Abstract
We investigated a family whose proband has a severe bleeding disorder and factor V antigenic and functional levels of 8% and less than 1% of control values, respectively. Molecular analysis of the factor V gene revealed a novel homozygous mutation in the last nucleotide of exon 10. 1701G>T causes activation of a cryptic exonic splice site in exon 10, which encodes part of the factor V heavy chain (A2 domain). This leads to the deletion of 35 nucleotides and results in a frameshift with a premature stop codon at amino acid position 498. The G1701 and corresponding Gln509 are conserved in murine, bovine, and porcine factor V and in human factor VIII. Few factor V deficiency mutations have been identified as yet. Several are present in the heterozygous form in combination with factor V Leiden (Arg506Gln). This is the first reported homozygous splice site mutation in a patient with factor V deficiency.
Introduction
Autosomal recessive factor V deficiency is a rare coagulation defect, equally affecting both sexes and diagnosed in multiple ethnic backgrounds.1 In most patients, the phenotype is clinically benign and is characterized by mild bleeding only.1-4 The factor V gene is located on chromosome 1q23,5 spans more than 80 kb, and contains 25 exons.6 The 5′ untranslated sequence, a signal peptide, and the factor V heavy chain (protein domains A1 and A2) are encoded by exons 1 to 12. Exon 13 encodes the B domain, which is posttranslationally removed during activation by thrombin. The light chain (protein domains A3, C1, and C2) and the 3′ untranslated region are encompassed by exons 14 to 25.6 Few mutations associated with factor V deficiency have been reported.7We investigated the genetic basis of factor V deficiency in a patient with umbilical bleeding at birth, gingival bleeding, and repeated spontaneous central nervous system hemorrhage. She requires weekly prophylactic fresh frozen plasma transfusions. The parents may be distantly related. Both parents and the patient's siblings are clinically unaffected.
Study design
Factor V activity and antigen assays
Factor V coagulant activity was determined in the proband and 5 family members (Figure 1) by a clotting assay using an MLA 1200 instrument (Medical Laboratory Automation, Pleasantville, NY). Factor V antigen concentrations were investigated with a paired antibody enzyme-linked immunosorbent assay as recommended by the manufacturer (Cedarlane Laboratories, Hornby, BC, Canada).
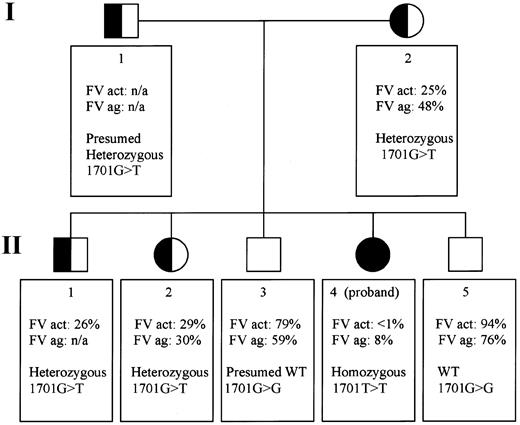
Pedigree of a Mexican family with exon 10 splice site mutation: genotypes and factor V phenotypes.
The pedigree portrays 2 generations (I and II) in which all members are numbered according to age (I-1 and I-2, II-1 to 5). FV act indicates Factor V activity; FV ag, factor V antigen; het, heterozygous; n/a, not available; WT, wild-type.
Pedigree of a Mexican family with exon 10 splice site mutation: genotypes and factor V phenotypes.
The pedigree portrays 2 generations (I and II) in which all members are numbered according to age (I-1 and I-2, II-1 to 5). FV act indicates Factor V activity; FV ag, factor V antigen; het, heterozygous; n/a, not available; WT, wild-type.
Polymerase chain reaction amplification and sequencing
Genomic DNA was isolated from the index patient and 5 relatives. Amplification of 24 factor V exons was achieved by the use of intronic primers. Exon 13 was amplified in 7 overlapping segments because of its large size. All primers and polymerase chain reaction (PCR) conditions are available on www.stanford.edu/∼zehnder.
After identification of the 1701G>T mutation, only RNA from the proband was available for further studies. RNA was extracted from peripheral blood mononuclear cells according to standard procedures. Reverse transcription (RT) was carried out with random hexamer priming.8 Primers for subsequent PCR were designed from the factor V cDNA sequence9 to investigate illegitimate splicing. The forward primer in exon 9 (5′-GTGACCTTCTCGCCTTATGAA-3′) and the reverse primer in exon 12 (5′-ACAGCCTGCTGTTCGATGT-3′) generate a 293-bp amplicon from the wild-type factor V cDNA sequence.9 PCR products were purified directly after amplification or through gel extraction after the excision of bands from an agarose gel. These purifications were performed according to the manufacturer's guidelines (Qiagen, Valencia, CA). Purified samples were then sequenced with fluorescent dideoxy chain terminators (Applied Biosystems, Foster City, CA) on an ABI 377 sequencing instrument (Applied Biosystems). Sequence analyses were aided by GCG Sequence Analysis (www.gcg.com).
Results and discussion
Factor V activity levels were determined in the proband, her mother, and 4 siblings (Figure 1). Factor V antigen levels were assayed in the same persons except sibling II-1. Only the proband (II-4) has severely reduced levels: less than 1% activity (normal range, 60%-120% of the control value) and 8% of factor V antigen compared with the standard. Siblings II-3 and II-5 do not appear to be factor V deficient. Siblings II-1 and II-2 and their mother, I-2, have intermediate levels, consistent with heterozygous factor V deficiency (Figure 1). In the proband, we identified 2 novel homozygous genetic alterations and 17 previously described, clinically innocent polymorphisms (Whitehead cSNP database,http://waldo.wi.mit.edu/cvar_snps/).6,9 10
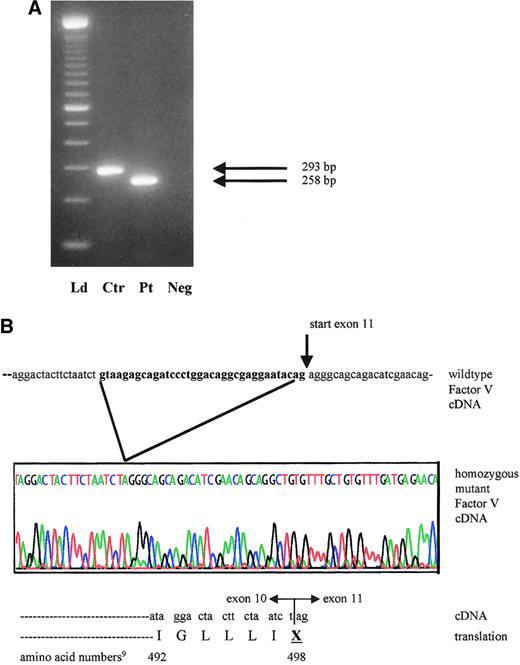
The patient's severe phenotype, together with the markedly reduced factor V antigenic and activity levels, point to a type 1 deficiency. One of the 2 novel sequence variants is a single nucleotide variant (4300C>T),9 which is inconsistent with type 1 factor V deficiency because it is located in exon 13. The entire protein domain (B) encoded by this exon is removed during factor V activation. Thus, unless the mutation interfered with activation, it would not be expected to result in a deficiency phenotype. Additionally, the 4300C>T mutation is located downstream from the homozygous splice site mutation and consequent premature stop codon in this family. Therefore, the most plausible disease causing mutation is a G>T transversion at position 17019 in the exon 10 splice site (GenBank accession number, AY046060). We postulated that this mutation could induce altered RNA splicing. The genotypes in this family are compatible with this hypothesis: the patient's mother I-2 and sister II-2 are heterozygous at position 1701, whereas her brother II-5 is homozygous wild type. To demonstrate use of another splice site and to determine the sequence of the consequent transcript, RNA studies were performed. The RT-PCR product of the patient is smaller than that of normal controls (Figure 2A). No normal-sized band was detected. Sequence analysis of the proband's amplified cDNA revealed deletion of the last 35 bases of exon 10 (Figure 2B). The cryptic splice site activation results in a shift of the reading frame. This frameshift ultimately introduces a premature stop codon wherein the first nucleotide of amino acid 498 in exon 10 (U) fuses to the first 2 nucleotides of exon 11 (AG) (Figure 2B).
mRNA analysis of proband with activation of cryptic exon 10 splicing from 1701G>T.
(A) Reverse transcription–PCR products of factor V cDNA spanning exon 10 and including part of exons 9 and 11. The expected wild-type product in the control (Ctr) measures 293 bp, whereas the patient (Pt) has a 35-bp deletion and a product size of only 258 bp. Neg indicates reagent control in RT-PCR; Ld, ladder. (B) Sequence tracing of the affected area in the proband's mutant cDNA and protein product, compared with the wild-type factor V cDNA from the same region. The sequence shown in bold reflects the 3′ exon 10 segment, which is illegitimately spliced out in the patient.
mRNA analysis of proband with activation of cryptic exon 10 splicing from 1701G>T.
(A) Reverse transcription–PCR products of factor V cDNA spanning exon 10 and including part of exons 9 and 11. The expected wild-type product in the control (Ctr) measures 293 bp, whereas the patient (Pt) has a 35-bp deletion and a product size of only 258 bp. Neg indicates reagent control in RT-PCR; Ld, ladder. (B) Sequence tracing of the affected area in the proband's mutant cDNA and protein product, compared with the wild-type factor V cDNA from the same region. The sequence shown in bold reflects the 3′ exon 10 segment, which is illegitimately spliced out in the patient.
The normal exon → intron (donor) splice site consensus sequence is [C/A]AGgt[a/g]agt,11 where capital letters indicate exonic nucleotides and lowercase letters correspond to the sequence in the intron. The bold g and t nucleotides at the beginning of the intron are nearly invariant. As in the donor splice site of factor V exon 10,9 the last exonic nucleotide is a G in 78% of 5′ splice sites.12 Guanine replacements at positions +1 and +5 in an intron, or at the last base of the preceding exon, are predicted to significantly reduce base pairing stability between the splice site and the complementary region of the U1 small nuclear (sn)RNA.12 This attachment forms the first step in the assembly of the spliceosome.13
An estimated 15% of the point mutations that cause human disease change the conventional splicing pattern of the primary RNA transcript.14 Skipping the preceding exon is the most frequently observed consequence, but illegitimate splice sites may also be used.15 Use of such a cryptic splice site may be preferred over exon skipping if the site is located in the direct vicinity of the expected splice site. It must also be sufficiently similar to the splice site consensus sequence to compete successfully with the affected splice site.12 In our patient, the sequenced mRNA product indicates the use of such an illegitimate splice site.
In conclusion, the observed 35-bp deletion in the patient's cDNA causes a reading-frame shift that predicts a truncated factor V molecule, missing the distal part of the A2 domain and the B, A3, C1, and C2 domains. This finding provides a compelling explanation for the deficiency phenotype in this family and is the first report of a homozygous splice site mutation in the factor V gene resulting in severe factor V deficiency and bleeding.
We thank Dr Uta Francke for helpful discussion and advice and Dr Samuel Parks for technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James L. Zehnder, Department of Pathology, L235, Stanford University Medical Center, Stanford, CA 94305; e-mail:zehnder@stanford.edu.