM EDTA; 0.3 M sucrose; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% SDS) for 10 minutes. The lysates were centrifuged at 14 000g for 15 minutes at 4°C and kept frozen. Cell lysates were immunoprecipitated by monoclonal antibodies as described, eluted into reduced sample buffer, and separated by SDS-PAGE. NuPage 4% to 12% Bis Tris gels in 2-N-morpholinoethanesulfonic acid running buffer were used according to the manufacturer's instructions (Invitrogen). The gels were fixed in isopropanol/acetic acid/water (25:10:65), vacuum-dried, and exposed to Biomax MS film with the use of Biomax Transcreen-LE (Kodak, Rochester, NY) at −70°C.
Immunostaining of intracellular factor XI
Transfected cells were grown on glass slides to subconfluence and then washed in PBS, dried for 24 hours, and fixed in cold acetone for 15 minutes. The cells were then incubated in TBS (pH 7.6) containing mAb-3 or mAb-5 (10 to 20 μg/mL) and stained with an alkaline phosphatase–antialkaline phosphatase kit (APAAP) (Dako, Glostrup, Denmark). Microscopy and photography were done by Duet (BioView, Rehovot, Israel).
Results
Bleeding manifestations
None of the 16 patients with severe or mild factor XI deficiency exhibited spontaneous bleeding. Hemostatic challenges occurred in 10 patients, 2 of whom with mild factor XI deficiency displayed (1) a hematoma following cervical disc surgery and (2) mild postpartum bleeding, respectively (Table1). Two siblings with severe factor XI deficiency underwent uneventful surgical procedures with plasma replacement therapy.
Identification of candidate mutations
Table 1 displays the origin of the 12 families studied, the individuals' factor XI activities, and the mutations that were identified. In 8 of the 12 families, a nucleotide (nt) 209T>C transition in exon 3 was detected, which predicts a Cys38Arg substitution. Patient 1 was homozygous for Cys38Arg, and 8 patients from 6 families were heterozygous for Cys38Arg. The twins of family VIII who had severe factor XI deficiency were compound heterozygotes for the Cys38Arg and a novel mutation, nt 1574T>C transition in exon 13, which predicts a Tyr493His substitution.
The 2 siblings of family IX were of mixed Basque and Jewish origin and had factor XI activity of less than 1 U/dL. Both were compound heterozygotes for the type II nonsense mutation observed in Jews (Glu117Stop) and for a novel nt 807G>A change in exon 8 predicting Cys237Tyr substitution. Both siblings also harbored a rare 1059G>T polymorphism in exon 9, predicting Cys321Phe substitution,32 which was phased to the Cys237Tyr-bearing allele (data not shown).
Patient 14 was heterozygous for a novel mutation, a deletion of nt G in codon 285, which causes a frameshift, leads to a stop codon at 5 codons downstream, and predicts a truncated protein. Since the patient was of mixed Basque and non-Basque origin and other family members were unavailable for examination, it is uncertain whether the mutation originated in the Basque side of the family.
In patient 15, the only sequence alteration found was a substitution of the third nucleotide of intron 6 (IVS6 + 3A>G). Conceivably, this change affects the acceptor splice site of intron 6 because the calculated splice site consensus score (reflecting similarity of an observed splice site to the consensus sequence) decreased from 0.98 in the normal sequence to 0.56 in the altered sequence.34 Unfortunately, messenger RNA was not available from this patient to demonstrate alternative splicing. This sequence alteration is probably not a polymorphism since it was not found in 114 alleles of a mixed population surveyed for single-nucleotide polymorphisms of the factor XI gene.32
In patient 16, who was of mixed Basque and non-Basque origin, no candidate mutation was found. Collectively, 5 novel alterations in the factor XI gene were identified in 12 families with factor XI deficiency; 1 of these alterations, Cys38Arg, is common in French Basques.
Expression of mutated recombinant factor XI in BHK cells
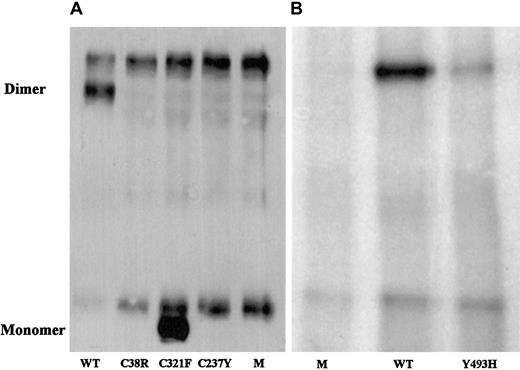
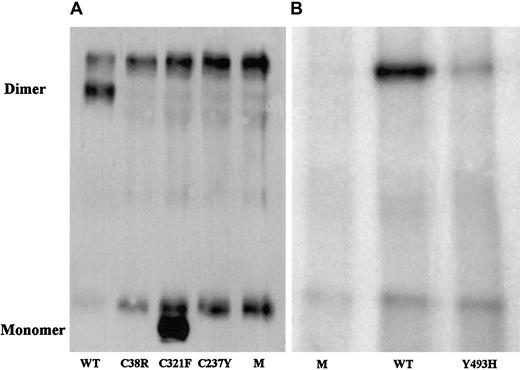
To evaluate the correlation of the identified candidate missense mutations with the factor XI phenotype, mutated and normal recombinant factor XI was expressed in BHK cells, and the secretion and intracellular contents of factor XI were assessed. In comparison with the medium of cells transfected with normal recombinant factor XI, the medium of cells transfected with factor XI Cys38Arg contained neither factor XI antigen, which was separately assayed by 3 monoclonal antibodies, nor factor XI activity (Table2). The medium of cells transfected with recombinant Cys237Tyr contained 3% of factor XI antigen secreted by normal cells but no demonstrable factor XI activity. The medium of cells transfected with Tyr493His contained approximately 12% of factor XI antigen and activity secreted by normal cells. Immunoprecipitation of the media from cells containing the 3 mutations and immunoblotting with a polyclonal antibody revealed no factor XI for Cys38Arg, a small amount for Tyr493His (Figure 1), and trace amounts for Cys237Tyr after long exposure (data not shown).
Western blot analysis of immunoprecipitated factor XI secreted by BHK cells transfected with mutant factor XI constructs.
Twenty-microliter samples of WT, mutants, and mock (M) were size-fractionated in 2 experiments (panels A and B) under nonreducing conditions on a 7.5% SDS-PAGE and reacted with polyclonal rabbit antihuman factor XI antibodies. Factor XI dimer was detected only in WT and Tyr493His samples, and factor XI monomer was observed only in the Cys321Phe mutant. A fast-migrating nonspecific band was observed in all samples, including the 2 mocks. A nonspecific slow-migrating band present in panel A most probably corresponds to mouse IgG, which is recognized by the secondary peroxidase-conjugated antibody. This band is absent from samples shown in panel B, probably because the secondary antibody used in this experiment was preadsorbed with mouse IgG, thereby eliminating cross-reactivity.
Western blot analysis of immunoprecipitated factor XI secreted by BHK cells transfected with mutant factor XI constructs.
Twenty-microliter samples of WT, mutants, and mock (M) were size-fractionated in 2 experiments (panels A and B) under nonreducing conditions on a 7.5% SDS-PAGE and reacted with polyclonal rabbit antihuman factor XI antibodies. Factor XI dimer was detected only in WT and Tyr493His samples, and factor XI monomer was observed only in the Cys321Phe mutant. A fast-migrating nonspecific band was observed in all samples, including the 2 mocks. A nonspecific slow-migrating band present in panel A most probably corresponds to mouse IgG, which is recognized by the secondary peroxidase-conjugated antibody. This band is absent from samples shown in panel B, probably because the secondary antibody used in this experiment was preadsorbed with mouse IgG, thereby eliminating cross-reactivity.
The Cys321Phe alteration did not contribute to factor XI deficiency; the recombinant Cys321Phe protein had activity similar to that of WT although it migrated as a monomer on nonreduced SDS-PAGE (Table 2; Figure 1).
Since no factor XI antigen was detected in the medium of BHK cells transfected with Cys38Arg and only trace amounts were present in the medium of cells transfected with Cys237Tyr factor XI, we addressed the question of whether the protein was normally produced but not secreted by the cells. As shown in Table 2, comparable amounts of factor XI antigen were observed in lysates of cells transfected with either WT, Cys38Arg, or Cys237Tyr factor XI. In contrast, measurements of Tyr493His cell lysates revealed only one third of the amount of factor XI produced by WT cells.
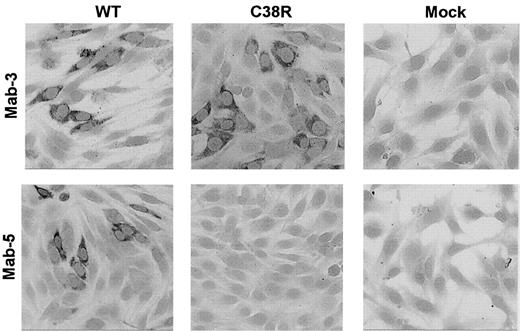
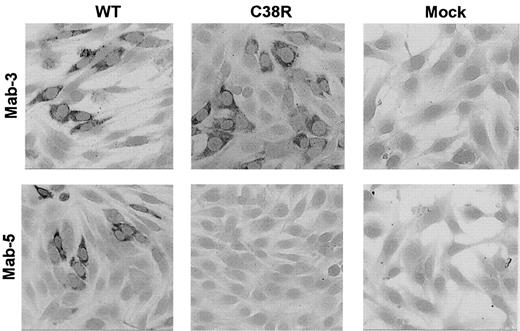
The presence of Cys38Arg factor XI in BHK transfected cells was evaluated by immunostaining of cells grown on glass slides with 2 different monoclonal antibodies. The mAb-3 directed against an epitope on apple 2 clearly detected Cys38Arg factor XI in the cells (Figure 2). In contrast, mAb-5 directed against an epitope on apple 1 did not recognize the Cys38Arg factor XI, suggesting that the mutation, which is localized in apple 1, probably interfered with recognition of the epitope. Both mAb-3 and mAb-5 stained Cys237Tyr factor XI in transfected cells, indicating that the production of the protein was not impaired by this mutation (data not shown).
Immunostaining of BHK cells transfected with WT or Cys38Arg factor XI.
Cells grown on glass slides were reacted with 2 different anti–factor XI monoclonal antibodies, and immune complexes were detected by antimouse IgG coupled to alkaline phosphatase. The mAb-3 directed against an epitope on apple 2 detected Cys38Arg factor XI in the cells, whereas the mAb-5 directed against an epitope on apple 1 (the site of the mutation) failed to detect Cys38Arg factor XI. Original magnification × 200.
Immunostaining of BHK cells transfected with WT or Cys38Arg factor XI.
Cells grown on glass slides were reacted with 2 different anti–factor XI monoclonal antibodies, and immune complexes were detected by antimouse IgG coupled to alkaline phosphatase. The mAb-3 directed against an epitope on apple 2 detected Cys38Arg factor XI in the cells, whereas the mAb-5 directed against an epitope on apple 1 (the site of the mutation) failed to detect Cys38Arg factor XI. Original magnification × 200.
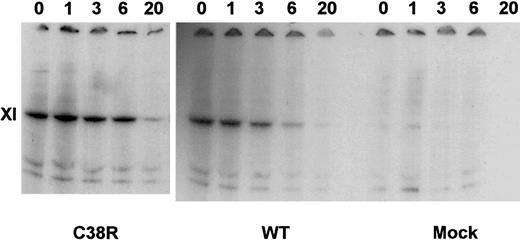
The intracellular degradation of factor XI in transfected BHK cells was assessed in pulse-chase experiments. Figure3 demonstrates that similar amounts of factor XI were produced by WT and Cys38Arg-transfected cells. Moreover, the preservation of the factor XI band of the Cys38Arg cells at 1 to 6 hours of chase indicated that there was no accelerated degradation of factor XI. Similar results were obtained in Cys237Tyr- and Tyr493His-transfected cells (data not shown).
Pulse-chase experiments of Cys38Arg and WT factor XI.
Transfected cells were radiolabeled by 35S-methionine and then chased by cold methionine for 1 to 20 hours. Cell lysates were precipitated by monoclonal antibodies against factor XI, electrophoresed on gradient 4% to 12% SDS-PAGE gels under reducing conditions, and exposed to autoradiography. Note that Cys38Arg was produced similarly to WT and did not undergo accelerated degradation.
Pulse-chase experiments of Cys38Arg and WT factor XI.
Transfected cells were radiolabeled by 35S-methionine and then chased by cold methionine for 1 to 20 hours. Cell lysates were precipitated by monoclonal antibodies against factor XI, electrophoresed on gradient 4% to 12% SDS-PAGE gels under reducing conditions, and exposed to autoradiography. Note that Cys38Arg was produced similarly to WT and did not undergo accelerated degradation.
Prevalence of Cys38Arg mutation in the French Basque population
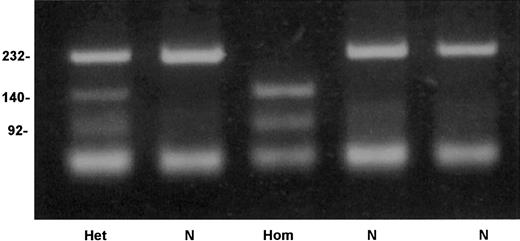
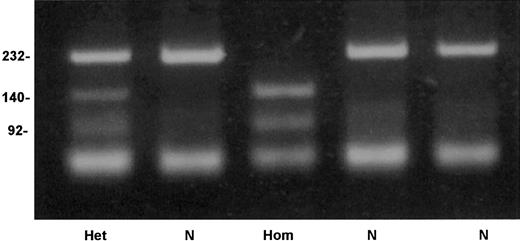
An allele-specific restriction analysis was devised for identification of the nt 209T>C substitution, which is responsible for the Cys38Arg mutation. The mutant allele 209C was cleaved by the enzymeTaiI (New England Biolabs), whereas the normal 209T allele was resistant to this enzyme (Figure 4). A survey for the mutation in 206 French Basque subjects detected 2 heterozygotes, predicting an allele frequency of 0.005 in this population (95% confidence interval, 0.0006-0.0174).
Detection of factor XI Cys38Arg mutation by allele-specific restriction analysis.
Depicted are a normal genotype (N), a homozygote for Cys38Arg (Hom), and a heterozygote (Het). Numbers on the left of the gel indicate the size in base pairs of the bands.
Detection of factor XI Cys38Arg mutation by allele-specific restriction analysis.
Depicted are a normal genotype (N), a homozygote for Cys38Arg (Hom), and a heterozygote (Het). Numbers on the left of the gel indicate the size in base pairs of the bands.
Identification of a possible founder haplotype for Cys38Arg
To discern whether the Cys38Arg substitution observed in 10 unrelated Basque individuals (8 patients and 2 subjects from the general Basque population) stemmed from a single mutational event (a common founder), we examined a series of 10 polymorphisms spanning the factor XI gene. This analysis enabled us to make a distinction between alleles bearing the mutation and those not bearing the mutation among carriers and controls, respectively. Table3 demonstrates that all subjects bearing the Cys38Arg mutation carried a unique intron B polymorphism (12 CA repeats), which was not detected in French Basque controls (P < .0001). Interestingly, the prevalence of this polymorphic allele was 2% in Ashkenazi Jews.25 The frequency of intron A −138A allele in carriers of Cys38Arg was also significantly different from controls (P = .002). For the remaining 8 polymorphisms, the frequency in controls and carriers of the Cys38Arg mutation was not statistically different. These data define a unique haplotype bearing the Cys38Arg mutation and suggest that the mutation occurred in a founder individual.
Discussion
The presented data established the molecular basis for factor XI deficiency in 11 of 12 unrelated families residing in the Basque area of France. In 8 Basque families, we identified a novel 209T>C nucleotide alteration, which predicts a Cys38Arg substitution. A survey among 206 French Basques revealed that the Cys38Arg allele frequency is 0.005. Haplotype analysis based on examination of 10 intragenic polymorphic sites in 10 carriers of the Cys38Arg mutation (only 1 being a homozygote) was consistent with a founder effect and indicated that the mutation probably occurred in an individual who had an haplotype that is extremely rare among Basques (Table 3).
The French Basques are thus the third population in which an ancestral mutation causing severe factor XI deficiency is prevalent. In previous studies, we identified the type II mutation (a stop in exon 5) in Ashkenazi Jews, Iraqi Jews, and Palestinian Arabs23,25,35and estimated that it occurred in a founder approximately 2500 years ago.36 The type II mutation was also reported in 2 Portuguese families, and haplotype analysis demonstrated that it was of the same ancestry.19 Type III missense mutation, also shown to have a distinct founder, was estimated to have evolved more recently and is confined to Ashkenazi Jews.25 36
Patients with severe factor XI deficiency are at significant risk of injury-related bleeding, particularly during operations involving tissues containing fibrinolytic activity such as the urinary tract, oral cavity, and nasal mucosa.35 37 In view of the high prevalence of factor XI deficiency in Israel, it has become mandatory to screen for factor XI deficiency prior to surgery by testing the aPTT. The prevalence of Cys38Arg heterozygotes is 1:100 in French Basques, which predicts a prevalence of severe factor XI deficiency of 1:40 000. From the limited clinical information presented in Table 1, it is difficult to determine whether homozygotes or heterozygotes for the Cys38Arg mutation are at significant risk of bleeding; 1 homozygote had no bleeding following breast surgery and 2 heterozygotes had mild bleeding following cervical disc surgery and delivery, respectively.
In 2 of the 3 mutations described in this study, highly conserved cysteine residues in the apple domain region (heavy chain) were substituted. The cysteine residues in the 4 apple domains are characteristic for the PAN module superfamily, which includes prekallikrein, a number of Caenorhabditis elegans proteins, and the N-terminal domains of the plasminogen/hepatocyte growth factor family.38 The Cys38Arg mutation disrupts the C32-C38 bond in apple domain 1, and the Cys237Tyr mutation disrupts the C208-C237 bond in apple domain 3. Our expression studies showed that although both mutant proteins were synthesized in BHK transfected cells, they were not secreted (Table 2, Figures 1-2) and also did not rapidly degrade intracellularly (Figure 3). Interestingly, in 6 of 7 previously described missense mutations, all localized in the apple domains, expression studies also demonstrated significantly impaired secretion.20 21 For the type III mutation (Phe283Leu), the impaired secretion was attributed to defective factor XI dimerization, but for the remaining mutations the mechanism has not been established.
In the 2 siblings who carried the Cys237Tyr mutation, a rare Cys321Phe polymorphism was also detected. The unpaired C321 was previously shown to play a role in stabilization of factor XI dimers by formation of an intermonomer disulfide bond.14 However, a Cys321Ser substitution did not impair factor XI dimer formation, secretion, or activity.14 In this study, we expressed the naturally occurring Cys321Phe polymorphism in BHK cells and also found normal factor XI secretion and specific activity (Table 2); however, on nonreducing denaturing SDS-PAGE, the protein appeared as a monomer rather than dimer (Figure 1). Interestingly, rabbit factor XI contains a histidine instead of cysteine at the equivalent position and displays similar features: a monomer on SDS-PAGE, but a dimer by size-exclusion chromatography.39
The third Tyr493His mutation is located in the catalytic domain of factor XI. The tyrosine residue is conserved in human, bovine, and rabbit factor XI and human prekallikrein. Interestingly, in mouse factor XI, this residue is replaced by histidine similarly to the mutation detected. The Tyr493His mutation caused a 90% reduction of factor XI measured in the medium of transfected BHK cells. This reduced amount probably stems from decreased synthesis and secretion of factor XI since (1) the amount of factor XI in cell lysates was approximately a third of the amount in WT cells (Table 2); (2) there was no accelerated intracellular degradation; and (3) the secreted amount of factor XI was disproportionally low (Table 2).
The Basque population is one of the oldest populations in Europe.40 Several unique genetic features have been observed in Basques. Examples are the following: (1) The Rh-negative blood group was found in about 20% of Basques; this frequency surpasses the frequency of most populations.40 (2) The frequency of delta 508 mutation among cystic fibrosis–bearing alleles in Basques is 87%, which by far exceeds the 53% frequency observed in Spaniards.41,42 (3) Factor V Leiden, which is confined to whites, appears to be absent in Basques.43 The present study identified a new characteristic genetic feature: the Cys38Arg mutation in the factor XI gene among French Basques. However, since a recent study demonstrated significant differences in genetic markers among autochthonous Basque subgroups,44 it remains to be established whether the Cys38Arg mutation is evenly distributed among all Basque subgroups.
We thank Nurit Kornbrot, Aviya Ifrah, and Sali Usher for expert technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Uri Seligsohn, Institute of Thrombosis and Hemostasis, The Chaim Sheba Medical Center, Tel-Hashomer 52621, Israel; e-mail: seligson@sheba.health.gov.il.