CD43 is an abundant, heavily glycosylated molecule expressed specifically on the surface of leukocytes and platelets. When leukocytes are at rest, CD43 acts to prevent both homotypic and heterotypic interactions. However, during leukocyte activation CD43 expression is repressed, facilitating the intercellular contact required for chemotaxis, phagocytosis, aggregation, adhesion to endothelium, and transendothelial migration. Consequently, CD43 repression plays a vital role both in innate and acquired immunity. Here we report that a dramatic down-regulation of CD43 mRNA levels occurs during activation of the leukocytic cell line K562. This repression coincides with repression of the transcriptional activity of the CD43 gene promoter. We have determined that heterogeneous nuclear ribonucleoprotein K (hnRNP-K) and Purα act together to mediate repression of the CD43 promoter during K562 activation. The hnRNP-K molecule and Purα bind single-stranded DNA. Therefore, exposure of single-stranded structures within theCD43 promoter probably plays a major role in effectingCD43 repression.
Introduction
CD43 is a heavily glycosylated transmembrane molecule that plays a critical role in leukocyte activation and adhesion.1-3 The importance of CD43 is demonstrated by 2 immunodeficiency diseases, Wiskott-Aldrich syndrome (WAS) and the early stages of HIV infection.4-9 The etiology of both diseases involves the development of defects in CD43. Patients with WAS, which is an X chromosome–linked, recessive disorder, express defective CD43 on the surface of their T lymphocytes. Affected males are subject to recurring opportunistic infections and do not respond to carbohydrate antigens, reflecting defects in T lymphocyte function. In addition, patients suffer from eczema and thrombocytopenia with platelets of reduced size and function. With regard to HIV infection, the finding that all affected individuals have circulating anti-CD43 antibodies has led to the suggestion that these autoantibodies contribute to severe immunodeficiency.8
CD43 is composed of 381 amino acids divided between a 235-residue extracellular region, a 23-residue transmembrane region, and a 123-amino acid C-terminal intracellular region.10,11 The extracellular region contains approximately 84 sialylated O-linked carbohydrate units and appears by electron microscopy to be a rodlike structure extending 45 nm from the cell surface.12Comparison of the rat, mouse, and human sequences indicates that the intracellular domain has been highly conserved during evolution, suggesting a critical function. The intracellular domain anchors CD43 to the cytoskeleton by binding actin, ezrin, and moesin.13 14
When leukocytes are at rest, CD43 maintains their circulation within the bloodstream by preventing intercellular adhesion. This function is achieved by virtue of the size and strong negative charge of the extracellular domain.15-18 During leukocyte activation, CD43 expression is dramatically down-regulated, allowing intercellular interactions mediated by molecules such as the β2integrins. Intercellular interactions are also actively facilitated by CD43, which, due to changes in glycosylation, switches from being an antiadhesion to a proadhesion molecule.19-28
The proadhesive function of CD43 is indicated by its identification as a counterreceptor for galectin-1, intercellular adhesion molecule 1 (ICAM-1), and the macrophage adhesion molecule sialoadhesin.29-32 In addition, antibodies to CD43 have been shown to activate monocytes, B lymphocytes, and dendritic, mast, and natural killer cells.33-39 CD43 binds major histocompatibility complex (MHC) class I molecules and activates T lymphocytes in a manner independent of both the T lymphocyte receptor/CD3 complex and CD28.40-43 The activation signals of this pathway are mediated through phosphorylation of the intracellular domain of CD43 and its physical interaction with the tyrosine kinases Fyn and Lck and the serine/threonine kinase STANK.44-47 Activation signals transduced by CD43 lead to phosphorylation of Shc, induction of the formation of a Shc/Grb2 complex, tyrosine phosphorylation of Vav, mitogen-activated protein kinase activation, and nuclear translocation of ERK2.48Ultimately, these CD43-mediated signals induce the DNA-binding activity of the transcription factors AP-1, nuclear factor of activated T cells (NF-AT), and nuclear factor-κB (NF-κB) and activate the genes encoding interleukin 2, CD69, and CD40-L.49
CD43 is clearly vital to the function of leukocytes. Of particular importance is CD43 repression because this mediates leukocyte adhesion under both physiologic and pathologic circumstances. Regulation of the transcriptional activity of the CD43 gene is the first level at which CD43 expression is controlled. Therefore, we have investigated the possibility that CD43 repression is mediated by transcriptional events. We have previously reported that transcriptional repression of the CD43 promoter occurs during activation of the monocytic cell line U937 and that this is mediated by the transcription factor Purα.50 Here we report that transcriptional repression of the CD43 promoter also occurs during activation of the pre-erythroid/premegakaryocytic cell line K562. We further demonstrate that this repression is mediated by Purα in combination with heterogeneous nuclear ribonucleoprotein K (hnRNP-K). The hnRNP-K molecule as well as Purα interact with single-stranded DNA.51 52 Consequently, CD43repression appears regulated by the interplay of Purα and hnRNP-K with the secondary structure of the CD43 promoter.
Materials and methods
Cells and culture media
Jurkat T lymphocytic cells and K562 pre-erythroid/premegakaryocytic cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mMl-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. K562 cells were activated by the addition of phorbol 12-myristate 13-acetate (PMA; Sigma Chemical, St Louis, MO) to a final concentration of 100 ng/mL. K562 cells stably transfected with the −2/+99 CD43 promoter linked to the luciferasereporter gene were selected using 300 μg/mL zeocin (Invitrogen, Carlsbad, CA). The DNA methyltransferase inhibitor 5-azacytidine (Sigma) was used at a concentration of 10 μM.
Northern blot analysis
Total RNA was isolated from K562 cells by guanidinium isothiocyanate lysis and centrifugation through a cesium chloride cushion. Then, 20 μg of total RNA was subjected to electrophoresis through a 1% agarose/2.2 M formaldehyde gel and subsequently transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech, Piscataway, NJ). This membrane was then hybridized with radiolabeled probes that specifically interact with the coding region of CD43 mRNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The GAPDH- and CD43-coding region probes have been described previously.11 53 Probes were labeled with α[32P]-deoxycytidine triphosphate (dCTP) using a NonaPrimer Kit (Appligene, Illkirch, France). Following hybridization, membranes were washed and subjected to autoradiography.
Southern blot analysis
DNA was subjected to electrophoresis through a 1% agarose gel and transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech). This membrane was then hybridized with the oligonucleotide LUC-2, which had been radiolabeled at its 5′ end and which specifically interacts with the coding region of the firefly luciferase gene. LUC-2 was labeled using T4 polynucleotide kinase and γ[32P]-adenosine triphosphate (ATP). The nucleotide sequence of LUC-2 is: 5′-ATAGCCTTATGCAGTTGCTCT-3′. Following hybridization, membranes were washed and subjected to autoradiography.
Plasmid construction
The activity of the CD43 promoter was assessed using the expression vector pATLuc54 that contains a promoterless firefly luciferase reporter gene. The polymerase chain reaction (PCR) was used to generate a fragment of theCD43 gene representing nucleotides −2 to +99 relative to the most 5′ of the 2 major transcription initiation sites.55 This fragment was then subcloned into the filled-in HindIII site of pATLuc to generate p43Wt.50,56 The correct orientation and nucleotide sequence of the CD43 promoter within p43Wt was verified by DNA sequencing.57 K562 cells carrying within their genome the −2/+99 region of the CD43 gene linked to theluciferase reporter were produced using the plasmid p43Wt/Zeo. This plasmid was generated by inserting between theSalI and PstI sites of p43Wt theXhoI/PstI fragment of pCMV/Zeo (Invitrogen Life Technologies) containing the zeocin-resistance gene. Prior to transfection, p43Wt/Zeo was linearized by digestion withPstI such that the zeocin gene lay downstream and head-to-tail relative to the luciferase gene. The hnRNP-K expression constructs, full-length hnRNP-K, glutathione S-transferase (GST)–RNP-K, and the equivalent vectors empty ofhnRNP-K–coding sequences were kindly provided by David Levens (National Institutes of Health, Bethesda, MD).52The Purα expression construct, pHAPur1, was kindly provided by Edward Johnson (Mount Sinai School of Medicine, New York, NY) and the empty vector equivalent, pHA, produced by religation following liberation of the Purα sequence by RsrII andEcoRI digestion.
Reverse transcriptase-PCR
Total RNA was prepared from K562 cells carrying within their genome the CD43 promoter fused to the luciferasereporter. A GeneRacer Kit (Invitrogen Life Technologies) was used to ligate the RNA oligonucleotide GeneRacer RNA Oligo to the 5′ end specifically of full-length mRNA within the total RNA mixture. This ligated mRNA was then converted to cDNA using reverse transcriptase (RT) and the GeneRacer Oligo dT Primer. Next, this cDNA was used as the template in a PCR with the oligonucleotides GeneRacer 5′ Primer, which represents the DNA equivalent of the 5′ end of the GeneRacer RNA Oligo, and LUC-4, which hybridizes to the coding strand of theluciferase gene. The resulting PCR products were then used as templates in a second round of PCR using the oligonucleotides GeneRacer 5′ Nested Primer, which represents the DNA equivalent of the 3′ end of the GeneRacer RNA Oligo, and LUC-2, which hybridizes to the coding strand of theluciferase gene 5′ to the LUC-4 hybridization site. The products of this second reaction were then cloned using the TOPO TA Cloning Kit for Sequencing (Invitrogen Life Technologies). Clones containing the luciferase coding region were identified by filter hybridization. The radiolabeled probe used in this analysis was a 720 bp EcoRI/PstI fragment isolated from the plasmid p43Wt/Zeo, which spans the CD43promoter and the 5′ end of the luciferase reporter gene. Clones containing the luciferase-coding region were analyzed by DNA sequencing. The nucleotide sequences of the oligonucleotides used in RT-PCR were:
GeneRacer RNA Oligo: 5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′; GeneRacer Oligo dT Primer: 5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18-3′; GeneRacer 5′ Primer: 5′-CGACTGGAGCACGAGGACACTGA-3′; GeneRacer 5′ Nested Primer: 5′-GGACACTGACATGGACTGAAGGAGTA-3′; LUC-2: 5′-ATAGCCTTATGCAGTTGCTCT-3′; and LUC-4: 5′-CACTACGGTAGGCTGCGAAATGTTCATACTGTT-3′.
Transfection
K562 cells were transfected by electroporation as previously described.58 59 Cells were transiently transfected with 23 μg of p43Wt together with 2 μg of the plasmid pRSV-β (Promega, Madison, WI), which contains the lacZ gene encoding β-galactosidase. Each transfection of p43Wt was performed in parallel with a transfection of the promoterless luciferase plasmid pATLuc. Transfected cells were then either left untreated or treated with PMA for 12 hours before harvesting and lysis. Luciferase and β-galactosidase activities were subsequently determined using reagents purchased from Promega and Tropix (Bedford, MA), respectively. Luciferase and β-galactosidase activities, assessed as light output, were measured using a Moonlight Luminometer (Analytical Luminescence Laboratory, San Diego, CA), which integrated peak luminescence 10 seconds after injection of assay buffer. The levels of β-galactosidase activity resulting from different transfections were taken as reflective of relative transfection efficiency and used to correct the measurements of luciferase activity. Transrepression by hnRNP-K in PMA-treated K562 cells was assessed by transient transfections in which 8 μg of pATLuc or p43Wt were mixed with 1 μg of pRSV-β and 16 μg of either full-length hnRNP-K or the equivalent vector empty of hnRNP-K–coding sequences. Full-length hnRNP-K contains the human hnRNP-K–coding region downstream of the cytomegalovirus (CMV) promoter. After correction for transfection efficiency, the level of luciferase activity directed by pATLuc in the presence of full-length hnRNP-K was used to divide the levels of luciferase activity directed by p43Wt also in the presence of full-length hnRNP-K. This calculation yielded the fold above background activity of p43Wt in the presence of hnRNP-K. Equivalent calculations of luciferase activity in the presence of the CMV vector empty of thehnRNP-K–coding region assessed nonspecific effects caused by the vector backbone. The figures resulting from this second set of calculations represented the fold above background activity of p43Wt in the absence of hnRNP-K. K562 cells carrying stably within their genome the CD43 promoter linked to the luciferasereporter were transfected as described above except p43Wt was omitted and the concentration of pRSV-β, full-length hnRNP-K, and its parent varied as described in Figure 6 and Figure 8. In experiments assessing the role of Purα, 6 μg of the Purα expression construct pHAPur1 or 6 μg of its empty parent pHA were used.
Affinity purification
Streptavidin MagneSphere paramagnetic particles (200 μg; Promega) were washed 2 times in 1 mL of phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA), then mixed on a rotating wheel for 30 minutes with 1 mL of 0.2 M NaCl containing 10 μg of a version of the oligonucleotide CD43 PyRo SS56 that was biotinylated at its 3′ end. The particles were then washed twice in 1 × washing buffer (75 mM NaCl, 20 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.5, 1 mM EDTA [ethylenediaminetetraacetic acid], 15% glycerol, 0.05% IGEPAL CA-630). DNA affinity particles were mixed for 30 minutes at room temperature on a rotating wheel with 500 μL of a binding mixture consisting of 1 × binding buffer (70 mM KCl, 5 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 mM dithiothreitol [DTT] and 10% glycerol), 300 μg of nuclear extract prepared from Jurkat cells, and 0.2 μg/μL of poly(dI-dC). The particles were captured with a magnetic stand and washed 3 times with 1 mL of 2 × binding buffer containing 0.2 μg/μL of poly(dI-dC). Bound protein was then eluted by adding 50 μL of washing buffer containing 1 M NaCl. The eluate was then subjected to electrophoretic mobility shift assay (EMSA) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Protein identification by MALDI-TOF-MS
Affinity purified protein from Jurkat nuclear extracts was subjected to SDS-PAGE and stained with Coomassie blue. The stained band was excised from the gel and then cut into small but uniform pieces. The gel was dehydrated with acetonitrile and rehydrated with 100 mM ammonium bicarbonate. Protein was protected from oxidation by incubation at 56°C for 1 hour with 10 mM DTT and the amino terminus protected by treatment with 10 mM iodoacetamide in 100 mM ammonium bicarbonate. Next, gel pieces were subjected to 2 rounds of washing with ammonium bicarbonate and subsequent drying with acetonitrile followed by a 12-hour incubation at 37°C with 12.5 ng/μL of trypsin in 50 mM ammonium bicarbonate.60 61 The masses of the trypsin-digested peptides were determined by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF-MS) using a Voyager DE-PRO instrument (Perseptive Biosystems, Framingham, MA). Protein identification was achieved by mass fingerprinting using the Mascot database created by Matrix Science (London, United Kingdom).
Expression and purification of recombinant GST proteins
The construct GST-RNP-K and its parent vector empty ofhnRNP-K sequences were introduced into the Escherichia coli strain XL-2 (Stratagene, La Jolla, CA). Bacteria were grown in 50 mL of Lauria broth at 37°C until they reached an optical density at 600 nm of 0.6, then recombinant protein expression was induced for 7 hours with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG). Bacteria were washed twice in PBS and resuspended in 1 mL of 1 × binding buffer (70 mM KCl, 5 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 mM DTT, and 10% glycerol) containing complete proteinase inhibitors (Roche Diagnostics, Indianapolis, IN). Bacterial lysates were prepared by sonication and clarified by centrifugation. These preparations were then incubated for 30 minutes at room temperature with 200 μL of glutathione-sepharose beads (Amersham Pharmacia Biotech). Beads were washed 3 times with 1 mL of PBS containing complete inhibitors and bound proteins eluted by adding 800 μL of 50 mM Tris-HCl, pH 8.0, 5 mM reduced glutathione. Finally, proteins were concentrated by centrifugation on a centricon 30 filter unit (Millipore, Bedford, MA) for 15 minutes at 5000g.
EMSA
Oligonucleotides were labeled using T4 polynucleotide kinase and purified through Micro Bio-Spin 6 columns (Bio-Rad Laboratories, Hercules, CA). DNA/protein–binding reactions were carried out in a 20 μL volume. Protein preparations were incubated with or without a molar excess of unlabeled specific or nonspecific competitor oligonucleotides at 4°C for 10 minutes in 1 × binding buffer (70 mM KCl, 5 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 mM DTT, and 10% glycerol) with 2.4 μg/μL of poly(dI-dC). Radiolabeled oligonucleotides were then added and the incubation continued for 20 minutes. DNA/protein complexes were resolved by electrophoresis through 4% or 6% polyacrylamide gels with 4.5 mM Tris-HCl, pH 7.5, 4.5 mM boric acid, 0.1 mM EDTA (0.5 × TBE) and visualized by autoradiography. The oligonucleotides used in the EMSA analyses were: CD43 PyRo SS56: 5′-GGGCCCACTTCCTTTCCCCTTG-3′; CD43 PyRo SSUB: 5′-GGGCCCACUUCCUUUCCCCUUGB-3′ (U is bromouracil; B, biotin); CD43 Mut-1156: 5′-GGGCCCACTTCCTTCATATATG-3′; and NS-SS56: 5′-GAGTTAGCTCACTCATTAGG-3′.
UV cross-linking
The UV cross-linking was performed using the oligonucleotide CD43 PyRo SSUB. This oligonucleotide was the same as the PyRo1-binding probe CD43 PyRo SS56 except that it was biotinylated at its 3′ end and its deoxythymidines were substituted with 5-bromo-2′-deoxyuridine. Then 30 μg of Jurkat nuclear extract were preincubated for 5 minutes in 50 μL of EMSA-binding buffer containing 2.4 μg/μL of poly(dI-dC), 105 cpm of radiolabeled CD43 PyRo SSUB were added and binding performed for 20 minutes on ice in 1.5 mL microcentrifuge tubes. The tubes were then opened and placed in a UV cross-linker (Stratagene) and exposed to 254 nm UV light for 10 minutes. Next, 100 μL of washed streptavidin-coated magnetic particles were added and cross-linked CD43 PyRo SSUB-protein complexes were captured by rotation for 20 minutes at room temperature. Streptavidin particles were sedimented with a magnetic stand, washed 5 times with 500 μL of EMSA-binding buffer, and then resuspended in 25 μL of EMSA buffer containing 2% SDS and 5% 2-mercaptoethanol. Samples were boiled for 5 minutes and centrifuged to pellet the magnetic particles. The supernatant was then analyzed by electrophoresis through a 12% SDS-polyacrylamide gel and subsequent autoradiography.
Results
Down-regulation of CD43 mRNA levels during K562 activation
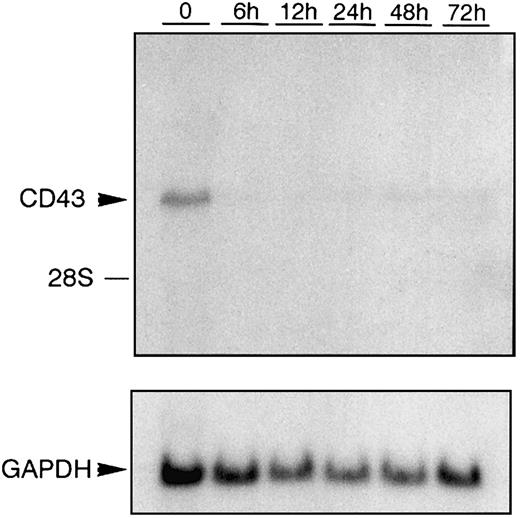
During activation, leukocytes increase in their adhesive capacity mediated in part by a down-regulation of CD43 expression. We have shown that 48 hours after activation of the monocytic cell line U937 with phorbol ester there is a 69% reduction in CD43 mRNA levels.50 A more dramatic reduction in CD43 mRNA levels follows activation of the pre-erythroid/premegakaryocytic cell line K562 (Figure 1). Within 6 hours of K562 cells being activated with phorbol ester the steady-state levels of CD43 mRNA become barely detectable.
Down-regulation of CD43 mRNA during K562 activation.
Total RNA was prepared from K562 cells treated with PMA for the hours indicated. RNA was then subjected to Northern blot analysis using anEcoRI/NcoI fragment (CEM-E/N) isolated from theCD43 cDNA clone pCEM1.7, which has previously been used to detect CD43 mRNA.11 As a control for RNA loading, the same Northern blot hybridized with the CD43-specific probe was subsequently hybridized with a probe that specifically recognizes GAPDH mRNA.53 The position of migration of 28S ribosomal RNA is marked.
Down-regulation of CD43 mRNA during K562 activation.
Total RNA was prepared from K562 cells treated with PMA for the hours indicated. RNA was then subjected to Northern blot analysis using anEcoRI/NcoI fragment (CEM-E/N) isolated from theCD43 cDNA clone pCEM1.7, which has previously been used to detect CD43 mRNA.11 As a control for RNA loading, the same Northern blot hybridized with the CD43-specific probe was subsequently hybridized with a probe that specifically recognizes GAPDH mRNA.53 The position of migration of 28S ribosomal RNA is marked.
Repression of CD43 promoter activity during K562 activation
Previous studies have indicated that the cis-acting elements responsible for directing the leukocytic expression of theCD43 gene are located in the proximal promoter region.50,56,62 63 Transfection experiments using the expression construct p43Wt, which contains the CD43 promoter spanning nucleotides −2 to +99, indicate that the proximal promoter also contains the elements required for CD43 repression during K562 activation. Specifically, we found that expression of p43Wt is repressed by 39% on PMA-mediated activation of K562 cells (Figure2).
Repression of the CD43 promoter during K562 activation.
The construct p43Wt containing nucleotides −2 to +99 of theCD43 gene promoter was transfected into K562 cells along with the control plasmid pRSV-β encoding β-galactosidase. Transfected cells were then either left untreated (−PMA) or treated for 12 hours with PMA (+PMA) prior to harvesting. Luciferase values were measured and normalized against β-galactosidase levels to correct for transfection efficiency. Expressed as histograms are the levels of luciferase activity directed by p43Wt in untreated and PMA-treated K562 cells after division by the background activity conferred by the control plasmid pATLuc. Each histogram represents the mean ± SD of 3 independent transfection experiments.
Repression of the CD43 promoter during K562 activation.
The construct p43Wt containing nucleotides −2 to +99 of theCD43 gene promoter was transfected into K562 cells along with the control plasmid pRSV-β encoding β-galactosidase. Transfected cells were then either left untreated (−PMA) or treated for 12 hours with PMA (+PMA) prior to harvesting. Luciferase values were measured and normalized against β-galactosidase levels to correct for transfection efficiency. Expressed as histograms are the levels of luciferase activity directed by p43Wt in untreated and PMA-treated K562 cells after division by the background activity conferred by the control plasmid pATLuc. Each histogram represents the mean ± SD of 3 independent transfection experiments.
Identification of a putative repressor of the CD43promoter
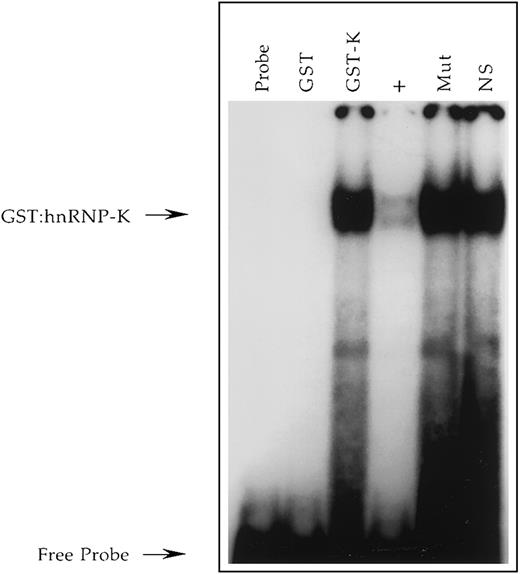
We have previously determined that the uncloned single-stranded DNA-binding activity PyRo1 interacts with the CD43 promoter and is induced concomitant with CD43 repression during U937 activation.56 Consequently, PyRo1 represents a potential means by which CD43 repression is effected. To determine the function of PyRo1 we undertook its molecular cloning. PyRo1 was purified from Jurkat T lymphocytic cells by affinity capture and digested with trypsin and its resulting peptides subjected to analysis by mass spectrometry. The fragmentation patterns produced from the PyRo1 peptides were then compared with those of known proteins deposited in the Mascot database created by Matrix Science. The most significant match to a known human protein was with heterogeneous nuclear ribonucleoprotein K (hnRNP-K).64 Next, a fusion protein of hnRNP-K and glutathione S-transferase (GST) was produced in bacteria. The fusion protein was purified and shown to bind a radiolabeled single-stranded oligonucleotide representing the PyRo1-binding site within the CD43 gene promoter (Figure3). This binding was effectively competed by an unlabeled excess of the PyRo1-binding site. However, binding failed to be competed with an identical molar excess of a mutant version of the binding site, which fails to support PyRo1 interaction.56 These studies demonstrate that the DNA-binding characteristics of PyRo1 and hnRNP-K are indistinguishable. In addition, UV cross-linking demonstrated that the apparent molecular mass of PyRo1 is in the range of 50 to 65 kDa (Figure4), which is comparable to that reported for hnRNP-K.65 Originally, hnRNP-K was identified as an RNA-binding protein and recently its consensus RNA-binding site was determined as 5-′UC3-4(U/A)2-3′.64,66 The single-stranded DNA equivalent of this consensus is present in the PyRo1-binding site within the CD43 promoter, and mutation analysis has demonstrated that this sequence is critical for PyRo1 binding.56 Consequently, by the criteria of molecular cloning, molecular mass analysis and nucleic acid–binding characteristics, PyRo1 and hnRNP-K are indistinguishable.
Demonstration in vitro that recombinant hnRNP-K binds the +18/+39 region of the CD43 promoter.
A radiolabeled single-stranded oligonucleotide, CD43 PyRo SS56, representing nucleotides +18 to +39 of the sense strand of the CD43 gene was incubated with no protein (Probe), purified GST (GST), or purified GST/hnRNP-K fusion protein (GST-K). Binding reactions containing GST/hnRNP-K were performed in the absence (GST-K) or presence (+) of a 100-fold molar excess of unlabeled CD43 PyRo SS, the presence of Mut-11 (Mut) representing a mutant version of CD43 PyRo SS, which fails to support PyRo1 binding56 or the presence of the unrelated oligonucleotide NS-SS (NS). The free probe and probe bound by the GST/hnRNP-K protein (GST:hnRNP-K) are denoted by arrows.
Demonstration in vitro that recombinant hnRNP-K binds the +18/+39 region of the CD43 promoter.
A radiolabeled single-stranded oligonucleotide, CD43 PyRo SS56, representing nucleotides +18 to +39 of the sense strand of the CD43 gene was incubated with no protein (Probe), purified GST (GST), or purified GST/hnRNP-K fusion protein (GST-K). Binding reactions containing GST/hnRNP-K were performed in the absence (GST-K) or presence (+) of a 100-fold molar excess of unlabeled CD43 PyRo SS, the presence of Mut-11 (Mut) representing a mutant version of CD43 PyRo SS, which fails to support PyRo1 binding56 or the presence of the unrelated oligonucleotide NS-SS (NS). The free probe and probe bound by the GST/hnRNP-K protein (GST:hnRNP-K) are denoted by arrows.
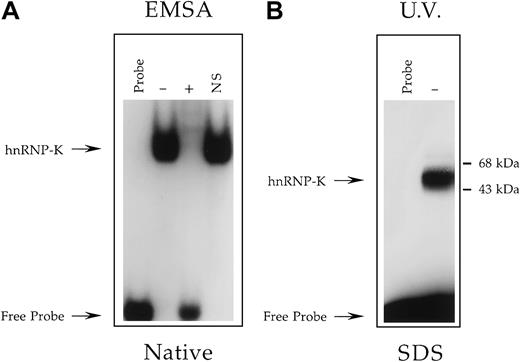
Determination of the molecular mass of the Jurkat nuclear factor that binds the +18/+39 region of the CD43promoter.
(A) EMSA analysis performed as described in Figure 3 except binding reactions contained the radiolabeled oligonucleotide CD43 PyRo SSUB, which is both bromouracil- and biotin-modified, and either no protein extract (Probe) or a nuclear extract prepared from Jurkat cells. Binding reactions containing Jurkat nuclear extract were performed in the absence (−) or presence (+) of a 100-fold molar excess of unlabeled CD43 PyRo SS56 or the presence of the unrelated oligonucleotide NS-SS (NS). Marked with arrows are the positions of migration through a native polyacrylamide gel of CD43 PyRo SSUB unbound by protein (Free Probe) and bound by hnRNP-K. This analysis established that the protein-binding characteristics of CD43 PyRo SSUB are indistinguishable from its equivalent CD43 PyRo SS, which is modified neither with bromouracil nor biotin56. (B) The oligonucleotide CD43 PyRo SSUB was radiolabeled, incubated with (−) or without (Probe) a nuclear extract prepared from Jurkat cells and then exposed to UV light. CD43 PyRo SSUB was captured by streptavidin-coated magnetic particles, washed, and subjected to electrophoresis through a 12% SDS-polyacrylamide gel and autoradiography.
Determination of the molecular mass of the Jurkat nuclear factor that binds the +18/+39 region of the CD43promoter.
(A) EMSA analysis performed as described in Figure 3 except binding reactions contained the radiolabeled oligonucleotide CD43 PyRo SSUB, which is both bromouracil- and biotin-modified, and either no protein extract (Probe) or a nuclear extract prepared from Jurkat cells. Binding reactions containing Jurkat nuclear extract were performed in the absence (−) or presence (+) of a 100-fold molar excess of unlabeled CD43 PyRo SS56 or the presence of the unrelated oligonucleotide NS-SS (NS). Marked with arrows are the positions of migration through a native polyacrylamide gel of CD43 PyRo SSUB unbound by protein (Free Probe) and bound by hnRNP-K. This analysis established that the protein-binding characteristics of CD43 PyRo SSUB are indistinguishable from its equivalent CD43 PyRo SS, which is modified neither with bromouracil nor biotin56. (B) The oligonucleotide CD43 PyRo SSUB was radiolabeled, incubated with (−) or without (Probe) a nuclear extract prepared from Jurkat cells and then exposed to UV light. CD43 PyRo SSUB was captured by streptavidin-coated magnetic particles, washed, and subjected to electrophoresis through a 12% SDS-polyacrylamide gel and autoradiography.
Repression of the CD43 gene promoter by hnRNP-K
Following identification of hnRNP-K as a candidate mediator ofCD43 repression, we sought to determine if this factor could indeed function in such a capacity. Transfection of 16 μg of a plasmid expressing hnRNP-K indicated that in PMA-treated K562 cells hnRNP-K does indeed repress the transcriptional activity of p43Wt. The level of this repression averaged 38% (Figure5). It has been reported that the effects of hnRNP-K on transcriptional activity are dependent on chromosomal structures.67 Consequently, we generated a pool of K562 cell lines in which the −2/+99 CD43 promoter linked to theluciferase reporter was randomly integrated into the genome. Transfection of the hnRNP-K expression plasmid into this stable cell line pool subsequently treated with PMA repressedluciferase reporter gene activity by an average of 62% (Figure 6). Consequently, hnRNP-K significantly represses the CD43 promoter either in a chromosomal or extrachromosomal context.
Repression by hnRNP-K of the CD43 promoter present within an extrachromosomal plasmid.
Eight micrograms of the luciferase reporter construct p43Wt were transfected into K562 cells mixed with either 16 μg of full-length hnRNP-K, which expresses hnRNP-K, or 16 μg of its parent vector empty of hnRNP-K–coding sequences. One microgram of the β-galactosidase expression plasmid pRSV-β was also included in each transfection to control for transfection efficiency. Transfected cells were treated with PMA for 12 hours and harvested and luciferase and β-galactosidase assays performed. The levels of β-galactosidase activity were taken as reflective of transfection efficiency and used to correct the luciferase assay results. Depicted as histograms are the levels of luciferase activity directed by p43Wt divided by those directed by the empty vector pATLuc in the presence of full-length hnRNP-K (+hnRNP-K) or the equivalent empty vector (−hnRNP-K). The means of these levels ± SD resulting from 3 independent experiments are displayed.
Repression by hnRNP-K of the CD43 promoter present within an extrachromosomal plasmid.
Eight micrograms of the luciferase reporter construct p43Wt were transfected into K562 cells mixed with either 16 μg of full-length hnRNP-K, which expresses hnRNP-K, or 16 μg of its parent vector empty of hnRNP-K–coding sequences. One microgram of the β-galactosidase expression plasmid pRSV-β was also included in each transfection to control for transfection efficiency. Transfected cells were treated with PMA for 12 hours and harvested and luciferase and β-galactosidase assays performed. The levels of β-galactosidase activity were taken as reflective of transfection efficiency and used to correct the luciferase assay results. Depicted as histograms are the levels of luciferase activity directed by p43Wt divided by those directed by the empty vector pATLuc in the presence of full-length hnRNP-K (+hnRNP-K) or the equivalent empty vector (−hnRNP-K). The means of these levels ± SD resulting from 3 independent experiments are displayed.
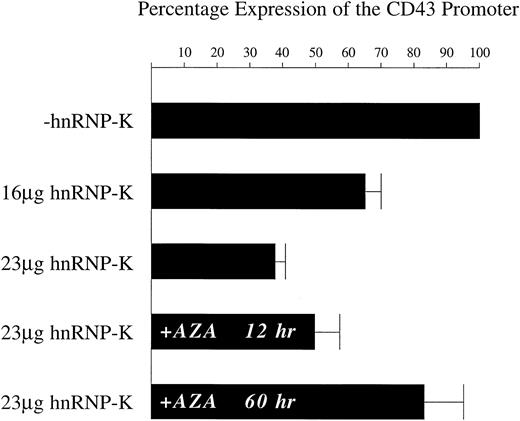
Repression by hnRNP-K of the CD43 promoter present within chromosomal DNA.
K562 cells were transfected with the linearized plasmid p43Wt/Zeo, which was derived from p43Wt by insertion of thezeocin-resistance gene. K562 cells in which the p43Wt/Zeo plasmid was stably integrated within the genome were selected by treatment with zeocin. The mixed pool of zeocin-resistant cells was then transfected with 2 μg of pRSV-β mixed with either 23 μg of full-length hnRNP-K or 23 μg of its parent vector empty ofhnRNP-K–coding sequences. Transfections were also performed using 9 μg of pRSV-β mixed with either 16 μg of full-length hnRNP-K or 16 μg of its parent. In those experiments in which the role of DNA methylation was assessed, cells were either untreated or pretreated for 48 hours with 5-azacytidine prior to transfection with 2 μg of pRSV-β mixed with either 23 μg of full-length hnRNP-K or 23 μg of its parent. Following all transfections, cells were treated with PMA for 12 hours prior to harvesting and assay of luciferase and β-galactosidase activities. In experiments assessing the influence of DNA methylation, transfected cells, untreated or pretreated with 5-azacytidine, were treated simultaneously with PMA and 5-azacytidine for 12 hours prior to harvesting. The levels of β-galactosidase activity were taken as reflective of transfection efficiency and used to correct the luciferase assay results. The level of luciferase activity directed by the CD43 promoter in the presence of the vector empty of hnRNP-K coding sequences (−hnRNP-K) was assigned a value of 100%. The luciferase levels directed by theCD43 promoter in matched, parallel transfections using full-length hnRNP-K (+hnRNP-K) were calculated as a proportion of the 100% value. The means of these proportional values ± SD resulting from 3 independent experiments are displayed.
Repression by hnRNP-K of the CD43 promoter present within chromosomal DNA.
K562 cells were transfected with the linearized plasmid p43Wt/Zeo, which was derived from p43Wt by insertion of thezeocin-resistance gene. K562 cells in which the p43Wt/Zeo plasmid was stably integrated within the genome were selected by treatment with zeocin. The mixed pool of zeocin-resistant cells was then transfected with 2 μg of pRSV-β mixed with either 23 μg of full-length hnRNP-K or 23 μg of its parent vector empty ofhnRNP-K–coding sequences. Transfections were also performed using 9 μg of pRSV-β mixed with either 16 μg of full-length hnRNP-K or 16 μg of its parent. In those experiments in which the role of DNA methylation was assessed, cells were either untreated or pretreated for 48 hours with 5-azacytidine prior to transfection with 2 μg of pRSV-β mixed with either 23 μg of full-length hnRNP-K or 23 μg of its parent. Following all transfections, cells were treated with PMA for 12 hours prior to harvesting and assay of luciferase and β-galactosidase activities. In experiments assessing the influence of DNA methylation, transfected cells, untreated or pretreated with 5-azacytidine, were treated simultaneously with PMA and 5-azacytidine for 12 hours prior to harvesting. The levels of β-galactosidase activity were taken as reflective of transfection efficiency and used to correct the luciferase assay results. The level of luciferase activity directed by the CD43 promoter in the presence of the vector empty of hnRNP-K coding sequences (−hnRNP-K) was assigned a value of 100%. The luciferase levels directed by theCD43 promoter in matched, parallel transfections using full-length hnRNP-K (+hnRNP-K) were calculated as a proportion of the 100% value. The means of these proportional values ± SD resulting from 3 independent experiments are displayed.
Repression mediated by hnRNP-K is dependent on DNA methylation
Previous studies have demonstrated that in nonhematopoietic cells the CD43 promoter is maintained in an inactive state by DNA methylation.75,76 This finding suggested that methylation may also influence CD43 repression in hematopoietic cells as they undergo differentiation. Therefore, we tested whether hnRNP-K repression of the CD43 promoter was dependent on methylation. The pool of stable cell lines containing theCD43 promoter linked to luciferase was treated for 48 hours with the DNA methyltransferase inhibitor 5-azacytidine.77 These treated cells were then transfected with the hnRNP-K expression plasmid and incubated for 12 hours in the presence of PMA and 5-azacytidine. Under these circumstances, theCD43 promoter was repressed by only 17% (Figure 6). Without a 48-hour pretreatment but with a 12-hour treatment with 5-azacytidine and PMA immediately after transfection, hnRNP-K repressed theCD43 promoter by 50%. In cells treated only with PMA after transfection, hnRNP-K represses the CD43 promoter by 62% (Figure 6). These results indicate that exposure of the hnRNP-K expression plasmid to 5-azacytidine for 12 hours influences its ability to effect repression by only 12%. In contrast, exposure of theCD43 promoter to 5-azacytidine for 48 hours reduces the repressive capacity of hnRNP-K by 33%. Consequently, repression of theCD43 promoter by hnRNP-K is substantially dependent on this promoter being methylated.
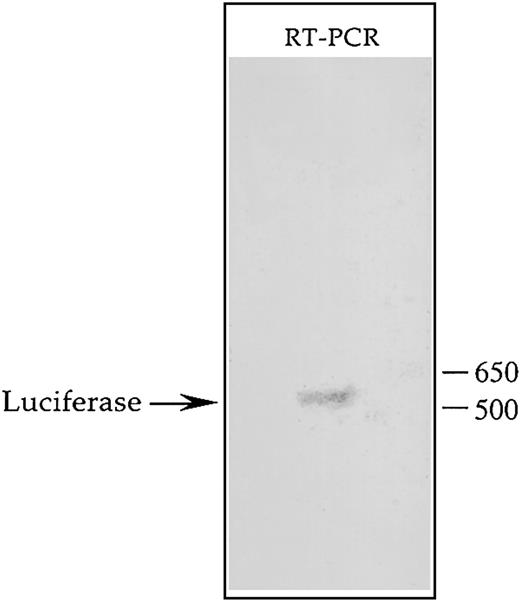
Determination of the transcription start site of theCD43 gene when linked to luciferase and integrated within the genome of K562
Transcription of the CD43 gene has been shown to be initiated at 2 major sites between which lies the binding site for hnRNP-K.55 This factor is known to bind RNA and has been implicated in mediating RNA stability and transport.68-74 Consequently, if transcription of theCD43/luciferase gene is initiated at the upstream of the 2 potential start sites, the effect of hnRNP-K on luciferase activity may reflect mechanisms mediated by RNA binding rather than transcriptional control mechanisms. Due to this reasoning, we identified the transcription start site of theCD43/luciferase gene present in the genome of K562 using RT-PCR coupled with rapid amplification of cDNA ends (RACE) technology. Southern blot analysis of the RT-PCR products resulting from this procedure revealed a single discrete cDNA containing theluciferase-coding region (Figure7). Cloning and sequencing of this cDNA determined that the downstream of the 2 potential transcription start sites is used by the CD43 promoter when linked to luciferase and integrated within K562. Therefore, the transcript produced from this promoter does not contain a binding site for hnRNP-K. Consequently, repression of theCD43/luciferase fusion gene by hnRNP-K is likely mediated by transcriptional mechanisms as opposed to mechanisms involving RNA binding such as decreased RNA stability or transport.
Southern blot analysis of the transcript produced from the CD43 promoter linked to luciferase and integrated within the K562 genome.
The mixed pool of K562 cells containing within their genome theCD43/luciferase fusion gene was treated for 24 hours with PMA and then total RNA isolated using an RNeasy Maxi Kit (Qiagen, Valencia, CA). A GeneRacer Kit (Invitrogen Life Technologies) was used to ligate the GeneRacer RNA Oligo specifically to the 5′ end of full-length mRNA, which was then reverse-transcribed using the GeneRacer Oligo dT Primer. The resulting products were used as templates in a PCR containing the primer LUC-4, which hybridizes to the coding strand of the luciferase gene, and the GeneRacer 5′ Primer, which represents the DNA equivalent of the 5′ end of the GeneRacer RNA Oligo. PCR products were then subjected to Southern blot analysis using as probe the radiolabeled oligonucleotide LUC-2, which hybridizes to the coding strand of the luciferasegene further 5′ than does LUC-4. The hybridization signal detected by autoradiography is depicted.
Southern blot analysis of the transcript produced from the CD43 promoter linked to luciferase and integrated within the K562 genome.
The mixed pool of K562 cells containing within their genome theCD43/luciferase fusion gene was treated for 24 hours with PMA and then total RNA isolated using an RNeasy Maxi Kit (Qiagen, Valencia, CA). A GeneRacer Kit (Invitrogen Life Technologies) was used to ligate the GeneRacer RNA Oligo specifically to the 5′ end of full-length mRNA, which was then reverse-transcribed using the GeneRacer Oligo dT Primer. The resulting products were used as templates in a PCR containing the primer LUC-4, which hybridizes to the coding strand of the luciferase gene, and the GeneRacer 5′ Primer, which represents the DNA equivalent of the 5′ end of the GeneRacer RNA Oligo. PCR products were then subjected to Southern blot analysis using as probe the radiolabeled oligonucleotide LUC-2, which hybridizes to the coding strand of the luciferasegene further 5′ than does LUC-4. The hybridization signal detected by autoradiography is depicted.
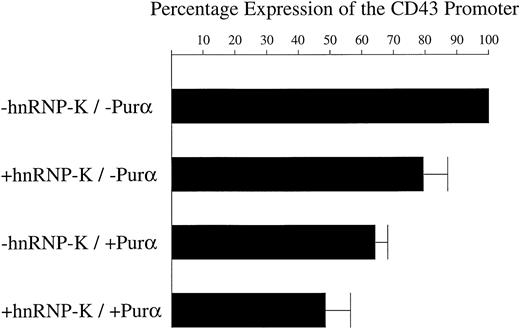
hnRNP-K acts together with Purα to effect repression of the CD43 promoter
We have previously demonstrated that Purα interacts with theCD43 promoter immediately upstream of the region that we show here binds hnRNP-K.50 In the promonocytic cell line U937 treated with PMA, Purα represses the CD43 promoter. Therefore, it was of interest to determine whether Purα also mediates repression within K562 cells and whether it works with or against hnRNP-K. In transfection experiments using expression plasmids constitutively expressing hnRNP-K or Purα, we found that alone, Purα represses the CD43/luciferase gene present within the K562 genome by 36% (Figure 8). However, when recombinant Purα is expressed together with recombinant hnRNP-K, repression of the CD43 promoter is increased to 51%. Consequently, hnRNP-K and Purα appear to work together to effect repression.
hnRNP-K and Purα act together to repress theCD43 promoter.
The β-galactosidase expression construct pRSV-β was mixed with 1 of 4 different combinations of plasmids: (1) pHAPur1 and full-length hnRNP-K, which express Purα and hnRNP-K, respectively; (2) pHAPur1 and the empty vector equivalent of full-length hnRNP-K; (3) full-length hnRNP-K and pHA, which represents the empty vector equivalent of pHAPur1; and (4) both empty vectors. These 4 DNA mixtures were then transfected into the mixed pool of K562 cells containing theCD43/luciferase fusion and treated for 12 hours with PMA. Cells were harvested, lysed, and luciferase and β-galactosidase assays performed. The levels of β-galactosidase activity were taken as reflective of transfection efficiency and used to correct the luciferase assay results. Using these corrected values, the level of luciferase activity directed by the CD43 promoter in the presence of the empty vectors was assigned a value of 100%. The level of luciferase activity directed by the promoter in the presence of constructs expressing Purα or hnRNP-K was calculated as a proportion of the 100% value. The means of these levels ± SD resulting from 3 independent experiments are displayed as histograms.
hnRNP-K and Purα act together to repress theCD43 promoter.
The β-galactosidase expression construct pRSV-β was mixed with 1 of 4 different combinations of plasmids: (1) pHAPur1 and full-length hnRNP-K, which express Purα and hnRNP-K, respectively; (2) pHAPur1 and the empty vector equivalent of full-length hnRNP-K; (3) full-length hnRNP-K and pHA, which represents the empty vector equivalent of pHAPur1; and (4) both empty vectors. These 4 DNA mixtures were then transfected into the mixed pool of K562 cells containing theCD43/luciferase fusion and treated for 12 hours with PMA. Cells were harvested, lysed, and luciferase and β-galactosidase assays performed. The levels of β-galactosidase activity were taken as reflective of transfection efficiency and used to correct the luciferase assay results. Using these corrected values, the level of luciferase activity directed by the CD43 promoter in the presence of the empty vectors was assigned a value of 100%. The level of luciferase activity directed by the promoter in the presence of constructs expressing Purα or hnRNP-K was calculated as a proportion of the 100% value. The means of these levels ± SD resulting from 3 independent experiments are displayed as histograms.
Discussion
On the surface of resting leukocytes CD43 prevents cellular interaction. However, during leukocyte activation the glycosylation pattern of CD43 changes and there is a down-regulation of the overall amount of CD43 expressed on the cell surface. These alterations in both the qualitative and quantitative expression of CD43 result in the promotion of both homotypic and heterotypic leukocyte interactions. Using in vitro models of leukocyte activation we have found that down-regulation of CD43 is mediated by repression of the transcriptional activity of the CD43 gene promoter. Previously, we demonstrated that the transcription factor Purα mediates repression of the CD43 promoter.50Here we demonstrate that hnRNP-K also mediates CD43 gene repression. The binding sites for Purα and hnRNP-K lie adjacent to one another within the CD43 promoter. Purα interacts with the −2/+17 region of the CD43 gene and hnRNP-K interacts with the +18/+39 region. Both factors are induced during U937 activation concomitant with repression of CD43 promoter activity.50 56
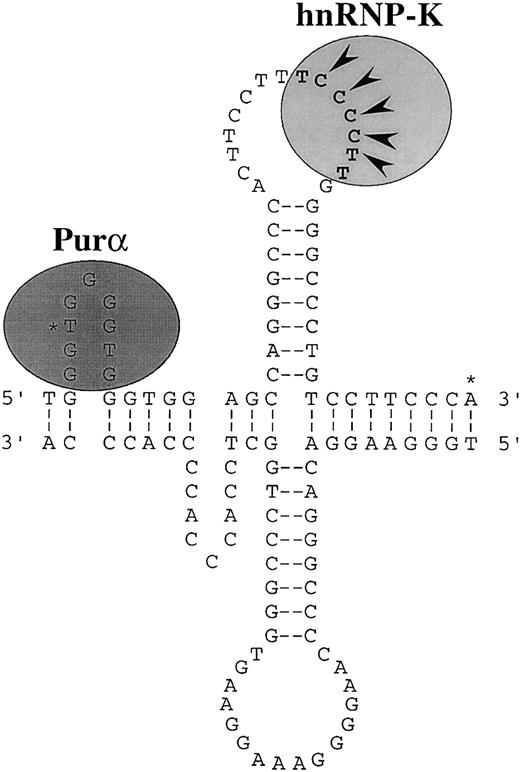
Purα and hnRNP-K interact preferentially with single-stranded DNA indicating that single-stranded structures within the CD43promoter are probably important in regulating its activity. Nuclease S1 analysis has established that single-stranded structures can exist within the CD43 promoter.56Furthermore, analysis of the primary sequence of the promoter reveals repeats that could form slippage or cruciform structures.78 The single-stranded loop of a slippage structure within the −2/+17 region would support efficient Purα binding, whereas the loop of a cruciform structure within the +18/+39 region would support efficient binding of hnRNP-K (Figure 9). The hnRNP-K protein was originally characterized as an RNA-binding protein.64Recently, the optimal RNA structures that bind hnRNP-K have been determined and a loop containing the consensus 5′-UC3-4(U/A)2-3′ has been identified as being of primary importance.66 The single-stranded DNA equivalent of this consensus is present within the loop of the potential cruciform structure of the CD43 promoter (Figure9).
Putative secondary structure of the CD43promoter.
Misalignment of direct repeats of the sequence GGTGG may form slippage structures at the 5′ end of the promoter. The single-stranded region on the sense strand of these structures would support efficient binding of Purα. Two types of slippage structures are possible. One is illustrated. Pairing of an inverted repeat of the sequence CAGGGCCC could form a cruciform structure immediately downstream of the slippage structures. This possible cruciform is illustrated. The single-stranded loop region on the sense strand of the cruciform would support efficient binding of hnRNP-K. The DNA equivalent of the consensus RNA binding site of hnRNP-K is marked by bold characters.66 The nucleotides defined by mutation analysis as being critical for hnRNP-K binding are marked with arrowheads.56 The 2 major transcription initiation sites of the CD43 gene are marked with asterisks.55
Putative secondary structure of the CD43promoter.
Misalignment of direct repeats of the sequence GGTGG may form slippage structures at the 5′ end of the promoter. The single-stranded region on the sense strand of these structures would support efficient binding of Purα. Two types of slippage structures are possible. One is illustrated. Pairing of an inverted repeat of the sequence CAGGGCCC could form a cruciform structure immediately downstream of the slippage structures. This possible cruciform is illustrated. The single-stranded loop region on the sense strand of the cruciform would support efficient binding of hnRNP-K. The DNA equivalent of the consensus RNA binding site of hnRNP-K is marked by bold characters.66 The nucleotides defined by mutation analysis as being critical for hnRNP-K binding are marked with arrowheads.56 The 2 major transcription initiation sites of the CD43 gene are marked with asterisks.55
The site within the CD43 promoter that binds hnRNP-K lies adjacent to a site that interacts with Sp1.50,63,76 This Sp1-binding site overlaps the region that binds Purα. Transient transfections have indicated that, unlike hnRNP-K and Purα, Sp1 functions to activate the CD43 promoter.63Because Sp1 binds double-stranded DNA, a mechanism of repression can be envisioned in which a double-stranded CD43 promoter supporting Sp1 binding and transcriptional activation acquires a single-stranded conformation supporting hnRNP-K and Purα binding. Such opening of the CD43 promoter may in fact be mediated by Purα and hnRNP-K themselves. This possibility is supported by the finding that Purα is capable of displacing an oligonucleotide annealed to single-stranded M13 DNA.79 In addition, hnRNP-K has been shown to impart a single-stranded conformation to the CT element of the c-myc promoter contained within a supercoiled plasmid.80 However, in the context of thec-myc promoter, hnRNP-K acts as a transcriptional activator not a repressor as it does within the CD43promoter.81 82 Therefore, although DNA unwinding may be intrinsic properties of Purα and hnRNP-K, the functional consequences of this activity appear dependent on promoter architecture and probably also cellular environment.
In addition to DNA remodeling, Purα and hnRNP-K may repress theCD43 promoter by inhibiting the factors that induce its transcriptional activity. The hnRNP-K molecule binds TATA binding protein (TBP) and, therefore, may inhibit the basal transcription machinery.81 Purα has been shown to bind Sp1 and hnRNP-K inhibits Sp1 binding to the promoter of the gene encoding the β4 subunit of the nicotinic acetylcholine receptor.83 84 Consequently, hnRNP-K and Purα may inhibit CD43 expression not only by altering the secondary structure of the Sp1-binding site but also by inhibiting the functional activity of the Sp1 protein.
A third mechanism by which hnRNP-K and Purα may inhibitCD43 expression is by the recruitment of repressor molecules. In this regard, it has been shown in vitro that hnRNP-K can bind the murine repressors Zik1 and Eed expressed in B lymphocytes.85 86 Our search of the human genome database failed to identify a known or potential human protein with homology to Eed. However, we did identify a hypothetical human protein that exhibits 44% amino acid identity to Zik1 (accession no. AC003682). Whether, this protein is expressed, interacts with hnRNP-K in vivo, or acts as a repressor is unknown.
Previous studies have shown that in nonhematopoietic cells theCD43 gene is maintained in a transcriptionally inactive state by methylation of CpG dinucleotides within the promoter.75 These methylated dinucleotides are then able to bind the methyl-CpG–binding protein MeCP2.76 We have shown that repression of the CD43 promoter by hnRNP-K is dependent on DNA methylation. This finding suggests that repression of the CD43 promoter during hematopoietic differentiation may be mediated by an increase in its degree of methylation. Such an increase could result in the recruitment of methyl-CpG–binding proteins or the more effective recruitment of hnRNP-K or both.
Beyond CD43 repression, leukocyte adhesion is induced by up-regulation of the β2 integrin family of proadhesion molecules. We have shown that Purα activates expression of the CD11c β2 integrin promoter.87 In addition, PyRo1, which we have now identified as hnRNP-K, interacts not only with the CD43 gene promoter but also the promoters of the CD11a, CD11b, CD11c, andCD11d genes.56 Deletion of the PyRo1/hnRNP-K–binding site within the CD11c promoter severely impairs its transcriptional activity in PMA-treated U937 cells. Therefore, whereas Purα and hnRNP-K repress CD43expression during leukocyte activation, they activate expression of theβ2 integrins. Consequently, hnRNP-K and Purα appear to help coordinate the changes in gene expression that mediate leukocyte adhesion.
We would like to thank David Levens for providing full-length hnRNP-K, GST-RNP-K, and the equivalent vectors empty of hnRNP-K coding sequences. In addition, we would like to thank Edward Johnson and Sharon Barr for providing the expression vector pHAPur1.
Supported by National Institutes of Health grants DK43351 and DK50779. N.D.S. was supported by a postdoctoral grant awarded by the Association pour la Recherche sur le Cancer. Additional support was provided by grant DAMD17-00-1-0255. In regard to this grant the US Army Medical Research Acquisition Activity (820 Chandler St, Fort Detrick, MD 21702) is the awarding and administering acquisition office.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Carl Simon Shelley, Renal Unit, Massachusetts General Hospital, 149 13th St, Charlestown, MA 02129; e-mail: shelley@receptor.mgh.harvard.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal