Abstract
Graft rejection or graft-versus-host (GVH) disease after HLA-identical stem cell transplantation is the result of recognition of minor histocompatibility antigens (mHags) by immunocompetent T lymphocytes from recipient or donor origin, respectively. Cytolytic T lymphocyte (CTL) clones can be isolated during graft rejection and GVH disease to identify mHags and their corresponding genes. Thus far, all human mHags identified appeared to be HLA class I–restricted. Here, we report the characterization of the first human HLA class II–restricted sex-linked mHag involved in GVH disease. Previously, we isolated an HLA-DQ5–restricted CD4+ CTL clone from a male patient with chronic myeloid leukemia who developed acute GVH disease grade III-IV after transplantation of HLA genotypically identical female stem cells. Using a panel of female HLA-DQ5+ EBV cells that we stably transfected with Y chromosome–specific genes, we determined that the HLA class II male-specific mHag (H-Y) was encoded by the Y chromosome–specific gene DBY. The H-Y epitope was localized in the DBY protein using female HLA-DQ5+peripheral blood mononuclear cells loaded with DBY protein fragments. The minimal peptide sequence leading to maximal recognition by the specific HLA-DQ5–restricted CTL clone was characterized as the 12–amino acid sequence HIENFSDIDMGE. Although the epitope differed by 3 amino acids from its X-homolog DBX, only 2 polymorphisms were shown to be essential for recognition by the CTL clone.
Introduction
HLA-identical stem cell transplantation can be complicated by graft-versus-host (GVH) disease or graft rejection. Both complications are initiated by T lymphocytes that recognize minor histocompatibility antigens (mHags) presented by HLA molecules.1-3 These mHags are peptides derived from polymorphic proteins that differ between donor and recipient.4 T-lymphocyte clones recognizing mHags can be isolated from patients who developed GVH disease or graft rejection.2,5-7 A number of mHags have been identified. The HLA-A2–restricted mHag HA-1 was found to be a nonapeptide derived from the KIAA0223 gene, and HA-2 originated from a gene that was a member of the nonfilament-forming class I myosin family.8,9 Both mHags were exclusively expressed on hematopoietic cells. The immunogenicity of mHag HA-8 resulted from differential antigen processing due to a polymorphic residue at the N-terminus of the antigen.10 HB-1 was an HLA-B44–restricted mHag specific for B-cell acute lymphoblastic leukemia.11 These identified mHags were derived from polymorphic proteins that contained one amino acid difference compared with their homologous allelic counterparts.
Male-specific mHags were shown to be involved in HLA-identical sex-mismatched stem cell transplantation.12,13SMCY was the first gene identified encoding human HLA-B7– and HLA-A2–restricted H-Y mHags.14,15 A systematic search of the nonrecombining region of the human Y chromosome identified other ubiquitously expressed genes that were potential candidates to encode H-Y antigens.16 Recently, the DFFRY andUTY genes were identified as genes coding for human major histocompatibility complex (MHC) class I–restricted H-Y antigens, demonstrating that human H-Y antigens are encoded by multiple Y-specific genes.17-20 Amino acid differences between the male-specific mHags and their X-homologs varied from 1 to 4 amino acids.
All human mHags identified to date are MHC class I–restricted and are recognized by cytolytic T lymphocytes (CTLs). However, efficient in vivo priming of these CTLs during graft rejection or GVH disease generally requires the participation of CD4+ T-helper lymphocytes, which recognize epitopes in MHC class II molecules.21,22 This help for CTLs may either be mediated by cytokine production by T-helper lymphocytes or by activation of the antigen-presenting cell via CD40-CD40L interaction.23Furthermore, the involvement of CD4+ T lymphocytes in graft rejection or GVH disease is not solely restricted to provide help to CTLs. In MHC class I–deficient mice, CD4+ CTLs induced and maintained GVH disease after allogeneic mHag-mismatched stem cell transplantation.24 Moreover, transgenic female mice that expressed the T-cell receptor (TCR) from a MHC class II–restricted H-Y–specific T-cell clone rapidly rejected male skin grafts in a CD4-dependent fashion.25 Recently, it was demonstrated that this T-cell clone recognized an epitope derived from the murine DBY protein.26
In this study, we report the identification of the first human MHC class II–restricted male-specific mHag. The mHag was recognized by a CD4+ CTL clone that was isolated from a male patient who developed acute GVH disease grade III-IV after transplantation of HLA genotypically identical female stem cells.27 The H-Y antigen presented by the HLA-DQ5 molecule was encoded by theDBY gene. The minimal peptide sequence for maximal recognition was characterized as the 12-residue peptide HIENFSDIDMGE.
Materials and methods
CTLs and cell lines
The H-Y–specific HLA-DQ5–restricted CD4+ CTL clone HLA-DQ5 HY CTL was derived by limiting dilution from peripheral blood mononuclear cells (PBMCs) of a male patient with chronic myeloid leukemia who developed acute GVH disease grade III-IV after transplantation of HLA genotypically identical female stem cells.27 HLA-DQ5 HY CTL was cultured by stimulation with irradiated allogeneic PBMCs and patient-derived Epstein-Barr virus–transformed B (EBV) cells in Iscoves modified Dulbecco medium (IMDM) (BioWhittaker, Verviers, Belgium) containing 10% human serum, 3 mM l-glutamine, and 300 U/mL recombinant interleukin-2 (Roussel Uclaf, Paris, France). EBV cells were maintained in RPMI 1640 medium (BioWhittaker) containing 10% fetal bovine serum (BioWhittaker).
Cloning of Y-specific genes
Of the 30 known Y chromosome–specific genes, 8 have been shown to be ubiquitously expressed.16 Of these 8 ubiquitously expressed genes, DBY, EIF-1AY, RPS4Y, and TB4Ywere first analyzed. Total RNA was isolated from male EBV cells with Trizol (Gibco, Gaithersburg, MD) according to the manufacturer's procedure. Complementary DNA (cDNA) was prepared from RNA using Moloney murine leukemia virus BRL reverse transcriptase (Gibco BRL, Gaithersburg, MD) for 60 minutes at 37°C as described.17One fiftieth of the cDNA reaction was amplified using specific primers for DBY, EIF-1AY, RPS4Y, and TB4Y. All genes were amplified using the expanded long template polymerase chain reaction (PCR) system (Boehringer Mannheim, Mannheim, Germany). The amplification was started with a denaturation step of 2 minutes at 92°C, followed by 30 cycles, with each cycle consisting of 20 seconds at 92°C, 1 minute at 60°C, and 1 minute at 68°C. After amplification, each Y-specific cDNA was visualized on an ethidium bromide–stained low melt agarose gel (Sigma Chemical, St Louis, MO). The cDNA was isolated from the low melt agarose gel using agarase (Boehringer Mannheim) according to the manufacturer's procedure.
Retroviral transduction of female HLA-DQ5+ EBV cells
The Y-specific cDNAs were cloned into a retroviral vector. The Moloney murine leukemia virus–based retroviral vector LZRS and packaging cells φ-NX-A used for this purpose were kindly provided by G. Nolan.28 A bicistronic retroviral vector was constructed in which the multiple cloning site is linked to the downstream internal ribosome entry sequence and the marker gene green fluorescent protein (GFP).29 After cloning of the Y-specific cDNAs in this vector, the constructs were transfected into φ-NX-A cells using calcium phosphate (Life Technologies, Gaithersburg, MD). After 2 days at 37°C, 2 μg/mL puromycin (Clontech Laboratories, Palo Alto, CA) was added to the cells. Ten to 14 days after transfection, 6 × 106 cells were plated per 10-cm Petri dish (Becton Dickinson, San Jose, CA) in 10 mL IMDM supplemented with 10% fetal calf serum without puromycin. The next day the medium was refreshed, and on the following day retroviral supernatant was harvested, centrifuged, and frozen in aliquots at −70°C. The Y-specific cDNAs were transduced into female HLA-DQ5+ EBV cells using the method developed by Hanenberg et al.30 Briefly, 5 × 106 EBV cells were cultured on recombinant human fibronectin fragments CH-296 (RetroNectin, Takara, Otzu, Japan) coated Petri dishes together with 1 mL thawed retroviral supernatant for 18 hours at 37°C, washed, and transferred to 24-well culture plates. After 3 to 5 days, the transduction efficiency, measured by the expression of the GFP marker, was analyzed by flow cytometry. Then, Y gene–transduced female EBV cells were FACS-sorted based on their GFP positivity.
Recognition assay of Y gene–transduced EBV cells
51Cr-release assays were used to determine lysis of target cells by the HLA-DQ5 HY CTL. Male, female, or Y gene–transduced female EBV target cells were labeled with 100 μCi (3.7 MBq) Na51CrO4. After 1 hour at 37°C, the cells were washed 3 times with RPMI supplemented with 2% fetal calf serum. Na51CrO4-labeled EBV cells were plated in a 96-well V-bottom microtiter plate in 100 μL IMDM plus 10% human serum at 2000 cells per well. In cold target inhibition assays, 20 000 Y gene–transduced EBV cells were added to the Na51CrO4-labeled male EBV. Then, 20 000 HLA-DQ5 HY CTLs were added in 100 μL IMDM plus 10% human serum.51Cr release was measured after incubation at 37°C for 4 hours.
DBY protein synthesis and CTL recognition assay
DBY proteins were synthesized using the pCRT7 TOPO TA cloning kit according to the manufacturer's procedure (Invitrogen, Carlsbad, CA). The DBY PCR product was ligated into the pCRT7/NT-TOPO vector. DBY fragments were obtained by amplification of the cloned DBY construct with specific primers annealing at internal DBY sequences. The overlapping DBY PCR products were also ligated into the pCRT7/NT-TOPO vector. The cloning reactions were transformed into chemically competent TOP10F' Escherichia coli cells, which were spread on Luria-Bertani (LB) plates containing 100 μg/mL ampicillin. Then, colonies were selected and analyzed for insert and correct orientation. DBY-pCRT7/NT-TOPO constructs were transformed into chemically competent BL21(DE3)pLysS cells, which were spread on LB plates containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Three colonies of each construct were separately added to 10 mL complete LB medium containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol and grown for 6 hours at 37°C. After overnight incubation at 4°C, the cultures of each construct were collected and added to 750 mL complete LB medium. After incubation for 2 hours at 37°C, expression of the DBY proteins was induced by adding isopropyl β-D-thiogalactoside to the culture medium at a final concentration of 0.5 mM. After incubation for 4 hours at 37°C, cultures were centrifuged for 10 minutes at 3000g. Then, inclusion bodies were isolated from the cell pellets using B-PER bacterial protein extraction reagent according to the manufacturer's procedure (Pierce, Rockford, IL). The isolated inclusion bodies that contained the DBY proteins were dissolved in a buffer containing 20 mM Tris-HCl, 100 mM NaCl, 50 mM imidazole, and 8 M ureum. Correct expression of the DBY constructs was evaluated by loading samples on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and staining with Coomassie blue (Bio-Rad Laboratories, Hercules, CA).
Irradiated (20 Gy) HLA-DQ5+ PBMCs were plated at 50 000 cells per well in a 96-well flat-bottom microtiter plate in 100 μL IMDM plus 10% human serum. DBY proteins were added to the cells at a final concentration of 10 μg/mL. After incubation of 24 hours at 37°C, 10 000 HLA-DQ5 HY CTLs were added in 100 μL IMDM plus 10% human serum supplemented with 120 U/mL interleukin-2. After 16 hours at 37°C, supernatant was harvested and the amount of interferon γ (IFN-γ) secreted into the supernatant was measured using a standard enzyme-linked immunosorbent assay (CLB, Amsterdam, The Netherlands) according to the manufacturer's procedure.
Peptide synthesis and peptide recognition assays
Peptides were synthesized by solid-phase strategies on an automated multiple peptide synthesizer (SyroII, MultiSynTech, Witten, Germany) and characterized by mass spectrometry. The purity of the peptides was determined by analytical reversed-phase high-performance liquid chromatography and proved to be at least 80%. Irradiated (50 Gy) HLA-DQ5+ female EBV cells were plated at 50 000 cells per well in a 96-well flat-bottom microtiter plate in 200 μL acidified RPMI 1640, pH 5.0, containing various concentrations of peptides. After 2 hours of incubation at 37°C, the cells were washed 3 times with RPMI 1640. Then, 25 000 HLA-DQ5 HY CTLs were added in 200 μL IMDM plus 10% human serum supplemented with 120 U/mL interleukin-2. After 16 hours at 37°C, supernatant was harvested and IFN-γ content was measured.
DBY real-time quantitative reverse transcriptase–PCR analysis
PCR was performed using the ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems, Foster City, CA). Double dye fluorogenic probes (Eurogentec, Seraing, Belgium) were designed using Primer Express software (PE Applied Biosystems). The DBY probe was labeled at the 3′ end with the quencher dye TAMRA (6-carboxy-tetramethylrhodamine) and at the 5′ end with the reporter dye TET (4,7,2′,7′-tetrachloro-6-carboxyfluorescein). The forward DBY primer was located in exon 2 (5′-AAC TGG ACC AGC AGC TTG CTA AT-3′) and the reverse primer in exon 3 (5′-TTC ACT GAA ATA ACC AGG CTT TCC T-3′), generating a PCR product of 150 base pairs. The double dye probe was chosen between the forward and reverse primers (DBY 5′-(FAM)-CCT GAA CTG TCT TTA TCA TGG AAT CCT TTA GAT GCT-(TAMRA)-3′).
For amplification, the qPCR core kit was used according to the manufacturer's procedure (Eurogentec). PCR amplifications were performed in duplicate experiments in 96-well 0.2-mL reaction plates (Thermofast 96 AB0600, ABgene, Epsom, United Kingdom), sealed with Clear Strong Seal (ABgene), and run on an ABI PRISM 7700 (PE Applied Biosystems). The PCR was started after activating the Hot GoldStar polymerase for 10 minutes at 95°C, followed by 55 cycles consisting of 15 seconds at 95°C, 30 seconds at 60°C, and elongation for 30 seconds at 60°C. A real-time fluorescence plot was obtained based upon normalized fluorescence signals. The threshold cycle was determined, ie, the fractional cycle number at which the amount of amplified target reached a fixed threshold. This threshold was defined as 10 times the SD of the baseline fluorescent signal, ie, the normalized fluorescence signal of PCR cycles 3 to 15. As a negative control, milliQ water was used. DBY messenger RNA expression was analyzed in human multiple tissue culture (MTC) panel I containing first-strand cDNA generated from the human tissues of heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas and human MTC panel II containing cDNA from spleen, thymus, prostate, testis, ovary, small intestine, colon, and peripheral blood leukocytes (Clontech Laboratories). Skin tissue was derived from a male individual, total RNA was isolated with Trizol (Gibco) according to the manufacturer's procedure, and cDNA was prepared from RNA using Moloney murine leukemia virus BRL reverse transcriptase (Gibco) for 60 minutes at 37°C.
Results
Transduction of female HLA-DQ5+ EBV cells with Y genes
Female HLA-DQ5+ EBV cells were transduced with theDBY, EIF-1AY, RPS4Y, and TB4Y genes using the retroviral vector LZRS that contained the marker gene GFP. Transduction efficiency as measured by fluorescence-activated cell sorter (FACS) analysis was 20% (data not shown). Y gene–transduced female EBV cells were FACS sorted based on their GFP positivity, resulting in a 99% GFP+ cell population (data not shown).
Identification of the gene encoding the HLA-DQ5–restricted H-Y T-cell epitope
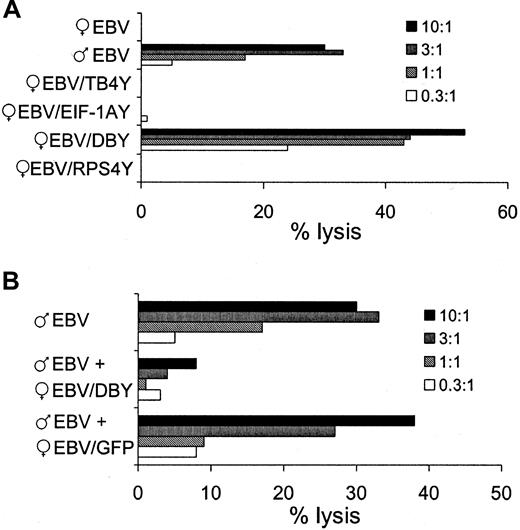
To determine whether one of the Y genes coded for the HLA-DQ5–restricted H-Y T-cell epitope, the Y gene–transduced EBV cells were tested for recognition by the specific CTL clone in a51Cr-release assay. As shown in Figure1A, HLA-DQ5+ female EBV cells transduced with the DBY gene (EBV/DBY) were recognized by the HLA-DQ5 HY CTL clone. The lysis was comparable to HLA-DQ5+ male EBV cells. To analyze whether the epitope recognized on EBV/DBY was identical to the H-Y epitope presented by male EBV cells, we measured the CTL-induced lysis of 51Cr-labeled male EBV cells after addition of a 10-fold excess of unlabeled female EBV/DBY cells. As demonstrated in Figure 1B, lysis of male EBV cells could be blocked by EBV/DBY, whereas unlabeled EBV cells transduced with the control vector containing the GFP did not affect the CTL-induced lysis of male EBV cells. This illustrated that the TCR from the CTLs binds to HLA/epitope complexes expressed on both female EBV/DBY cells as well as male EBV cells.
Specific lysis of DBY-transduced female EBV target cells by the HLA-DQ5 HY CTL.
Target cells were added to the HLA-DQ5 HY CTL at indicated effector-target ratios, and 51Cr release was measured after 4 hours. (A) Lysis of male EBV and DBY-transduced female EBV cells by the HLA-DQ5 HY CTL. (B) Lysis of male EBV cells could be blocked by a 10-fold excess of unlabeled DBY-transduced female EBV cells, whereas unlabeled EBV cells transduced with the control vector containing the GFP did not affect the CTL-induced lysis of male EBV cells.
Specific lysis of DBY-transduced female EBV target cells by the HLA-DQ5 HY CTL.
Target cells were added to the HLA-DQ5 HY CTL at indicated effector-target ratios, and 51Cr release was measured after 4 hours. (A) Lysis of male EBV and DBY-transduced female EBV cells by the HLA-DQ5 HY CTL. (B) Lysis of male EBV cells could be blocked by a 10-fold excess of unlabeled DBY-transduced female EBV cells, whereas unlabeled EBV cells transduced with the control vector containing the GFP did not affect the CTL-induced lysis of male EBV cells.
Localization of the HLA-DQ–restricted H-Y epitope in the DBY gene
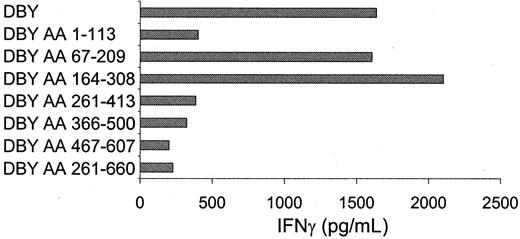
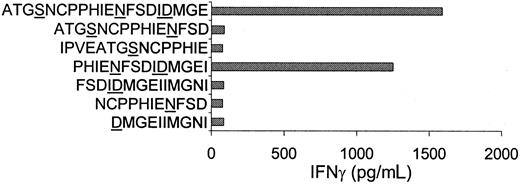
Because no HLA-DQ5–binding motifs were available, the DBY protein could not be screened for HLA-DQ5–binding peptides. Therefore, DBY protein fragments were used to localize the H-Y epitope in the DBY protein. To analyze whether the DBY-derived H-Y epitope could be exogenously processed by antigen presenting cells, the DBY protein was synthesized and added to HLA-DQ5+ female PBMCs. As shown in Figure 2, the HLA-DQ5 HY CTL clone released high amounts of IFN-γ when stimulated with PBMCs loaded with the DBY protein. Overlapping DBY protein fragments were synthesized and added to PBMCs. Two overlapping DBY protein fragments were recognized by the HLA-DQ5 HY CTL, illustrating that the H-Y epitope was situated on the DBY protein between amino acid 164 and 209 (Figure 2). This region contained only 4 amino acids that differed from the DBX-homolog protein. Peptides spanning this region were synthesized. Two peptides containing the common amino acids PHIENFSDIDMGE were recognized by the HLA-DQ5 HY CTL (Figure 3).
Localization of the H-Y epitope on the DBY protein between amino acids 164 and 209.
DBY protein and the DBY protein fragments indicated were added to irradiated HLA-DQ5+ female PBMCs. After incubation of 24 hours at 37°C, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. Protein fragments containing residues at positions 67 to 209 and 164 to 308 in the DBY protein induced IFN-γ release by the HLA-DQ5 HY CTL.
Localization of the H-Y epitope on the DBY protein between amino acids 164 and 209.
DBY protein and the DBY protein fragments indicated were added to irradiated HLA-DQ5+ female PBMCs. After incubation of 24 hours at 37°C, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. Protein fragments containing residues at positions 67 to 209 and 164 to 308 in the DBY protein induced IFN-γ release by the HLA-DQ5 HY CTL.
HLA-DQ5 HY CTL recognized DBY amino acid sequence PHIENFSDIDMGE.
The DBY peptides indicated were added to irradiated HLA-DQ5+ female PBMCs. After incubation of 24 hours at 37°C, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. Two peptides containing the common amino acids PHIENFSDIDMGE were recognized by the HLA-DQ5 HY CTL.
HLA-DQ5 HY CTL recognized DBY amino acid sequence PHIENFSDIDMGE.
The DBY peptides indicated were added to irradiated HLA-DQ5+ female PBMCs. After incubation of 24 hours at 37°C, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. Two peptides containing the common amino acids PHIENFSDIDMGE were recognized by the HLA-DQ5 HY CTL.
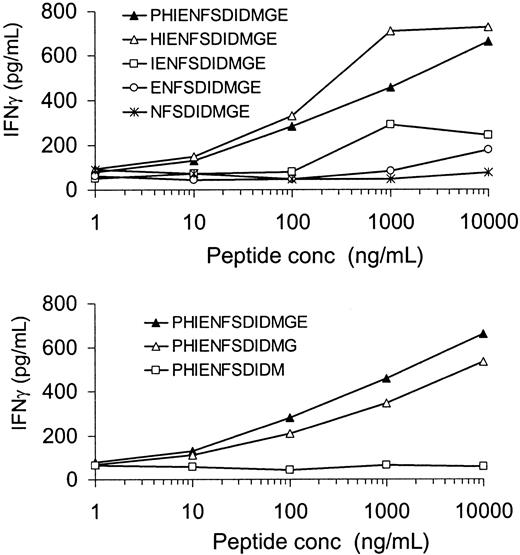
Identification of minimal epitope recognized by HLA-DQ5 HY CTL
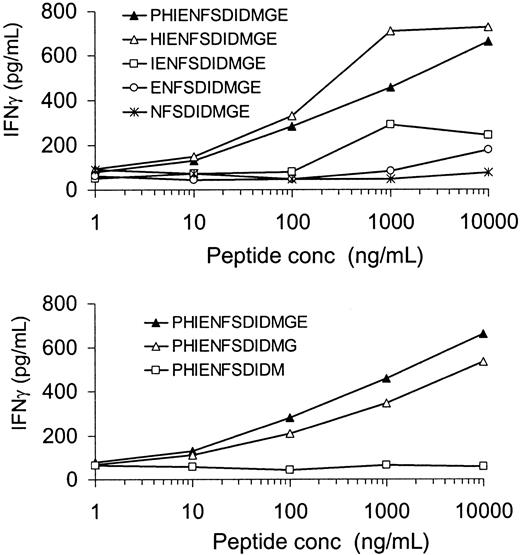
The peptide PHIENFSDIDMGE was loaded on female HLA-DQ5+ EBV cells and tested for recognition by the HLA-DQ5 HY CTL. As shown in Figure 4, PHIENFSDIDMGE induced IFN-γ production by the HLA-DQ5 HY CTL in a dose-dependent manner. N- and C-terminal–trimmed variants of this peptide were used to characterize the minimal H-Y epitope (Figure 4). Recognition considerably decreased after removal of the histidine residue on position 2, and further removal of N-terminal amino acids resulted in a further reduction of IFN-γ release by HLA-DQ5 HY CTL. Trimming of the C-terminal glutamic acid residue led to a slightly diminished recognition, while removal of the glycine residue completely abolished IFN-γ release by HLA-DQ5 HY CTL. These results indicated that the minimal peptide leading to recognition by the HLA-DQ5 HY CTL clone was the 10–amino acid sequence IENFSDIDMG, while the minimal peptide for maximal recognition was the 12-residue peptide HIENFSDIDMGE.
Characterization of the 12-mer peptide HIENFSDIDMGE as the minimal peptide sequence leading to maximal recognition by the HLA-DQ5 HY CTL.
Irradiated HLA-DQ5 female EBV cells were incubated with various concentrations (conc) of the indicated peptides for 2 hours. After washing, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. Recognition considerably decreased after removal of the histidine residue on position 2, and further removal of N-terminal amino acids resulted in a further reduction of IFN-γ release by HLA-DQ5 HY CTL. Trimming of the C-terminal glutamic acid residue led to a slightly diminished recognition, while removal of the glycine residue completely abolished IFN-γ release by HLA-DQ5 HY CTL.
Characterization of the 12-mer peptide HIENFSDIDMGE as the minimal peptide sequence leading to maximal recognition by the HLA-DQ5 HY CTL.
Irradiated HLA-DQ5 female EBV cells were incubated with various concentrations (conc) of the indicated peptides for 2 hours. After washing, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. Recognition considerably decreased after removal of the histidine residue on position 2, and further removal of N-terminal amino acids resulted in a further reduction of IFN-γ release by HLA-DQ5 HY CTL. Trimming of the C-terminal glutamic acid residue led to a slightly diminished recognition, while removal of the glycine residue completely abolished IFN-γ release by HLA-DQ5 HY CTL.
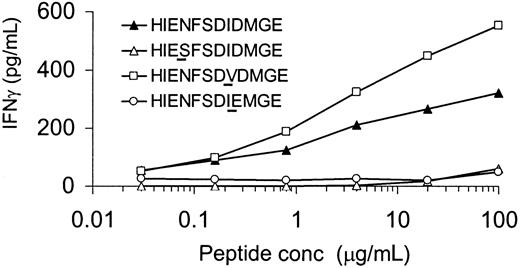
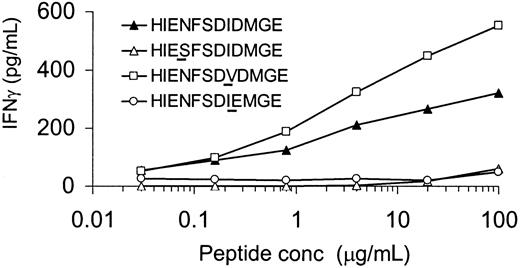
Two polymorphisms in H-Y epitope are essential for T-cell recognition
The H-Y epitope HIENFSDIDMGE differed by 3 amino acids from its X-homolog HIESFSDVEMGE. To determine whether all 3 polymorphisms were equally important for recognition by the HLA-DQ5 HY CTL, 3 peptides each containing 1 DBX-homolog amino acid at positions P4, P8, or P9 were tested. As shown in Figure 5, IFN-γ release by HLA-DQ5 HY CTL was completely abrogated when the amino acid at P4 or P9 was substituted by the X-homolog residue. In contrast, substitution of the isoleucin residue at P8 by the DBX-derived valine resulted in an even enhanced cytokine release by the specific CTL clone. The X-homolog peptide HIESFSDVEMGE and DBY-derived peptides containing 2 X-homolog amino acids were not recognized by HLA-DQ5 HY CTL (data not shown).
Determination of residues at positions P4 and P9 of HIENFSDIDMGE as critical residues for HLA-DQ5 HY CTL recognition.
Irradiated HLA-DQ5 female EBV cells were incubated with various concentrations of the indicated peptides for 2 hours. After washing, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. IFN-γ release by HLA-DQ5 HY CTL was completely abrogated when the amino acid at P4 or P9 was substituted by the X-homolog residue. In contrast, substitution of the isoleucin residue at P8 by the DBX-derived valine resulted in an even enhanced cytokine release by the specific CTL clone.
Determination of residues at positions P4 and P9 of HIENFSDIDMGE as critical residues for HLA-DQ5 HY CTL recognition.
Irradiated HLA-DQ5 female EBV cells were incubated with various concentrations of the indicated peptides for 2 hours. After washing, HLA-DQ5 HY CTLs were added; 16 hours later, supernatant was harvested and IFN-γ content was measured. IFN-γ release by HLA-DQ5 HY CTL was completely abrogated when the amino acid at P4 or P9 was substituted by the X-homolog residue. In contrast, substitution of the isoleucin residue at P8 by the DBX-derived valine resulted in an even enhanced cytokine release by the specific CTL clone.
Ubiquitous expression of the DBY gene
To determine whether DBY was expressed in multiple tissues, we performed real-time quantitative reverse transcriptase–PCR analysis on human MTC panels containing first-strand cDNA derived from multiple tissues. The expression level of DBY was similar in all tissues tested with the exception of ovary tissue (data not shown).
Discussion
T-lymphocyte clones isolated from patients who developed graft rejection or GVH disease after HLA-identical stem cell transplantation can be used to identify mHags and their corresponding genes. This led to the isolation of a number of MHC class I–restricted human mHags.8-11 These mHags were derived from polymorphic genes, which encoded proteins that contained one amino acid difference compared with the proteins derived from their homologous allelic counterparts. CTL clones recognizing male-specific mHags were mainly isolated during graft rejection. Male-specific mHags or H-Y epitopes were shown to be encoded by the ubiquitously expressed Y-chromosome–specific genes SMCY, DFFRY, andUTY.14,15,17-20 The epitopes presented in different MHC class I molecules contained 1 to 4 amino acid differences compared with their X-homologs. Previously, we isolated an H-Y–specific MHC class II–restricted CD4+ CTL clone during severe GVH disease.27 In this paper, we report the identification of DBY as the gene encoding the HLA-DQ5–restricted H-Y epitope. DBY belongs to a well-conserved family of genes coding for RNA helicases. This family of proteins shares a group of conserved motifs, including the sequence Asp-Glu-Ala-Asp or the so-called DEAD motif. These proteins are involved in diverse cellular functions, including splicing, ribosomal assembly, and translation.
The DBY-derived H-Y epitope leading to maximal recognition by the HLA-DQ5 HY CTL clone was characterized as the 12–amino acid sequence HIENFSDIDMGE. The epitope contained 3 different amino acids—at positions P4, P8, and P9—compared with the X-homolog sequence. The residues at positions P4 and P9 were shown to be critical residues for CTL recognition. Substitution of the asparagine residue at P4 led to a complete abrogation of CTL recognition even at high peptide concentrations. The terminal amide group of the asparagine residue that is absent from the DBX-derived serine residue may interact with the TCR, and abolition of this interaction may result in the dramatic loss of CTL recognition. Substitution of the aspartic acid residue at position P9 by the DBX-derived glutamic acid residue also led to a complete abrogation of cytokine production by the CTL clone. Both residues contain 2 nearly identical acidic side chains. The larger glutamic acid residue contained an additional methylene group that may disturb the 3-dimensional structure of the HLA-DQ5, H-Y epitope, and TCR complex. This finding illustrates that TCRs have high specificities for their epitopes and that small variations at critical sites in these epitopes are readily detected. Alternatively, residue substitutions may negatively affect peptide binding, resulting in decreased recognition by the CTL clone.
There are no HLA-DQ5–binding motifs known, and it is therefore difficult to point out the anchor residues in this epitope. However, a peptide binding motif for HLA-DQ6 has been characterized by determining the influence of amino acid sustitutions on HLA-DQ6 binding of a peptide derived from insulin B.31 The α chain of HLA-DQ6 is identical to HLA-DQ5, and only the β chain is different. The minimal insulin B–derived peptide for binding to HLA-DQ6 contained 9 residues, while the minimal peptide for maximal binding contained 11 residues. Single arginine substitutions at each position revealed that positions P1, P3, P4, P6, and P9 of the 9 residue–long peptide were important for binding. Amino acids P6 and P9 exhibited the highest specificity. A similar binding motif was present in the DBY-derived peptide HIENFSDIDMGE, in which the amino acids at positions P3, P5, P6, P8, and P11 resembled the anchor residues in the insulin B peptide and/or were well tolerated in amino acid substitution assays. The observation that removal of the glycine residue at position P11 of the H-Y epitope resulted in a complete abolition of recognition by the HLA-DQ5 HY CTL, even at high peptide concentrations, may point to an important residue for binding to the HLA-DQ5 molecule. Substitution of the isoleucin residue at position P8 by a valine residue did not cause reduction of cytokine release by the specific CTL clone, indicating that this residue may be imbedded in the HLA-DQ5–binding pocket rather than interact with the TCR. Although the N-terminal histidine residue may lie outside the MHC anchor residues, recognition by the HLA-DQ5 HY CTL clone was significantly decreased after removal of this residue. It has been demonstrated that most MHC class II–restricted T cells derived from hen egg lysozyme–immunized mice were completely dependent on, and specific for, the C-terminal peptide-flanking residues, lying outside MHC anchor residues.32 Thus, peptide-flanking residues of naturally processed MHC class II–bound peptides can be recognized by TCRs and markedly influence immunogenicity of the peptide.
The identification of DBY as a gene encoding a MHC class II–restricted H-Y epitope demonstrates that multiple male antigens complexed with MHC class I or class II molecules derived from different Y-specific genes are involved in GVH disease and graft rejection. We demonstrated that DBY was expressed at similar levels in all tissues tested, including GVH disease target organs. BecauseDBY is ubiquitously expressed, it may be the expression of the HLA-DQ5 molecule that determines whether or not tissues become GVH disease target organs. These cells may process the DBY protein and may present the DBY-derived epitope in HLA-DQ5, resulting in activation of male-specific T lymphocytes.
The role of different H-Y antigens in GVH disease needs to be further evaluated. HLA class I–restricted H-Y–specific T cells present in male patients during GVH disease may be monitored by using HLA/HY peptide tetrameric complexes. Tetrameric complexes of HLA-A2 or HLA-B7 with SMCY-derived H-Y epitopes were used to monitor H-Y–specific T cells during GVH disease of male patients transplanted with female stem cells.33 A significant increase of H-Y–specific CTLs during acute and chronic GVH disease could be visualized. The involvement of MHC class II–restricted H-Y–specific CD4+T lymphocytes during GVH disease may also be monitored if tetrameric complexes with sufficient binding affinity to the TCR can be generated.34 Immunodominancy of the HLA-DQ5–associated DBY-derived H-Y epitope may be evaluated by determining the presence of T lymphocytes specific for this epitope during GVH disease in other male HLA-DQ5+ patients who received female stem cells.
In conclusion, we report the identification of the first human HLA class II–restricted male-specific mHag. The mHag was recognized by a CD4+ CTL clone that was isolated from a male patient who developed acute GVH disease grade III-IV after transplantation of HLA genotypically identical female stem cells.27 The H-Y mHag presented by the HLA-DQ5 molecule was encoded by the DBYgene. The minimal peptide sequence leading to maximal recognition by the specific HLA-DQ5–restricted CTL clone was characterized as the 12–amino acid sequence HIENFSDIDMGE.
Supported by the Dutch Cancer Society (grant RUL 99-2028) and the JA Cohen Institute for Radiopathology and Radiation Protection.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mario H. J. Vogt, Leiden University Medical Center, Dept of Hematology, C2-R, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: m.h.j.vogt@lumc.nl.