Abstract
It is unclear how a paroxysmal nocturnal hemoglobinuria (PNH) clone expands in bone marrow, although immune mechanisms involving cytotoxic T lymphocytes, autosomal proliferation, and apoptosis resistance have been hypothesized. To clarify aspects of immune mechanisms and proliferation of PNH cells, we investigated HLA-DRB1, -DQA1, and -DQB1 alleles by polymerase chain reaction (PCR)–based genotyping and expression of the Wilms' tumor gene, WT1, by real-time reverse transcriptase–PCR (RT-PCR) in 21 PNH and 21 aplastic anemia (AA) patients. HLA genotyping indicated that the frequency of DRB1*1501, DQA1*0102, and DQB1*0602 alleles in PNH patients and of DQB1*0602 allele in AA patients was significantly higher than in 916 Japanese controls, and that the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype, found in 13 of 21 PNH patients, 5 of 7 AA-PNH syndrome patients, and 7 of 21 AA patients showed significant differences compared with healthy individuals. RT-PCR analysis showed that the mean values of WT1 RNA were 3413, 712, and 334 copies/μg RNA in PNH, AA, and healthy individuals, respectively. The values for PNH patients were significantly higher than for AA patients and healthy volunteers and were correlated with the proportion of CD16b−granulocytes. The high frequency of HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype in PNH, including AA-PNH syndrome, and AA patients suggests that linkage exists between the disorders and that immune mechanisms in an HLA-restricted manner play an important role in the pathogenesis of these disorders. In addition, high expression of WT1 RNA in PNH patients is related to a PNH clone, but it remains unclear whether this causes expansion of a PNH clone.
Introduction
Recently, paroxysmal nocturnal hemoglobinuria (PNH) has been considered to be an acquired stem cell disorder affecting all hematopoietic lineages, which lack glycosylphosphatidylinositol (GPI)-anchored membrane proteins because of abnormalities in the phosphatidylinositol glycan–class A (PIG-A) gene.1 It is well known that deficiencies in CD55 and CD59 expression on erythrocytes cause complement-mediated hemolysis in PNH.2,3 It has been hypothesized that expansion of a PNH clone with a PIG-A gene abnormality is associated with an immunologic mechanism involving cytotoxic T cells,4 autosomal proliferation of a PNH clone,5 and resistance to apoptosis in a PNH clone.6 Some reports7,8 support the concept that loss of GPI-anchored proteins results in less efficient killing of GPI-deficient hematopoietic target cells by putative autoimmune T-cell clones, causing expansion of a PNH clone. In addition, some researchers also reported abnormalities of T cells derived from PNH patients.9,10 Nevertheless, a wide variety of immunologic assays have not demonstrated a difference between normal and PIG-A–mutated target cells.11 Although resistance of PNH cells to apoptosis has been reported,12,13 this is a property of both GPI-deficient cells with PIG-A gene abnormalities and GPI-positive cells, suggesting that apoptosis resistance does not underlie the expansion only of PNH clones. In fact, it is well known that there is no known molecular mechanism by which GPI-anchored proteins influence apoptosis, and there are conflicting data concerning the apoptosis resistance of PNH cells.6,11,14 Moreover, in chimeric knockout mice, the cells without the PIG-A gene constitute a minor, static fraction of the bone marrow or circulating hematopoietic compartments, and PIG-A–negative and normal cells behave similarly in paired tissue culture assays,5 15 negating autosomal proliferation of a PNH clone. At present, it is unclear how a PNH clone in bone marrow expands.
The transformation of aplastic anemia (AA) into PNH is well known as the AA-PNH syndrome, described by Lewis and Dacie.16Recently, several reports17-22 have indicated that PNH eventually develops in 29% to 45% of patients with AA who have been treated with immunosuppressive agents and/or androgens and that some patients with PNH respond to immunosuppressive therapy. These facts suggest an immunologic linkage between AA and PNH, with the common feature of bone marrow failure.4 Several studies on HLA class I and II antigens and alleles in AA patients have been reported,23-27 but the relationship between AA and HLA class I and II antigens or alleles is controversial because of conflicting data, mainly due to differences in statistical analysis.28 Ustariz et al29 reported that there was no significant increase in any of the 26 HLA antigens corresponding to loci A and B in 10 Cuban patients with PNH. Nevertheless, from many clinical and basic studies of AA patients, it is certain that cytotoxic T lymphocytes and/or helper/inducer T lymphocytes are implicated in some of the immune mechanisms involved in the occurrence of AA.28 Recently, Zeng et al30found sequence identity for complementarity determining region 3 among a majority of CD4 clones from a patient with AA, from which the dominant clone showed a Th1 secretion pattern, lysed autologous CD34+ cells, and inhibited their hematopoietic colony formation. Thus, we considered the possibility that HLA class II alleles and haplotypes may be related to the immunologic pathogenesis of AA and PNH because it is known that antigen-specific T helper/inducer cells, when present at very low levels, are capable of rapid clonal expansion during antigenic challenge, and the specificity of this response provides for the activation and expansion of a very select cohort of T cells.31
The Wilms' tumor gene, WT1, is responsible for the childhood renal neoplasm, Wilms' tumor, and is located at chromosome 11p13.32 Although the kind of hematopoietic cells expressing WT1 RNA in normal bone marrow is controversial, it is generally believed that the WT1 gene is expressed in CD34+ hematopoietic progenitor cells, but not in CD34− differentiated cells.33-35 TheWT1 gene encodes a transcription factor that is involved in the control of growth and differentiation of various cell types, including hematopoietic cells.36 It activates or suppresses the transcription of target genes depending on the structure of their promoters and the presence of other transcriptional regulators.37-39 In addition, several reports40-43 have indicated that immunization with HLA class I and II binding WT1 peptides induces WT1 peptide–specific cytotoxic T-lymphocyte and helper/inducer T-lymphocyte responses specific for the immunizing WT1 peptides in animal models and/or leukemic patients. Recently, it has been reported that WT1 mRNA is a novel tumor marker for leukemic blast cells, and its level of expression is a new prognostic factor for acute leukemias.44,45 In addition, Tamaki et al46reported that WT1 expression levels directly reflect the progression of myelodysplastic syndrome (MDS), which is one of the bone marrow failure syndromes.4 Thus, it would be of interest to know the expression levels of WT1 RNA in bone marrow hematopoietic cells from AA and PNH patients because AA and PNH are also thought to be bone marrow failure syndromes, and the WT1 gene may be related to various immune and proliferative mechanisms in hematopoietic progenitor cells, as described above.
In the present study, to clarify some characteristics in the pathophysiology of PNH and AA, we investigated HLA-DRB1, -DQA1, and -DQB1 alleles and haplotypes, and WT1 RNA expression in bone marrow, in 21 PNH patients and 21 AA patients, including 2 with pure red cell aplasia.
Patients, materials, and methods
Patients and controls
Peripheral blood and bone marrow samples were taken from 21 Japanese PNH patients after obtaining informed consent and approval from the institutional Human Research Committee. Generally, the samples were obtained from each patient at the same time. The diagnosis of PNH was made on the basis of the history, clinical and laboratory findings, and the results of CD59 expression on erythrocytes as determined by flow cytometry.3,47 A patient with a CD59−population of more than 1% was judged to have PNH erythrocytes.48 At the time of the study, all patients were clinically stable with no recent severe hemolytic attacks. The clinical, hematologic, and laboratory findings of the PNH patients are summarized in Table 1. Seven of the 21 patients had AA-PNH syndrome.16 Patient no. 17 transformed from PNH to acute leukemia (M0 according to the French-American-British classification49) 5 months after the examination and then died of acute pneumonia about 1 month later despite intensive chemotherapy.
Clinical, laboratory, and hematologic findings in 21 PNH patients
| Case no. . | Age/sex . | WBC, × 109/L . | Neutrophil count, × 109/L . | Hb, g/dL . | Reticulocyte count, × 109/L . | PLT, × 109/L . | LDH, IU/L . | Flow cytometry of erythrocytes (CD59), % . | Flow cytometry of granulocytes (CD16b), % . | Duration of illness, mo . | Treatment/transfusion requirement . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos* . | Int* . | Neg* . | Neg* . | ||||||||||
| 1 | 54/M | 2.9 | 1.769 | 15.6 | 100 | 55 | 800 | 96.9 | 0 | 3.1 | 12.8 | 92 | A/− |
| 2 | 60/M | 5.5 | 2.200 | 15.7 | 77 | 149 | 361 | 65.6 | 25.8 | 8.6 | 12.3 | 270 | P/− |
| 3† | 55/M | 6.5 | 4.160 | 13.5 | 98 | 156 | 673 | 90.7 | 0 | 9.3 | 3.0 | 180 | P/− |
| 4 | 27/M | 3.4 | 2.584 | 14.2 | 91 | 147 | 386 | 88.2 | 0 | 11.8 | 2.1 | 31 | ATG, Cy, G → P/− |
| 5 | 57/M | 6.4 | 3.712 | 11.4 | 108 | 183 | 1183 | 15.1 | 67.9 | 17.0 | 92.3 | 203 | P/+ |
| 6† | 35/F | 4.9 | 2.744 | 15.5 | 69 | 170 | 818 | 92.9 | 0 | 7.1 | 10.4 | 54 | A/− |
| 7 | 53/M | 3.2 | 1.088 | 10.8 | 153 | 98 | 2713 | 34.2 | 32.7 | 33.1 | 55.8 | 20 | P, continuous iron/+ |
| 8 | 82/M | 2.2 | 0.570 | 9.6 | 84 | 50 | 2028 | 1.7 | 89.4 | 8.9 | 62.7 | 254 | Intermittent iron/− |
| 9 | 52/F | 1.4 | 0.392 | 7.0 | 227 | 178 | 6070 | 68.7 | 0 | 31.3 | 89.3 | 115 | P → None/+ |
| 10 | 70/M | 3.1 | 1.612 | 9.4 | 104 | 269 | 1033 | 13.1 | 0 | 86.9 | 94.4 | 168 | A/+ |
| 11 | 29/F | 6.0 | 5.220 | 5.5 | 126 | 194 | 3094 | 61.6 | 9.8 | 28.6 | 95.0 | 114 | P/− |
| 12 | 20/M | 3.3 | 0.875 | 14.0 | 93 | 112 | 548 | 93.4 | 0 | 6.6 | 97.9 | 52 | A/− |
| 13† | 69/F | 3.1 | 0.992 | 11.1 | 75 | 52 | 995 | 85.5 | 0 | 14.5 | 37.4 | 164 | A/− |
| 14 | 23/F | 3.5 | 1.785 | 9.8 | 83 | 152 | 953 | 86.1 | 0 | 13.9 | 51.2 | 5 | None/− |
| 15 | 20/M | 4.1 | 3.034 | 7.9 | 108 | 106 | 2653 | 27.0 | 37.6 | 35.4 | 83.6 | 10 | None/− |
| 16† | 59/M | 2.8 | 1.064 | 12.7 | 44 | 54 | 348 | 96.5 | 0 | 3.5 | 3.5 | 0 | A/− |
| 17† | 71/M | 1.2 | 0.216 | 6.9 | 20 | 144 | 368 | 98.6 | 0 | 1.4 | 97.1 | 0 | None/− |
| 18† | 56/M | 2.6 | 1.612 | 9.9 | 205 | 56 | 3770 | 30.7 | 30.7 | 38.5 | 98.6 | 198 | P, continuous iron/− |
| 19† | 59/F | 2.4 | 1.944 | 5.6 | 24 | 13 | 558 | 88.4 | 0 | 11.6 | 11.6 | 0 | ATG, Cy, G → Cy, G, A+/− |
| 20 | 71/F | 9.2 | 6.656 | 10.1 | 261 | 126 | 1218 | < 1.0 | 99.6 | < 1.0 | 98.3 | 5 | P/− |
| 21 | 46/M | 2.4 | 0.936 | 13.1 | 74 | 96 | 1054 | 91.6 | 0 | 8.4 | 98.9 | 8 | None/− |
| Case no. . | Age/sex . | WBC, × 109/L . | Neutrophil count, × 109/L . | Hb, g/dL . | Reticulocyte count, × 109/L . | PLT, × 109/L . | LDH, IU/L . | Flow cytometry of erythrocytes (CD59), % . | Flow cytometry of granulocytes (CD16b), % . | Duration of illness, mo . | Treatment/transfusion requirement . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos* . | Int* . | Neg* . | Neg* . | ||||||||||
| 1 | 54/M | 2.9 | 1.769 | 15.6 | 100 | 55 | 800 | 96.9 | 0 | 3.1 | 12.8 | 92 | A/− |
| 2 | 60/M | 5.5 | 2.200 | 15.7 | 77 | 149 | 361 | 65.6 | 25.8 | 8.6 | 12.3 | 270 | P/− |
| 3† | 55/M | 6.5 | 4.160 | 13.5 | 98 | 156 | 673 | 90.7 | 0 | 9.3 | 3.0 | 180 | P/− |
| 4 | 27/M | 3.4 | 2.584 | 14.2 | 91 | 147 | 386 | 88.2 | 0 | 11.8 | 2.1 | 31 | ATG, Cy, G → P/− |
| 5 | 57/M | 6.4 | 3.712 | 11.4 | 108 | 183 | 1183 | 15.1 | 67.9 | 17.0 | 92.3 | 203 | P/+ |
| 6† | 35/F | 4.9 | 2.744 | 15.5 | 69 | 170 | 818 | 92.9 | 0 | 7.1 | 10.4 | 54 | A/− |
| 7 | 53/M | 3.2 | 1.088 | 10.8 | 153 | 98 | 2713 | 34.2 | 32.7 | 33.1 | 55.8 | 20 | P, continuous iron/+ |
| 8 | 82/M | 2.2 | 0.570 | 9.6 | 84 | 50 | 2028 | 1.7 | 89.4 | 8.9 | 62.7 | 254 | Intermittent iron/− |
| 9 | 52/F | 1.4 | 0.392 | 7.0 | 227 | 178 | 6070 | 68.7 | 0 | 31.3 | 89.3 | 115 | P → None/+ |
| 10 | 70/M | 3.1 | 1.612 | 9.4 | 104 | 269 | 1033 | 13.1 | 0 | 86.9 | 94.4 | 168 | A/+ |
| 11 | 29/F | 6.0 | 5.220 | 5.5 | 126 | 194 | 3094 | 61.6 | 9.8 | 28.6 | 95.0 | 114 | P/− |
| 12 | 20/M | 3.3 | 0.875 | 14.0 | 93 | 112 | 548 | 93.4 | 0 | 6.6 | 97.9 | 52 | A/− |
| 13† | 69/F | 3.1 | 0.992 | 11.1 | 75 | 52 | 995 | 85.5 | 0 | 14.5 | 37.4 | 164 | A/− |
| 14 | 23/F | 3.5 | 1.785 | 9.8 | 83 | 152 | 953 | 86.1 | 0 | 13.9 | 51.2 | 5 | None/− |
| 15 | 20/M | 4.1 | 3.034 | 7.9 | 108 | 106 | 2653 | 27.0 | 37.6 | 35.4 | 83.6 | 10 | None/− |
| 16† | 59/M | 2.8 | 1.064 | 12.7 | 44 | 54 | 348 | 96.5 | 0 | 3.5 | 3.5 | 0 | A/− |
| 17† | 71/M | 1.2 | 0.216 | 6.9 | 20 | 144 | 368 | 98.6 | 0 | 1.4 | 97.1 | 0 | None/− |
| 18† | 56/M | 2.6 | 1.612 | 9.9 | 205 | 56 | 3770 | 30.7 | 30.7 | 38.5 | 98.6 | 198 | P, continuous iron/− |
| 19† | 59/F | 2.4 | 1.944 | 5.6 | 24 | 13 | 558 | 88.4 | 0 | 11.6 | 11.6 | 0 | ATG, Cy, G → Cy, G, A+/− |
| 20 | 71/F | 9.2 | 6.656 | 10.1 | 261 | 126 | 1218 | < 1.0 | 99.6 | < 1.0 | 98.3 | 5 | P/− |
| 21 | 46/M | 2.4 | 0.936 | 13.1 | 74 | 96 | 1054 | 91.6 | 0 | 8.4 | 98.9 | 8 | None/− |
WBC indicates white blood cell count; Hb, hemoglobin concentration; PLT, platelet count; LDH, lactate dehydrogenase; A, androgens; P, prednisolone; ATG, antithymocyte globulin; Cy, cyclosporine; G, granulocyte colony-stimulating factor.
Pos, Int, and Neg refer to positive, intermediate, and negative populations by single-color flow cytometric analysis of erythrocytes or granulocytes using CD59 or CD16b monoclonal antibody, respectively.
Includes AA-PNH syndrome.
As controls, peripheral blood samples were obtained after informed consent from 20 healthy volunteers for single-color flow cytometric analysis of CD59 expression on erythrocytes and CD16b expression on granulocytes, and bone marrow samples were obtained from an additional 20 healthy volunteers for quantitation of WT1 RNA expression by real-time reverse transcriptase–polymerase chain reaction (RT-PCR). In addition, peripheral blood and bone marrow samples were taken from 21 AA patients, including 2 with pure red cell aplasia. Based on the criteria of the Research Committee of Idiopathic Hematopoietic Disturbances of the Ministry of Health and Welfare of Japan,50 the diagnosis and grading of the severity of AA were determined by peripheral blood and/or bone marrow cytomorphologic findings following exclusion of the other disorders presenting with pancytopenia. Severe, moderate, or mild AA was established when at least 2 of the following criteria were met: absolute numbers of reticulocytes, neutrophils, or platelets less than 20 × 109/L, 0.5 × 109/L, or 20 × 109/L; less than 60 × 109/L, 1 × 109/L, or 50 × 109/L; or more than 60 × 109/L, 1 × 109/L, or 50 × 109/L, respectively.
Clinical and hematologic parameters
Age, sex, white blood cell count, neutrophil count, lymphocyte count, hemoglobin concentration, reticulocyte count, platelet count, lactate dehydrogenase (LDH), bone marrow cellularity, time after diagnosis, treatment, and transfusion requirements were determined when peripheral blood and bone marrow samples were taken from the patients.
CD59 expression on erythrocytes and CD16b expression on granulocytes by flow cytometry
Immunofluorescent staining and flow cytometric analysis of CD59 expression on erythrocytes from all the patients and 20 healthy volunteers were performed as described previously.3,47Analysis of CD16b expression on granulocytes was also performed with cells from 21 PNH patients and healthy volunteers as described previously.51 Mouse monoclonal antibodies to CD59 (3E1, IgG1)52 and to CD16b (ID3, IgM, κ) labeled with fluorescein isothiocyanate (Immunotech, Marseille, France) were used, and irrelevant monoclonal antibodies of the same subclass were used as negative controls.3,47 51
Analyses of HLA-DRB1, -DQA1, and -DQB1 alleles and haplotypes
High-molecular-weight DNA was extracted from leukocytes of patients by standard methods.53 The second exons of the HLA-DRB1 and -DQB1 genes and the first 3 exons of the HLA-DQA1 gene were amplified using PCR with group-specific primers and sequence-specific primers, respectively. Each allele was typed using the restriction fragment length polymorphism method and/or the PCR sequence-specific primers method, as described previously,54-56 with the modification of adding several restriction enzymes or several primers to detect the DRB1 and DQB1 alleles or the DQA1 alleles, respectively, the nomenclature of which was officially updated by the World Health Organization Nomenclature Committee. An HLA class II haplotype in these patients was determined on the basis of the frequency of HLA class II haplotypes reported in unrelated Japanese individuals.57,58 The frequency of an HLA class II haplotype in these patients was compared with that in a control population consisting of 916 unrelated Japanese individuals.57 This control population was considered valid because both populations originated from the same area in Japan.
Quantitation of WT1 RNA by RT-PCR
WT1 RNA levels in bone marrow samples from 21 PNH and AA patients were quantified by RT-PCR. Total RNA was extracted from bone marrow mononuclear cells using a QIAamp RNA Blood Mini Kit (Qiagen GmbH, Hiden, Germany) according to the manufacturer's instruction. Two micrograms of total RNA was converted into cDNA in 45 μL reaction mixture containing 200 units of reverse-transcriptase SuperScript II (Gibco BRL, Rockville, MD), 1 mM of each deoxynucleotide triphosphate, 500 ng random hexamers (Gibco BRL), and 40 U RNase inhibitor (Gibco BRL). RT-PCR59,60 was performed in a MicroAmp optical 96-well plate with 2.3 μL of the above cDNA solution, 7.5 pmol forward and reverse primers, 5 pmol TaqMan probe, and 12.5 μL 2X TaqMan Universal PCR Master Mix (PE Biosystems, Foster City, CA). The sequences of forward and reverse primers and TaqMan probe for the quantitation of the expression levels of the WT1 and GAPDH genes were as follows: the WT1 gene, 5′-GATAACCACACAACGCCCATC-3′ (forward primer), 5′-CACACGTCGCACATCCTGAAT-3′ (reverse primer), 5′-FAM-ACACCGTGCGTGTGTATTCTGTATTGG-TAMRA-3′ (TaqMan probe); the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, 5′-CACCAGGGCTGCTTTTAACTC-3′ (forward primer), 5′-GAAGATGGTGATGGGATTTC-3′ (reverse primer), 5′-FAM-CATGGGTGGAATCATATTGGAA-TAMRA-3′ (TaqMan probe). The above reaction mixture was heated at 50°C for 2 minutes and then at 95°C for 10 minutes to activate the polymerase. PCR was then performed using an ABI Prism 7700 Sequence Detector System (PE Applied Biosystems) with 45 cycles, one of which consisted of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. Known copy numbers of WT1 cDNA (102, 104, and 106 copies/well) and GAPDH cDNA (104, 106, and 108 copies/well) were simultaneously amplified by PCR to make standard curves for measuring WT1 and GAPDH expression levels in the samples. These standard cDNAs were prepared by cloning the PCR-amplified cDNA fragments into PCR II vector (Invitrogen, Carlsbad, CA), amplifying the vector, and cutting out the cDNA fragments from the vector. The DNA contents of standard cDNA solutions were quantitated by their absorbance at 260 nm, and copy numbers were calculated by dividing the amounts of DNA by molecular weights. Copy numbers per microgram RNA of WT1 and GAPDH RNA in the patients' samples were calculated using a standard curve. To normalize the differences in RNA loading for RT-PCR and in RNA degradation for individual samples, the values (WT1/GAPDH) of the WT1 RNA levels (copy number/μg RNA) divided by the GAPDH RNA levels (copy number/μg RNA) were calculated according to a procedure described previously,61 and (WT1/GAPDH) × (6.1 × 109) (the mean copy numbers/μg RNA of GAPDH in 47 normal human peripheral blood mononuclear cells, which were determined to have the corrected values of WT1/GAPDH using copy numbers) were defined as copy numbers/μg RNA of WT1 RNA in the samples. To check the reproducibility of the precision of the WT1 RNA assay, we simultaneously PCR-amplified 103copies of GAPDH and WT1 cDNA with the samples and confirmed with every WT1 RNA assay that their copy numbers were within the mean ± 2 SDs.
Statistical analysis
The various hematologic and laboratory parameters of the 2 disorders were compared statistically by the Student ttest. The proportions of CD59− erythrocytes among PNH patients, AA patients, and healthy volunteers were examined statistically by a one-way analysis of variance (Bonferroni/Dunn) model. The proportions of CD16b− granulocytes in PNH patients and healthy volunteers were compared statistically by the Student t test. Differences in HLA class II allele or haplotype frequencies between patients with PNH or AA and controls57 or between the disorders were calculated by χ2 analysis or Fisher exact test. Various hematologic and laboratory findings, including the proportions of CD59−erythrocytes and CD16b− granulocytes, and the values for WT1 RNA levels, in the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype group and another haplotype group in the PNH patients were compared statistically by the Student t test. The WT1 RNA levels quantitated by RT-PCR were investigated statistically among PNH patients, AA patients, and healthy volunteers by a one-way analysis of variance (Bonferroni/Dunn) model. In addition, the relationship between the values for WT1 RNA and the proportion of CD59−erythrocytes or CD16b− granulocytes in 21 PNH patients was analyzed using correlation coefficients.
Results
Clinical, hematologic, and laboratory findings in patients with PNH and AA
Male-to-female ratio and mean age of the AA patients were 11:10 and 56.7 years (range, 18-80 years), respectively. Among 19 AA patients, there were 3 severe, 6 moderate, and 10 mild cases of disease at the time of examination. The clinical, hematologic, and laboratory findings described in “Materials and methods” at the time of examination were compared between patients with PNH and AA. The reticulocyte counts (mean, 106 × 109/L; range, 20-261 × 109/L) and LDH (1506 IU/L; range, 348-6070 IU/L) in PNH patients (Table 1) were significantly higher than those in AA patients (50 × 109/L; range, 3-116 × 109/L;P < .001; and 413 IU/L; range, 171-697 IU/L;P < .002, respectively), but there were no differences between them in the other parameters. The percentages of 21 PNH and AA patients with absolute neutrophil counts below 0.5 × 109/L or reticulocyte counts greater than 50 × 109/L were 9.5% and 9.5% or 85.7% and 47.6%, respectively.
The treatment and transfusion regimens of the PNH patients are summarized in Table 1. Twelve of 21 AA patients were treated with immunosuppressive therapy using antithymocyte globulin and/or cyclosporine prior to or at the time of examination. Three of 12 patients received granulocyte colony-stimulating factor (G-CSF) in addition to immunosuppressive therapy, and one received oxymetholone at the time of examination. Six of 21 AA patients received prednisolone and/or androgens, including metenolone and fluoxymesterone, and one of them also received G-CSF at the time of examination. Two and one of the 21 patients received only G-CSF or blood transfusions at the time of examination, respectively.
CD59 expression on erythrocytes and CD16b expression on granulocytes in patients with PNH and AA
The phenotypes and proportions of each population of erythrocytes in 21 PNH patients are summarized in Table 1. Flow cytometric profiles of CD59 expression on erythrocytes from 21 AA patients and 20 healthy volunteers showed that the erythrocytes consisted of a single positive population. The mean proportions of negative populations determined by flow cytometry were 19.0% (range, 1.4% to 86.9%), 0.43% (0.07% to 0.87%), and 0.39% (0.10% to 0.80%) in 20 PNH, 21 AA, and 20 healthy individuals, respectively. The proportion of CD59− populations in PNH patients was higher than that in AA (P < .0001) and healthy individuals (P < .0001). The proportion of CD16b−granulocytes in 21 PNH patients is summarized in Table 1. It was significantly higher in the PNH patients (mean ± SD, 57.5% ± 39.8%) than in the healthy volunteers (0.46% ± 0.38%,P < .001). Flow cytometric profiles of CD16b expression on granulocytes from 20 healthy volunteers showed a single positive population.
HLA-DRB1, -DQA1, and -DQB1 alleles and haplotypes in patients with PNH and AA
The frequencies of the HLA-DRB1, -DQA1, and -DQB1 alleles in 21 PNH and AA patients are presented in Table2. The frequencies of each allele were statistically compared between the disorders. The frequency of all the alleles did not show significant differences between PNH and AA patients. In addition, the frequency of each HLA-DRB1, -DQA1, and -DQB1 allele in the 2 disorders was statistically compared with that in 916 Japanese individuals.57 The frequencies of HLA-DRB1*1501, -DQA1*0102, and -DQB1*0602 alleles in PNH patients and of HLA-DQB1*0602 allele in AA patients were significantly different than in controls (P < .005, P < .005,P < .005, and P < .025, respectively).
Frequencies of HLA-DRB1, -DQA1, and -DQB1 alleles in PNH and AA patients and Japanese healthy controls
| Gene allele* . | . | Frequency of each allele, % . | ||
|---|---|---|---|---|
| PNH (n = 21) . | AA (n = 21) . | Healthy controls (n = 916) . | ||
| DRB1 | 0101 | 2.4 | 2.4 | 4.8 |
| 0406 | 7.1 | 2.4 | 3.2 | |
| 0901 | 9.5 | 16.7 | 12.4 | |
| 1101 | 2.4 | 4.8 | 2.9 | |
| 1201 | 4.8 | 7.1 | 3.9 | |
| 1302 | 7.1 | 7.1 | 5.3 | |
| 1401 | 7.1 | 0 | 4.5 | |
| 1501 | 33.3 | 16.7 | 6.1 | |
| 1502 | 14.3 | 14.3 | 8.7 | |
| 04051 | 7.1 | 14.3 | 15.5 | |
| 08032 | 4.8 | 4.8 | 7.6 | |
| DQA1 | 0101 | 2.4 | 2.4 | 12.3† |
| 0104 | 7.1 | 2.4 | 12.3† | |
| 0102 | 40.5 | 21.4 | 12.1 | |
| 0103 | 19.0 | 21.4 | 16.9 | |
| 03011 | 7.1 | 2.4 | 42.3† | |
| 0302 | 9.5 | 21.4 | 42.3† | |
| 0303 | 7.1 | 16.7 | 42.3† | |
| 0505 | 7.1 | 7.1 | 8.7† | |
| DQB1 | 0301 | 4.8 | 9.5 | 13.1 |
| 0302 | 9.5 | 2.4 | 11.8 | |
| 0401 | 7.1 | 16.7 | 15.1 | |
| 0501 | 2.4 | 4.8 | 5.4 | |
| 0601 | 19.0 | 19.0 | 16.1 | |
| 0602 | 33.3 | 16.7 | 6.0 | |
| 0604 | 7.1 | 4.8 | 5.0 | |
| 03032 | 9.5 | 21.4 | 11.2 | |
| 05031 | 7.1 | 2.4 | 5.0 | |
| Gene allele* . | . | Frequency of each allele, % . | ||
|---|---|---|---|---|
| PNH (n = 21) . | AA (n = 21) . | Healthy controls (n = 916) . | ||
| DRB1 | 0101 | 2.4 | 2.4 | 4.8 |
| 0406 | 7.1 | 2.4 | 3.2 | |
| 0901 | 9.5 | 16.7 | 12.4 | |
| 1101 | 2.4 | 4.8 | 2.9 | |
| 1201 | 4.8 | 7.1 | 3.9 | |
| 1302 | 7.1 | 7.1 | 5.3 | |
| 1401 | 7.1 | 0 | 4.5 | |
| 1501 | 33.3 | 16.7 | 6.1 | |
| 1502 | 14.3 | 14.3 | 8.7 | |
| 04051 | 7.1 | 14.3 | 15.5 | |
| 08032 | 4.8 | 4.8 | 7.6 | |
| DQA1 | 0101 | 2.4 | 2.4 | 12.3† |
| 0104 | 7.1 | 2.4 | 12.3† | |
| 0102 | 40.5 | 21.4 | 12.1 | |
| 0103 | 19.0 | 21.4 | 16.9 | |
| 03011 | 7.1 | 2.4 | 42.3† | |
| 0302 | 9.5 | 21.4 | 42.3† | |
| 0303 | 7.1 | 16.7 | 42.3† | |
| 0505 | 7.1 | 7.1 | 8.7† | |
| DQB1 | 0301 | 4.8 | 9.5 | 13.1 |
| 0302 | 9.5 | 2.4 | 11.8 | |
| 0401 | 7.1 | 16.7 | 15.1 | |
| 0501 | 2.4 | 4.8 | 5.4 | |
| 0601 | 19.0 | 19.0 | 16.1 | |
| 0602 | 33.3 | 16.7 | 6.0 | |
| 0604 | 7.1 | 4.8 | 5.0 | |
| 03032 | 9.5 | 21.4 | 11.2 | |
| 05031 | 7.1 | 2.4 | 5.0 | |
The alleles shown here were found in at least 2 of the 42 patients.
DQA1∗0101, DQA1∗0301, and DQA1∗0501 determined by Hashimoto et al57 include DQA1∗0101, 0104, and (0105); DQA1∗03011, 0302, and 0303; and DQA1∗(0501) and 0505, respectively.
The HLA class II haplotypes in 21 PNH patients are presented in Table3. Thirteen of 21 PNH patients showed the DRB1*1501-DQA1*0102-DQB1*0602 haplotype. Five of 7 AA-PNH syndrome patients and 8 of the 14 primary PNH patients showed this haplotype. In contrast, the same haplotype was found in 7 of 21 AA patients. The frequency of this haplotype in 21 PNH patients was significantly higher than in Japanese healthy volunteers (P < .005).57 The frequency of this haplotype in patients with primary PNH and AA-PNH syndrome was also higher than in healthy controls (P < .005 andP < .005, respectively). In addition, various hematologic and laboratory findings, including the proportions of CD59− erythrocytes and CD16b− granulocytes and the levels of WT1 RNA, were statistically compared between the specific haplotype group and another haplotype group in the PNH patients. No differences were found between the groups. Moreover, the frequency of HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype in AA patients was significantly different than in healthy volunteers (P < .01). The frequencies of all the other haplotypes were not significantly different between the disorders or between each of the disorders and healthy controls.
HLA class II haplotypes and the copy numbers of WT1 RNA in patients with PNH
| Case no. . | HLA class II allele3-150 . | Copy number of WT1 (copies/μg RNA) . | ||
|---|---|---|---|---|
| DRB1 . | DQA1 . | DQB1 . | ||
| 1 | 1302,1501 | 0102,0102 | 0602, 0604 | 2 100 |
| 2 | 0901, 0901 | 0302, 0302 | 03032, 03032 | 580 |
| 33-151 | 0406,1501 | 0102, 03011 | 0302,0602 | 360 |
| 4 | 0901, 1101 | 0302, 0505 | 0302, 03032 | 460 |
| 5 | 0101, 1501 | 0101,0102 | 0501, 0602 | 1 100 |
| 63-151 | 1201, 1502 | 0103, 0505 | 0301, 0601 | 650 |
| 7 | 04051, 1502 | 0103, 0303 | 0401, 0601 | 380 |
| 8 | 04051, 1502 | 0103, 0303 | 0401, 0601 | 2 000 |
| 9 | 08032,1501 | 0102, 0103 | 0601,0602 | 4 000 |
| 10 | 04051,1501 | 0102, 0303 | 0401,0602 | 8 300 |
| 11 | 1401,1501 | 0102, 0104 | 05031,0602 | 500 |
| 12 | 1501, 1502 | 0102, 0103 | 0601, 0602 | 17 000 |
| 133-151 | 1401, 1501 | 0102, 0104 | 05031, 0602 | 610 |
| 14 | 1302, 1502 | 0102, 0103 | 0601, 0604 | 1 900 |
| 15 | 0406,1501 | 0102, 03011 | 0302,0602 | 2 800 |
| 163-151 | 0901,1501 | 0102, 0302 | 03032,0602 | 510 |
| 173-151 | 1501,1501 | 0102,0102 | 0602, 0602 | 16 000 |
| 183-151 | 1201, 1501 | 0102, 0505 | 0301, 0602 | 790 |
| 193-151 | 0406, 1502 | 0103, 03011 | 0302, 0601 | < 250 |
| 20 | 1302,1501 | 0102,0102 | 0602, 0604 | 390 |
| 21 | 08032, 1401 | 0103, 0104 | 05031, 0601 | 11 000 |
| Case no. . | HLA class II allele3-150 . | Copy number of WT1 (copies/μg RNA) . | ||
|---|---|---|---|---|
| DRB1 . | DQA1 . | DQB1 . | ||
| 1 | 1302,1501 | 0102,0102 | 0602, 0604 | 2 100 |
| 2 | 0901, 0901 | 0302, 0302 | 03032, 03032 | 580 |
| 33-151 | 0406,1501 | 0102, 03011 | 0302,0602 | 360 |
| 4 | 0901, 1101 | 0302, 0505 | 0302, 03032 | 460 |
| 5 | 0101, 1501 | 0101,0102 | 0501, 0602 | 1 100 |
| 63-151 | 1201, 1502 | 0103, 0505 | 0301, 0601 | 650 |
| 7 | 04051, 1502 | 0103, 0303 | 0401, 0601 | 380 |
| 8 | 04051, 1502 | 0103, 0303 | 0401, 0601 | 2 000 |
| 9 | 08032,1501 | 0102, 0103 | 0601,0602 | 4 000 |
| 10 | 04051,1501 | 0102, 0303 | 0401,0602 | 8 300 |
| 11 | 1401,1501 | 0102, 0104 | 05031,0602 | 500 |
| 12 | 1501, 1502 | 0102, 0103 | 0601, 0602 | 17 000 |
| 133-151 | 1401, 1501 | 0102, 0104 | 05031, 0602 | 610 |
| 14 | 1302, 1502 | 0102, 0103 | 0601, 0604 | 1 900 |
| 15 | 0406,1501 | 0102, 03011 | 0302,0602 | 2 800 |
| 163-151 | 0901,1501 | 0102, 0302 | 03032,0602 | 510 |
| 173-151 | 1501,1501 | 0102,0102 | 0602, 0602 | 16 000 |
| 183-151 | 1201, 1501 | 0102, 0505 | 0301, 0602 | 790 |
| 193-151 | 0406, 1502 | 0103, 03011 | 0302, 0601 | < 250 |
| 20 | 1302,1501 | 0102,0102 | 0602, 0604 | 390 |
| 21 | 08032, 1401 | 0103, 0104 | 05031, 0601 | 11 000 |
HLA class II haplotypes DRB1∗1501, DQA1∗0102, and DQB1∗0602 are underlined.
Includes AA-PNH syndrome.
Quantitation of WT1 RNA in patients with PNH and AA
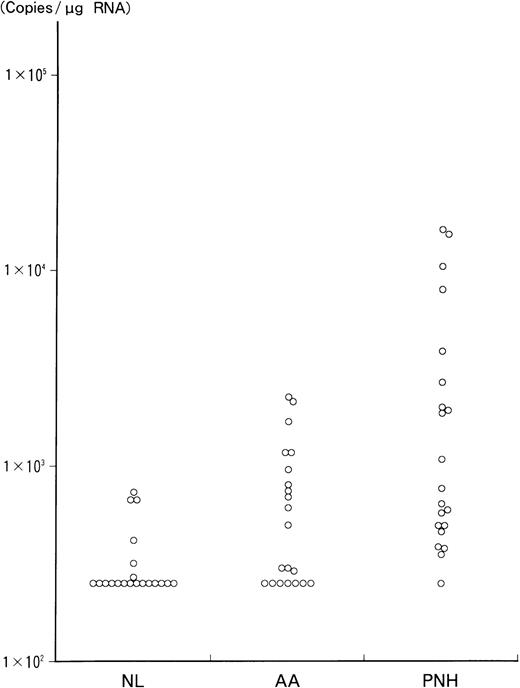
The results of RT-PCR analysis of WT1 RNA levels in bone marrow mononuclear cells from 21 PNH and AA patients and 20 healthy volunteers are presented in Figure 1, and the values for the PNH patients are shown in Table 3. The values were statistically compared among PNH patients, AA patients, and healthy individuals. The values in PNH patients (mean ± SD, 3413 ± 5149 copies/μg RNA) were significantly higher than in AA patients (712 ± 647 copies/μg RNA; P < .005) and in healthy individuals (333 ± 170 copies/μg RNA;P < .002). In contrast, the values in patients with AA did not show significant differences compared with those in healthy volunteers. Five AA patients with more than 1000 copies/μg of WT1 RNA had no specific clinical and hematologic features, including response to immunosuppressive therapy, compared with the other AA patients. In addition, the relationship between the WT1 RNA levels and the proportion of CD59− erythrocytes or CD16b−granulocytes in 21 PNH patients was statistically analyzed. The WT1 RNA levels were significantly correlated with the proportion of CD16b− granulocytes (r = 0.5207,P < .02), but not with that of CD59−erythrocytes.
Quantitation of WT1 RNA by RT-PCR in 21 PNH and AA patients and 20 normal individuals.
WT1 RNA was quantitated by RT-PCR, as described in “Materials and methods.” The values are presented in a logarithmic scale. Vertical axis shows the quantitative values (copies/μg RNA) of WT1 RNA in each individual. NL indicates normal controls.
Quantitation of WT1 RNA by RT-PCR in 21 PNH and AA patients and 20 normal individuals.
WT1 RNA was quantitated by RT-PCR, as described in “Materials and methods.” The values are presented in a logarithmic scale. Vertical axis shows the quantitative values (copies/μg RNA) of WT1 RNA in each individual. NL indicates normal controls.
Discussion
To our knowledge, this is the first investigation of HLA class II alleles or haplotypes in patients with PNH. We found that HLA-DRB1*1501, -DQA1*0102, and -DQB1*0602 alleles and HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype or HLA-DQB1*0602 allele and HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype, determining the presentation of HLA-DR2 in the Japanese, were characteristic in Japanese patients with PNH or AA, respectively, compared with those in healthy individuals. Nakao et al62 reported that the cyclosporine-dependent response of AA is closely related to an HLA class II haplotype of DRB1*1501, DQA1*0102, and DQB1*0602. In addition, the frequency of this haplotype found in 5 of 7 AA-PNH syndrome patients in our study appears to be similar to the frequency of occurrence of GPI-deficient cells in AA patients.63 These facts strongly suggest that a linkage exists in the pathophysiology or pathogenesis between PNH and AA. They also suggest that immune mechanisms in an HLA-restricted manner play an important role in the pathogenesis of primary PNH and AA-PNH syndrome, especially a relationship between the immune mechanisms involving cytotoxic T lymphocytes and negative selection of a PNH clone with a PIG-A gene mutation rather than proliferation of a PNH clone.4,11 Therefore, the specific haplotype in PNH patients was not related to the proportion of CD59−erythrocytes and CD16b− granulocytes. Also, it is possible that the characteristic haplotype or other genes linked to this haplotype tend to cause the circumstances of gene instability in bone marrow hematopoietic precursor cells by some immune mechanisms.64,65 Our views with respect to the HLA class II haplotypes in PNH suggest some important and relevant problems, including the possibility that CD4+ clone(s) elicited in an HLA-restricted manner are related to negative selection of hematopoietic precursor cells in PNH patients,30 and this should be resolved in the future.
We found that the expression of WT1 RNA in 21 PNH patients was significantly higher than in AA patients and healthy individuals. Recently, there has been increasing evidence that WT1 is strongly expressed in normal CD34+ bone marrow stem cells but is only weakly expressed or not expressed in normal mature blood cells.33-35 Moreover, a more recent report66indicated that WT1 expression in human bone marrow and peripheral blood cells is biphasic. WT1 RNA is present in CD34+CD38− cells, absent in lineage-committed precursors, and present again in a subset of the more differentiated populations, including CD11b+ cells and CD19+ B lymphocytes, but not in cells expressing erythroid or megakaryocytic markers. Thus, although it is possible that the increased WT1 expression in PNH patients reflects an altered bone marrow differential, for example one with a lymphocytic infiltrate, we did not find a correlation between the absolute numbers of lymphocytes in bone marrow and the copy numbers of WT1 RNA (data not shown). The expression pattern of the WT1 gene in normal bone marrow cells is unclear at this point, but it is thought that at least CD34+CD38− cells express theWT1 gene, suggesting that expression of the WT1gene is relevant to physical expansion and control of differentiation of immature CD34+ hematopoietic progenitors.35On the other hand, a possible role of WT1 in human leukemogenesis has been proposed. The WT1 gene is highly expressed in all subtypes of human acute leukemia,44 in the accelerated or blast phase of chronic myelogenous leukemia,44 and during MDS stages.46WT1 expression in leukemic blasts was confirmed by immunofluorescence using WT1-specific antibodies.45 These findings have raised the possibility that WT1 can be used as a diagnostic tool to detect minimal residual disease and imminent relapse in treated acute leukemia patients44,67 or to diagnose disease progression in MDS patients.46 In fact, 1 of 3 PNH patients (case 17), who had more than 10 000 copies/μg RNA of WT1 RNA, transformed to acute leukemia 5 months after the diagnosis of PNH.
Subsequently, we found that the levels of WT1 RNA in bone marrow cells from our PNH patients were correlated with the proportion of CD16b− granulocytes, which suggests that the expression of WT1 RNA is related to a PNH clone. This finding suggests the possibility that high expression of WT1 RNA is related to expansion of a PNH clone if the WT1 gene functions as an oncogene. The possibility indicates that expansion of a PNH clone requires a gene abnormality in addition to a PIG-A gene mutation due to gene instability.64,65 Also, a relationship between levels of WT1 RNA and the proportion of CD166− granulocytes may suggest that the expression is related to eliciting cytotoxic T lymphocytes forWT1-expressing hematopoietic stem cells and then to physiologic mechanisms of bone marrow recovery that are not specific to PNH if WT1 protein acts as an autoimmune target for cytotoxic T lymphocytes during selection.4,11,17,18 40-43 However, because of the functional diversity of the WT1 gene, as described above, there is no convincing evidence thatWT1 expression immediately causes or contributes to the expansion or selection of the PNH clone. As in MDS, increased expression of WT1 in PNH will become important when we have more data demonstrating its significance in the course of the disease.
In conclusion, the HLA-DRB1*1501-DQA1*0102-DQB1*0602 haplotype was found with high frequency in PNH patients, which suggests that immune mechanisms in an HLA-restricted manner play an important role in the pathogenesis of PNH and AA-PNH syndrome. In addition, the higher amounts of WT1 RNA in PNH patients compared with AA patients and healthy individuals were correlated with CD16−granulocytes, which suggests that WT1 gene expression is related to a PNH clone, but the role of WT1 gene expression remains unclear.
We are grateful to Dr Hideyoshi Noji, Dr Kazuei Ogawa, and Dr Toshiyuki Ishibashi (Fukushima Medical University, Japan), Dr Yoshiyuki Kamiyama, Dr Yurie Saitoh, Dr Hiroyuki Kambayashi, Dr Tetsugoroh Tanaka, and Dr Shin Matsuda (Ohta Nishino-uchi Hospital, Japan), Dr Yutaka Shiga and Dr Hideo Kimura (General Hobara-chuoh Hospital, Japan), Dr Rokuo Abe (Fukushima-ken Taiyo-no-kuni Hospital, Japan), Dr Masayuki Mita (Hoshi General Hospital, Japan), Dr Kenichi Nakamura (Shirakawa-Kohsei Hospital, Japan), Dr Toshiaki Sai (Iwaki-Kyoritsu Hospital, Japan), Dr Junichi Kameoka (Tohoku University, Japan), Dr Kazuyasu Endoh (Sendai-City Hospital, Japan), and Dr Kyoko Kaneda (National Miyagi Hospital, Japan) for providing the samples from patients with PNH and AA. Also, we wish to thank Dr Yuuji Sugita (Showa University, Japan), who provided the monoclonal antibody to CD59/membrane attack complex-inhibitory factor.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yukio Maruyama, First Department of Internal Medicine, Fukushima Medical University, 1 Hikariga-oka, Fukushima, Fukushima 960-1295, Japan; e-mail: t-shichi@fmu.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal