Gene patterns of expression in purified CD34+ bone marrow cells from 7 patients with low-risk myelodysplastic syndrome (MDS) and 4 patients with high-risk MDS were compared with expression data from CD34+ bone marrow cells from 4 healthy control subjects. CD34+ cells were isolated by magnetic cell separation, and high-density oligonucleotide microarray analysis was performed. For confirmation, the expression of selected genes was analyzed by real-time polymerase chain reaction. Class membership prediction analysis selected 11 genes. Using the expression profile of these genes, we were able to discriminate patients with low-risk from patients with high-risk MDS and both patient groups from the control group by hierarchical clustering (Spearman confidence). The power of these 11 genes was verified by applying the algorithm to an unknown test set containing expression data from 8 additional patients with MDS (3 at low risk, 5 at high risk). Patients at low risk could be distinguished from those at high risk by clustering analysis. In low-risk MDS, we found that the retinoic-acid–induced gene (RAI3), the radiation-inducible, immediate-early response gene (IEX1), and the stress-induced phosphoprotein 1 (STIP1) were down-regulated. These data suggest that CD34+cells from patients with low-risk MDS lack defensive proteins, resulting in their susceptibility to cell damage. In summary, we propose that gene expression profiling may have clinical relevance for risk evaluation in MDS at the time of initial diagnosis. Furthermore, this study provides evidence that in MDS, hematopoietic stem cells accumulate defects that prevent normal hematopoiesis.
Introduction
The underlying causes of primary myelodysplastic syndrome (MDS) are still being defined. A proposal for the multistep pathogenesis of MDS is that after initial damage to the progenitor cell by a toxin or a spontaneous mutation, several additional alterations may affect these cells and provide them with a growth advantage. These alterations can influence the expression of cell-cycle–related genes, including checkpoint and mismatch repair genes, transcription factors, and tumor-suppressor genes. In addition, early MDS was associated with an elevated ratio of apoptosis to proliferation,1 but the mechanisms for this finding are not yet established.
The defect of the hematopoietic stem cell in MDS is not well characterized. One technical problem is that most of the experiments that use clinical samples from MDS patients were performed with low-density, nonadherent cells from the bone marrow of these patients. This may be adequate for high-risk MDS and acute myeloid leukemia (AML) evolved from MDS because of the more uniform blast population in the bone marrow. In contrast, in low-risk MDS, bone marrow cells are heterogenous. Therefore, molecular abnormalities characteristic of malignant cells are more difficult to find in low-risk MDS than in high-risk MDS or AML. Because CD34 expression is a marker for hematopoietic stem cells, experiments to detect abnormalities have focused on the CD34+ cells from patients with MDS.2-4
To expand the understanding of genetic defects of the hematopoietic stem cell in MDS, we performed oligonucleotide microarray analysis on highly purified CD34+ cells from patients with MDS who were clinically well characterized. All MDS samples used for our analyses had the morphologic and immunologic features of MDS. In addition, the defects in proliferation and differentiation of these CD34+cells were demonstrated on a routine basis in suspension cultures (data not shown).2 As a control, we investigated the gene expression of CD34+ cells selected from the bone marrow of healthy control subjects. We used 2 different strategies for analysis of changes in gene expression. First, we singled out genes that are highly predictive for the risk classification of the disease. Using genes with a uniform expression pattern among subclasses, we were able to distinguish MDS samples from those of healthy controls and to differentiate low-risk from high-risk MDS patients according to their gene expression profiles. Second, we focused on highly differentially expressed genes in low-risk and high-risk MDS. Microarray results were confirmed by real-time polymerase chain reaction (PCR) for selected genes.
Patients, materials, and methods
Patients
CD34+ bone marrow cells selected from 18 MDS patients (10 men, 8 women) were studied at initial diagnosis (Table1). Patients were subgrouped according to the International Prognostic Scoring System (IPSS)5 into low-risk (including low and intermediate-1 risk) and high-risk (including intermediate-2 and high risk) subtypes as follows: low risk, 10 patients; high risk, 9 patients (all had refractory anemia with excess of blasts according to the World Health Organization [WHO] classification6). From one patient, paired samples were available at the time of initial diagnosis and during the progression of the disease. Approval was obtained from the institutional review board of the University of Frankfurt/Main for these studies; informed consent was provided according to the Declaration of Helsinki. CD34+ bone marrow cells from 4 healthy subjects were used as controls.
MDS patient characteristics
| No. . | Age, y . | Sex . | IPSS . | Karyotype . |
|---|---|---|---|---|
| 275 | 74 | F | IR1 | 46XY, del(5) |
| 276 | 72 | M | LR | 46XY |
| 277 | 70 | M | IR1 | 46XY, del(11) |
| 278 | 66 | M | IR1 | 46XY |
| 279 | 65 | M | LR | 46XY |
| 280 | 67 | F | IR1 | 46XX, del(5)(q13;q33) |
| 281 | 59 | F | IR1 | 46XX |
| 283 | 73 | F | IR2 | 46XX, del(22)(q11) |
| 284 | 71 | M | HR | 46XY, del(11) |
| 285 | 70 | M | IR2 | 46XY |
| 286 | 58 | M | HR | 46XY |
| 3000* | 50 | F | LR | 46XX, del(5) |
| 3853* | 50 | M | IR2 | NA |
| 3883* | 55 | M | IR2 | 46XY |
| 3923* | 52 | F | IR2 | NA |
| 4015* | 71 | F | HR | 46XX, t(3;5) |
| 5050* | 70 | M | IR1 | 46XY |
| 5324* | 54 | M | IR2 | 44-46XY, −5, −7, −21, +8 |
| 13591* | 63 | F | IR1 | 46XX, del(5), +8, del(12) |
| No. . | Age, y . | Sex . | IPSS . | Karyotype . |
|---|---|---|---|---|
| 275 | 74 | F | IR1 | 46XY, del(5) |
| 276 | 72 | M | LR | 46XY |
| 277 | 70 | M | IR1 | 46XY, del(11) |
| 278 | 66 | M | IR1 | 46XY |
| 279 | 65 | M | LR | 46XY |
| 280 | 67 | F | IR1 | 46XX, del(5)(q13;q33) |
| 281 | 59 | F | IR1 | 46XX |
| 283 | 73 | F | IR2 | 46XX, del(22)(q11) |
| 284 | 71 | M | HR | 46XY, del(11) |
| 285 | 70 | M | IR2 | 46XY |
| 286 | 58 | M | HR | 46XY |
| 3000* | 50 | F | LR | 46XX, del(5) |
| 3853* | 50 | M | IR2 | NA |
| 3883* | 55 | M | IR2 | 46XY |
| 3923* | 52 | F | IR2 | NA |
| 4015* | 71 | F | HR | 46XX, t(3;5) |
| 5050* | 70 | M | IR1 | 46XY |
| 5324* | 54 | M | IR2 | 44-46XY, −5, −7, −21, +8 |
| 13591* | 63 | F | IR1 | 46XX, del(5), +8, del(12) |
No. indicates sample number; M, male; F, female; IR1, intermediate-1 risk; IR2, intermediate-2 risk; and NA, not available.
Nos. 277 and 284 are paired samples from the same patient.
Verification sample group.
CD34+ cell selection and nucleic acid preparation
Heparinized bone marrow samples were obtained by aspiration from the posterior iliac crest at the time of initial diagnosis after informed consent. To lessen activation of the cells by any technical manipulation, fresh bone marrow was processed immediately after aspiration to select the CD34+ cells within the next 4 hours. Mononuclear cells were separated by density- gradient centrifugation through Ficoll-Hypaque (Biochrom, Berlin, Germany). After depletion of adherent cells by 1-hour adherence to plastic, CD34+ cells were purified by high-gradient magnetic cell separation (MACS) using superparamagnetic streptavidin-microparticles for labeling CD34+ cells (Miltenyi Biotec, Mönchengladbach, Germany). Positive cells were discharged from the column after removal from the magnetic field. Yield and purity of the positively selected CD34+ cells was evaluated by flow cytometry (FACScan; Becton Dickinson, Heidelberg, Germany).
Total RNA was extracted using TRIzol (Life Technologies, Grand Island, NY) according to the manufacturer's protocol with minor modifications. In addition, RNA was purified by the RNeasy system (Qiagen, Valencia, CA) following the manufacturer's advice.
To ensure that the gene expression we measured by microarray assay was not affected by degradation of the RNA extracted from the purified CD34+ cells, we used denaturing gel electrophoresis to evaluate the quality of the RNA. Furthermore, the expression level of β-actin, as determined by GeneChip assay, was required to be greater than 30 000 (raw data) in all of our MDS and control samples.
Oligonucleotide microarray
A detailed protocol for the sample preparation and microarray processing is available from Affymetrix (Santa Clara, CA). Because of the limited number of CD34+ cells and the low content of RNA in these hematopoietic stem cells, a double in vitro transcription technique (nanogram scale assay) was used. To assay 50 ng total RNA, the standard Affymetrix target amplification protocol was modified by using first-round cRNA product to generate double-stranded cDNA that was then used for a second round of in vitro transcription for synthesis of the biotinylated cRNA.
Fifteen micrograms fragmented, biotinylated cRNA was hybridized to an HG-U95Av2 microarray (Affymetrix) for 16 hours at 45°C with constant rotation at 60 rpm, according to the Affymetrix protocol. This high-density oligonucleotide array targets 9670 human genes as selected from the National Center for Biotechnology Information (NCBI) Gene Bank database, with a total of 12 000 oligonucleotide sets. Each microarray was used to assay a single sample. After hybridization, the microarray was washed and stained on an Affymetrix fluidics station and was scanned with an argon–ion confocal laser with 488-nm emission and detection at 570 nm. Fluorescence intensity normalized to the average fluorescence for the entire microarray.7 Data were imported into a Microsoft Excel 2000 (Microsoft, Redmond, WA) database.
Data analysis
GeneChip image analysis was performed using the Microarray Analysis Suites 4.0.6 and 5.0 (Affymetrix). Data analysis was performed with the GeneSpring software version 4.0 (Silicon Genetics, San Carlos, CA). To eliminate changes within the range of background noise and to select the most differentially expressed genes, restrictions to classify genes as up-regulated or down-regulated required that raw data values be greater than 1000, changes in expression be greater than 5-fold, and genes be present by Affymetrix data analysis. Statistical significance of changes was calculated by the nonparametrict test, with P < .05. We used the class membership prediction method8-10 to determine whether the pattern of gene expression could be used to classify MDS and healthy control samples into 3 classes (control, low-risk MDS, high-risk MDS). The maximum number of genes to predict the class membership was set to 50. Hierarchical clustering analysis with Spearman confidence correlation was used to identify gene clusters. The separation ratio was set at 0.5.
Real-time PCR
Quantification of RNA in CD34+ cells by real-time PCR was performed as described previously.11Briefly, 500 ng total RNA was processed directly to cDNA by reverse transcription. PCR primers and TaqMan (Applied Biosystems, Foster City, CA) probes were designed using software PRIMER3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) with published sequence data from the NCBI database. Amplification reactions contained 5 μL diluted cDNA and 12.5 μL Universal TaqMan 2× PCR mastermix (Applied Biosystems). Concentrations of primer and TaqMan probes were 300 nM and 100 nM, respectively, in a final volume of 25 μL. All reactions were performed in triplicate in an iCycler system (Bio-Rad, Hercules, CA), and the thermal cycling conditions were as follows: 2 minutes at 50°C and 10 minutes at 95°C, followed by 45 cycles at 95°C for 15 seconds and at 60°C for 1 minute.
Beta-actin (BAC) and 18S were used as active and endogenous references to correct for differences in the amount of total RNA added to a reaction and to compensate for different levels of inhibition during reverse transcription of RNA and during PCR.11-13No difference in relative expression was noted using either 18S or BAC for the normalization of gene expression.
Results
Low-risk MDS versus normal CD34+ cells
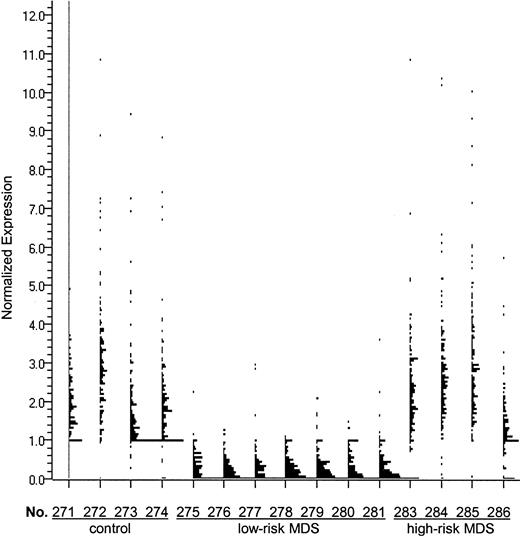
Analysis of the genes expressed in CD34+ marrow cells, which were more than 5-fold down-regulated in low-risk MDS than in those of healthy controls, resulted in a list of 161 highly differentially expressed genes (Figure1). We examined this gene list using public databases and noted that 32 of these genes were involved in cell growth and signaling (Table 2). In low-risk MDS, 117 genes were more than 5-fold up-regulated compared with healthy CD34+ cells. A detailed analysis of these genes identified 27 that were probably involved in the regulation of hematopoiesis (Table 2).
Genes expressed at least 5-fold lower in CD34+ marrow cells from patients with low-risk MDS than in healthy controls.
The 161 genes that were more than 5-fold down-regulated in CD34+ marrow cells from patients with low-risk MDS compared with the CD34+ cells from healthy controls includeIEX1 (radiation-inducible immediate-early response gene),RAI3 (retinoic acid–induced gene), H4M (H4 histone family), p63 (tumor protein 63), and TM4SF(transmembrane 4 superfamily). Expression of these genes is displayed by GeneSpring software for each of the samples. Each single dot represents the normalized expression level of one gene. “No.” indicates sample number.
Genes expressed at least 5-fold lower in CD34+ marrow cells from patients with low-risk MDS than in healthy controls.
The 161 genes that were more than 5-fold down-regulated in CD34+ marrow cells from patients with low-risk MDS compared with the CD34+ cells from healthy controls includeIEX1 (radiation-inducible immediate-early response gene),RAI3 (retinoic acid–induced gene), H4M (H4 histone family), p63 (tumor protein 63), and TM4SF(transmembrane 4 superfamily). Expression of these genes is displayed by GeneSpring software for each of the samples. Each single dot represents the normalized expression level of one gene. “No.” indicates sample number.
Genes differentially expressed in CD34+ cells from patients with low-risk MDS compared with healthy controls
| Gene . | Accession . | Map . | Description . | -fold . |
|---|---|---|---|---|
| Down-regulation | ||||
| ANXA2 | D00017 | 15q21 | Annexin A2 | >10 |
| BENE | U17077 | 2q13 | T-lymphocyte maturation factor | >10 |
| BTUB | X79535 | 6p21.3 | Beta tubulin | >10 |
| CAPN4 | X04106 | 19 | Calpain | 5 |
| CARD2 | X74929 | 12q13 | Keratin 8 | 8 |
| CCND1 | M73554 | 11q13 | Cyclin D1 | >10 |
| CDK2AP1 | AF089814 | 11q13 | CDK2-associated protein | 10 |
| H4M | AL021807 | 6p21.3 | H4 histone family | >10 |
| HRAS1 | J00277 | 11p15.5 | H-RAS 1 | 6 |
| HSP70 | L12723 | 5q31 | Heat shock protein 70 | 5 |
| IEX1 | S81914 | 6p21.3 | Radiation-inducible immediate-early response gene | 7 |
| ILF3 | U10324 | 19 | Interleukin enhancer binding factor 3 | >10 |
| MAGE3 | U03735 | Xq28 | Melanoma antigen 3 | 7 |
| MAGE4 | U10688 | Xq28 | Melanoma antigen 4 | >10 |
| MAGE6 | U10691 | Xq28 | Melanoma antigen 6 | 6 |
| MIG2 | Z24725 | 14 | Mitogen-inducible gene | 6 |
| MKPL | AF038844 | 17q12 | MKP1-like protein tyrosine phosphatase | 6 |
| NIN1P | D38047 | 19 | Proteasome 26S subunit 31 | 5 |
| OCT3 | Z11898 | 6p21 | Octamer-binding protein 3A | 6 |
| P63 | X69910 | 12 | Tumor protein 63 | 8 |
| AB000584 | 19p | Prostate-derived factor | >10 | |
| PSMD11 | AB003102 | 17 | Proteasome 26S subunit p44.5 | 5 |
| PSMD13 | AB009398 | 11p15.5 | Proteasome 26S subunit p40.5 | 10 |
| RAI3 | AF095448 | 12p13 | Retinoic acid-induced gene | 8 |
| STIP1 | M86752 | 11q13 | Stress-induced phosphoprotein 1 | >10 |
| STRAT | X57348 | 1p | Stratifin | >10 |
| TFAP2 | U85658 | 20q13.2 | Transcription factor AP-2 | 8 |
| TM4SF1 | AF065388 | 1 | Transmembrane 4 superfamily 1 | 5 |
| TXNR | X91247 | 12q23 | Thioredoxin reductase 1 | 6 |
| UBC12 | AF075599 | 19 | Ubiquitin-conjugating enzyme 12 | >10 |
| Up-regulation | ||||
| AML1 | D43968 | 21q22.3 | Acute myeloid leukemia 1 | 8 |
| ATF3 | L19871 | 1 | Activating transcription factor 3 | 8 |
| CDKI1A | U03106 | 6p21.2 | Cyclin-dependent kinase inhibitor 1A (p21) | >10 |
| COIL | U06632 | 17q22-q23 | Coilin | 7 |
| DLEC1 | AB020522 | 3p22 | Deleted in lung and esophageal cancer | 7 |
| DLK1 | U15979 | 14q32 | Delta-like homolog 1 | 7 |
| DNM2 | L36983 | 19 | Dynamin | 8 |
| EBAF | AF081507 | 1q42.1 | Endometrial bleeding-associated factor | 6 |
| GNA15 | M63904 | 19p13.3 | Guanine-nucleotide-binding protein alpha 15 | 5 |
| HOX7 | M97676 | 4p16 | Homeobox 7 | 6 |
| IL-1RAP | AB006537 | 3q28 | Interleukin-1 receptor accessory protein | >10 |
| LTK | D16105 | 15q15.1 | Leukocyte tyrosine kinase | 5 |
| MEOX1 | U10492 | 17q21 | Mesenchyme homeobox 1 | 6 |
| PKCB | X06318 | 16p11.2 | Protein kinase C, beta 1 | 6 |
| PTPN7 | M64322 | 1q32.1 | Protein tyrosine phosphatase nonreceptor type 7 | 5 |
| PTPRS | U35234 | 19p13.3 | Protein tyrosine phosphatase receptor S | >10 |
| SCYA18 | Y13710 | 17q11.2 | Small inducible cytokine subfamily A18 | 6 |
| TBR1 | U49250 | 2q23-q37 | T-box gene (transcription factor) | 6 |
| TOB2 | D64109 | 22q13 | Transducer of ERB B2 | 6 |
| TRAF6 | U78798 | 11 | TNF receptor-associated factor 6 | >10 |
| ZNF136 | U09367 | 19p13 | Zinc finger protein 136 | 7 |
| ZNF261 | X95808 | Xq13.1 | Zinc finger protein 261 | >10 |
| Gene . | Accession . | Map . | Description . | -fold . |
|---|---|---|---|---|
| Down-regulation | ||||
| ANXA2 | D00017 | 15q21 | Annexin A2 | >10 |
| BENE | U17077 | 2q13 | T-lymphocyte maturation factor | >10 |
| BTUB | X79535 | 6p21.3 | Beta tubulin | >10 |
| CAPN4 | X04106 | 19 | Calpain | 5 |
| CARD2 | X74929 | 12q13 | Keratin 8 | 8 |
| CCND1 | M73554 | 11q13 | Cyclin D1 | >10 |
| CDK2AP1 | AF089814 | 11q13 | CDK2-associated protein | 10 |
| H4M | AL021807 | 6p21.3 | H4 histone family | >10 |
| HRAS1 | J00277 | 11p15.5 | H-RAS 1 | 6 |
| HSP70 | L12723 | 5q31 | Heat shock protein 70 | 5 |
| IEX1 | S81914 | 6p21.3 | Radiation-inducible immediate-early response gene | 7 |
| ILF3 | U10324 | 19 | Interleukin enhancer binding factor 3 | >10 |
| MAGE3 | U03735 | Xq28 | Melanoma antigen 3 | 7 |
| MAGE4 | U10688 | Xq28 | Melanoma antigen 4 | >10 |
| MAGE6 | U10691 | Xq28 | Melanoma antigen 6 | 6 |
| MIG2 | Z24725 | 14 | Mitogen-inducible gene | 6 |
| MKPL | AF038844 | 17q12 | MKP1-like protein tyrosine phosphatase | 6 |
| NIN1P | D38047 | 19 | Proteasome 26S subunit 31 | 5 |
| OCT3 | Z11898 | 6p21 | Octamer-binding protein 3A | 6 |
| P63 | X69910 | 12 | Tumor protein 63 | 8 |
| AB000584 | 19p | Prostate-derived factor | >10 | |
| PSMD11 | AB003102 | 17 | Proteasome 26S subunit p44.5 | 5 |
| PSMD13 | AB009398 | 11p15.5 | Proteasome 26S subunit p40.5 | 10 |
| RAI3 | AF095448 | 12p13 | Retinoic acid-induced gene | 8 |
| STIP1 | M86752 | 11q13 | Stress-induced phosphoprotein 1 | >10 |
| STRAT | X57348 | 1p | Stratifin | >10 |
| TFAP2 | U85658 | 20q13.2 | Transcription factor AP-2 | 8 |
| TM4SF1 | AF065388 | 1 | Transmembrane 4 superfamily 1 | 5 |
| TXNR | X91247 | 12q23 | Thioredoxin reductase 1 | 6 |
| UBC12 | AF075599 | 19 | Ubiquitin-conjugating enzyme 12 | >10 |
| Up-regulation | ||||
| AML1 | D43968 | 21q22.3 | Acute myeloid leukemia 1 | 8 |
| ATF3 | L19871 | 1 | Activating transcription factor 3 | 8 |
| CDKI1A | U03106 | 6p21.2 | Cyclin-dependent kinase inhibitor 1A (p21) | >10 |
| COIL | U06632 | 17q22-q23 | Coilin | 7 |
| DLEC1 | AB020522 | 3p22 | Deleted in lung and esophageal cancer | 7 |
| DLK1 | U15979 | 14q32 | Delta-like homolog 1 | 7 |
| DNM2 | L36983 | 19 | Dynamin | 8 |
| EBAF | AF081507 | 1q42.1 | Endometrial bleeding-associated factor | 6 |
| GNA15 | M63904 | 19p13.3 | Guanine-nucleotide-binding protein alpha 15 | 5 |
| HOX7 | M97676 | 4p16 | Homeobox 7 | 6 |
| IL-1RAP | AB006537 | 3q28 | Interleukin-1 receptor accessory protein | >10 |
| LTK | D16105 | 15q15.1 | Leukocyte tyrosine kinase | 5 |
| MEOX1 | U10492 | 17q21 | Mesenchyme homeobox 1 | 6 |
| PKCB | X06318 | 16p11.2 | Protein kinase C, beta 1 | 6 |
| PTPN7 | M64322 | 1q32.1 | Protein tyrosine phosphatase nonreceptor type 7 | 5 |
| PTPRS | U35234 | 19p13.3 | Protein tyrosine phosphatase receptor S | >10 |
| SCYA18 | Y13710 | 17q11.2 | Small inducible cytokine subfamily A18 | 6 |
| TBR1 | U49250 | 2q23-q37 | T-box gene (transcription factor) | 6 |
| TOB2 | D64109 | 22q13 | Transducer of ERB B2 | 6 |
| TRAF6 | U78798 | 11 | TNF receptor-associated factor 6 | >10 |
| ZNF136 | U09367 | 19p13 | Zinc finger protein 136 | 7 |
| ZNF261 | X95808 | Xq13.1 | Zinc finger protein 261 | >10 |
These genes were selected because a greater than 5-fold difference in expression occurred between CD34+ cells from patients with low-risk MDS and healthy controls.
Accession indicates GenBank accession number; -fold, mean fold change.
High-risk MDS versus normal CD34+cells
Comparing gene expression in CD34+ marrow cells of patients with high-risk MDS versus healthy controls, 72 genes were more than 5-fold down-regulated. Twenty probably are important for hematopoiesis (Table 3), includingHLF (hepatic leukemia factor) and STAT2 (signal transducer and activator of transcription). In CD34+ marrow cells from patients with high-risk MDS, 53 genes were more than 5-fold up-regulated compared with CD34+ marrow cells from healthy controls. A more detailed analysis of these genes identified 10 of them (Table 3) associated with hematopoiesis.
Genes differentially expressed in CD34+ cells from patients with high-risk MDS compared with healthy controls
| Gene . | Accession . | Map . | Description . | -fold . |
|---|---|---|---|---|
| Down-regulation | ||||
| AZU | M96326 | 19p13.3 | Azurocidin | 6 |
| CD1E | X14975 | — | CD1e antigen (major histocompatibility complex related) | 5 |
| CD8 | X13444 | 2p12 | CD8 antigen | >10 |
| CLC | L01664 | 19q13.1 | Charcot-Leyden crystal | 7 |
| CRA | U78556 | 1 | Cisplatin resistance associated | >10 |
| DHH | U59748 | 12q12 | Desert hedgehog homolog (Drosophila) | >10 |
| DYRK2 | Y09216 | 12 | Dual tyrosine-phosphorylation-regulated kinase 2 | 6 |
| GPR12 | U18548 | 13q12 | G-protein–coupled receptor 12 | 6 |
| HLF | M95585 | 17q22 | Hepatic leukemia factor | 7 |
| H-PLK | M55422 | 7 | Krueppel-related zinc finger protein | 6 |
| IFNAR1 | J03171 | 21q22.1 | Interferon alpha receptor 1 | >10 |
| MBP | Z26248 | 11 | Eosinophil granule major basic protein | >10 |
| P63 | Y16961 | 3q27-q29 | Tumor protein 63 | 5 |
| PF4 | M25897 | 4q12-q21 | Platelet factor 4 | 6 |
| PIK3CG | X83368 | 7 | Phosphatidylinositol kinase 3 catalytic gamma | 9 |
| PRKAA1 | AB022017 | 5p12 | AMP-activated protein kinase alpha 1 | 6 |
| PTCH | U59464 | 9q22.3 | Patched homolog (Drosophila) | >10 |
| STAT2 | U18671 | 12 | Signal transducer and activator of transcription 2 | 10 |
| TGB1 | M54995 | 4q12-q13 | Source: Human connective tissue activation peptide III | >10 |
| UPK3 | AF085808 | 22q13.31 | Uroplakin | >10 |
| Up-regulation | ||||
| COIL | U06632 | 17q22-q23 | Coilin | 5 |
| ELK1 | M25269 | Xp11.2 | Tyrosine kinase (oncogene) | 5 |
| HOGG1 | U88620 | 3p26.2 | Alpha-hydroxyguanine DNA glycosylase 1 (homolog) | 7 |
| MEGF3 | D87469 | 1 | Multiple epidermal growth factor-like domain 3 | 5 |
| PLC1 | AL022394 | 20q12 | Phospholipase C148 | >10 |
| PRKAR2 | X14968 | 3p21 | Protein kinase, cAMP-dependent | >10 |
| PTPRS | U35234 | 19p13.3 | Protein tyrosine phosphatase receptor S | >10 |
| SP1LZTF | AI991531 | — | SP1-like zinc finger transcription factor | >10 |
| TRAF6 | U78798 | 11 | TNF receptor-associated factor 6 | >10 |
| ZNF261 | X95808 | Xq13.1 | Zinc finger protein 261 | 9 |
| Gene . | Accession . | Map . | Description . | -fold . |
|---|---|---|---|---|
| Down-regulation | ||||
| AZU | M96326 | 19p13.3 | Azurocidin | 6 |
| CD1E | X14975 | — | CD1e antigen (major histocompatibility complex related) | 5 |
| CD8 | X13444 | 2p12 | CD8 antigen | >10 |
| CLC | L01664 | 19q13.1 | Charcot-Leyden crystal | 7 |
| CRA | U78556 | 1 | Cisplatin resistance associated | >10 |
| DHH | U59748 | 12q12 | Desert hedgehog homolog (Drosophila) | >10 |
| DYRK2 | Y09216 | 12 | Dual tyrosine-phosphorylation-regulated kinase 2 | 6 |
| GPR12 | U18548 | 13q12 | G-protein–coupled receptor 12 | 6 |
| HLF | M95585 | 17q22 | Hepatic leukemia factor | 7 |
| H-PLK | M55422 | 7 | Krueppel-related zinc finger protein | 6 |
| IFNAR1 | J03171 | 21q22.1 | Interferon alpha receptor 1 | >10 |
| MBP | Z26248 | 11 | Eosinophil granule major basic protein | >10 |
| P63 | Y16961 | 3q27-q29 | Tumor protein 63 | 5 |
| PF4 | M25897 | 4q12-q21 | Platelet factor 4 | 6 |
| PIK3CG | X83368 | 7 | Phosphatidylinositol kinase 3 catalytic gamma | 9 |
| PRKAA1 | AB022017 | 5p12 | AMP-activated protein kinase alpha 1 | 6 |
| PTCH | U59464 | 9q22.3 | Patched homolog (Drosophila) | >10 |
| STAT2 | U18671 | 12 | Signal transducer and activator of transcription 2 | 10 |
| TGB1 | M54995 | 4q12-q13 | Source: Human connective tissue activation peptide III | >10 |
| UPK3 | AF085808 | 22q13.31 | Uroplakin | >10 |
| Up-regulation | ||||
| COIL | U06632 | 17q22-q23 | Coilin | 5 |
| ELK1 | M25269 | Xp11.2 | Tyrosine kinase (oncogene) | 5 |
| HOGG1 | U88620 | 3p26.2 | Alpha-hydroxyguanine DNA glycosylase 1 (homolog) | 7 |
| MEGF3 | D87469 | 1 | Multiple epidermal growth factor-like domain 3 | 5 |
| PLC1 | AL022394 | 20q12 | Phospholipase C148 | >10 |
| PRKAR2 | X14968 | 3p21 | Protein kinase, cAMP-dependent | >10 |
| PTPRS | U35234 | 19p13.3 | Protein tyrosine phosphatase receptor S | >10 |
| SP1LZTF | AI991531 | — | SP1-like zinc finger transcription factor | >10 |
| TRAF6 | U78798 | 11 | TNF receptor-associated factor 6 | >10 |
| ZNF261 | X95808 | Xq13.1 | Zinc finger protein 261 | 9 |
These genes were selected because a greater than 5-fold difference in expression occurred when comparing CD34+ cells from marrow samples from patients with high-risk MDS versus healthy controls.
Abbreviations are explained in Table 2.
High-risk versus low-risk MDS
Using the restrictions described and comparing high-risk and low-risk MDS, 210 genes showed higher expression in CD34+cells from patients with high-risk MDS, whereas 134 genes were down-regulated. Further analysis identified 24 genes up-regulated and 25 genes down-regulated in CD34+ cells from high- versus low-risk MDS (Table 4), which are likely involved in the regulation of hematopoietic proliferation and differentiation.
Genes differentially expressed in CD34+ cells from patients with low-risk and high-risk MDS
| Gene . | Accession . | Map . | Description . | -fold . |
|---|---|---|---|---|
| Down-regulation | ||||
| ADPRF | U52521 | — | Arfaptin (ADP-ribosylation factor) | 8 |
| ARD1 | U14575 | 1 | Activator of RNA decay 1 | 5 |
| ARHH | Z35227 | 4p13 | Ras homolog gene family H | 6 |
| CDH | D88797 | 5q31 | Cadherin | 6 |
| CRA | U78556 | 1 | Cisplatin resistance associated | >10 |
| DLEC1 | AB020522 | 3p22 | Deleted in lung and esophageal cancer | >10 |
| DYRK2 | Y09216 | 12 | Dual tyrosine-phosphorylation-regulated kinase 2 | 6 |
| H-PLK | M55422 | 7 | Krueppel-related zinc finger protein | 8 |
| IFNAR1 | J03171 | 21q22.1 | Interferon alpha receptor 1 | >10 |
| LCP2 | U20158 | 5q33.1 | Lymphocyte cytosolic protein | 7 |
| MNDA | M81750 | 1q22 | Myeloid cell nuclear differentiation antigen | 7 |
| NFE2 | S77763 | 12q13 | Nuclear factor, erythroid derived 2 | 8 |
| NKG7 | S69115 | 19 | Natural killer cell group 7 | 5 |
| P63 | Y16961 | 3q27-q29 | Tumor protein 63 | 7 |
| PAGE1 | AF058989 | X | Prostate-associated gene 1 | 6 |
| PF4 | M25897 | 4q12-q21 | Platelet factor 4 | 10 |
| PIK3CG | X83368 | 7 | Phosphatidylinositol kinase 3 catalytic gamma | >10 |
| PRKAA1 | AB022017 | 5p12 | AMP-activated protein kinase alpha 1 | 8 |
| RGS19 | X91809 | — | Regulator of G-protein signaling 19 | 7 |
| SDF1 | L36033 | 10q11.1 | Stromal-cell-derived factor 1 | >10 |
| STAT2 | U18671 | 12 | Signal transducer and activator of transcription 2 | >10 |
| TDGF3 | M96956 | Xq21-q22 | Teratocarcinoma-derived growth factor 3 | 5 |
| UPK3 | AF085808 | 22q13.31 | Uroplakin | >10 |
| Up-regulation | ||||
| ALTE | D31762 | 6p21 | AC-like transposable element | 6 |
| BENE | U17077 | 2q13 | T-lymphocyte maturation factor | >10 |
| CCND1 | M73554 | 11q13 | Cyclin D1 | >10 |
| CD9 | M38690 | 12q13 | CD9 antigen | 6 |
| CDK2AP1 | AF089814 | 11q13 | CDK2-associated protein | 10 |
| GA733-2 | M93036 | 4q | Carcinoma-associated antigen | >10 |
| HRAS1 | J00277 | 11p15.5 | H-RAS 1 | 6 |
| IEX1 | S81914 | 6p21.3 | Radiation-inducible immediate-early response gene | 9 |
| JUND | X56681 | 19p13.2 | Jun-D proto-oncogene | 5 |
| MAGE1 | M77481 | Xq28 | Melanoma antigen 1 | >10 |
| MAGE3 | U03735 | Xq28 | Melanoma antigen 3 | 7 |
| MAGE6 | U10691 | Xq28 | Melanoma antigen 6 | >10 |
| MIG2 | Z24725 | 14 | Mitogen-inducible gene | 8 |
| MKPL | AF038844 | 17q12 | MKP1-like protein tyrosine phosphatase | 6 |
| MLF2 | AF070539 | 12p13 | Myeloid leukemia factor 2 | 8 |
| NCOR1 | AF044209 | 17p11.2 | Nuclear receptor corepressor 1 | 7 |
| AB000584 | 19p | Prostate-derived factor | >10 | |
| PSMD11 | AB003102 | 17 | Proteasome 26S subunit p44.5 | 5 |
| PTPRN | L18983 | 2q35 | Protein tyrosine phosphatase receptor N | 5 |
| RAI3 | AF095448 | 12p13 | Retinoic-acid-induced gene | 8 |
| STIP1 | M86752 | 11q13 | Stress-induced phosphoprotein 1 | >10 |
| TK1 | M15205 | 17q23 | Thymidine kinase 1 | 5 |
| TM4SF3 | M35252 | 12 | Transmembrane 4 superfamily 3 | >10 |
| TOM1 | AJ006973 | 22q13.1 | Target of myb 1 | 5 |
| Gene . | Accession . | Map . | Description . | -fold . |
|---|---|---|---|---|
| Down-regulation | ||||
| ADPRF | U52521 | — | Arfaptin (ADP-ribosylation factor) | 8 |
| ARD1 | U14575 | 1 | Activator of RNA decay 1 | 5 |
| ARHH | Z35227 | 4p13 | Ras homolog gene family H | 6 |
| CDH | D88797 | 5q31 | Cadherin | 6 |
| CRA | U78556 | 1 | Cisplatin resistance associated | >10 |
| DLEC1 | AB020522 | 3p22 | Deleted in lung and esophageal cancer | >10 |
| DYRK2 | Y09216 | 12 | Dual tyrosine-phosphorylation-regulated kinase 2 | 6 |
| H-PLK | M55422 | 7 | Krueppel-related zinc finger protein | 8 |
| IFNAR1 | J03171 | 21q22.1 | Interferon alpha receptor 1 | >10 |
| LCP2 | U20158 | 5q33.1 | Lymphocyte cytosolic protein | 7 |
| MNDA | M81750 | 1q22 | Myeloid cell nuclear differentiation antigen | 7 |
| NFE2 | S77763 | 12q13 | Nuclear factor, erythroid derived 2 | 8 |
| NKG7 | S69115 | 19 | Natural killer cell group 7 | 5 |
| P63 | Y16961 | 3q27-q29 | Tumor protein 63 | 7 |
| PAGE1 | AF058989 | X | Prostate-associated gene 1 | 6 |
| PF4 | M25897 | 4q12-q21 | Platelet factor 4 | 10 |
| PIK3CG | X83368 | 7 | Phosphatidylinositol kinase 3 catalytic gamma | >10 |
| PRKAA1 | AB022017 | 5p12 | AMP-activated protein kinase alpha 1 | 8 |
| RGS19 | X91809 | — | Regulator of G-protein signaling 19 | 7 |
| SDF1 | L36033 | 10q11.1 | Stromal-cell-derived factor 1 | >10 |
| STAT2 | U18671 | 12 | Signal transducer and activator of transcription 2 | >10 |
| TDGF3 | M96956 | Xq21-q22 | Teratocarcinoma-derived growth factor 3 | 5 |
| UPK3 | AF085808 | 22q13.31 | Uroplakin | >10 |
| Up-regulation | ||||
| ALTE | D31762 | 6p21 | AC-like transposable element | 6 |
| BENE | U17077 | 2q13 | T-lymphocyte maturation factor | >10 |
| CCND1 | M73554 | 11q13 | Cyclin D1 | >10 |
| CD9 | M38690 | 12q13 | CD9 antigen | 6 |
| CDK2AP1 | AF089814 | 11q13 | CDK2-associated protein | 10 |
| GA733-2 | M93036 | 4q | Carcinoma-associated antigen | >10 |
| HRAS1 | J00277 | 11p15.5 | H-RAS 1 | 6 |
| IEX1 | S81914 | 6p21.3 | Radiation-inducible immediate-early response gene | 9 |
| JUND | X56681 | 19p13.2 | Jun-D proto-oncogene | 5 |
| MAGE1 | M77481 | Xq28 | Melanoma antigen 1 | >10 |
| MAGE3 | U03735 | Xq28 | Melanoma antigen 3 | 7 |
| MAGE6 | U10691 | Xq28 | Melanoma antigen 6 | >10 |
| MIG2 | Z24725 | 14 | Mitogen-inducible gene | 8 |
| MKPL | AF038844 | 17q12 | MKP1-like protein tyrosine phosphatase | 6 |
| MLF2 | AF070539 | 12p13 | Myeloid leukemia factor 2 | 8 |
| NCOR1 | AF044209 | 17p11.2 | Nuclear receptor corepressor 1 | 7 |
| AB000584 | 19p | Prostate-derived factor | >10 | |
| PSMD11 | AB003102 | 17 | Proteasome 26S subunit p44.5 | 5 |
| PTPRN | L18983 | 2q35 | Protein tyrosine phosphatase receptor N | 5 |
| RAI3 | AF095448 | 12p13 | Retinoic-acid-induced gene | 8 |
| STIP1 | M86752 | 11q13 | Stress-induced phosphoprotein 1 | >10 |
| TK1 | M15205 | 17q23 | Thymidine kinase 1 | 5 |
| TM4SF3 | M35252 | 12 | Transmembrane 4 superfamily 3 | >10 |
| TOM1 | AJ006973 | 22q13.1 | Target of myb 1 | 5 |
These genes were selected because a greater than 5-fold difference in expression occurred in CD34+ cells from marrow samples in patients with low-risk compared with high-risk MDS.
Abbreviations are explained in Table 2.
Class membership prediction and hierarchical clustering
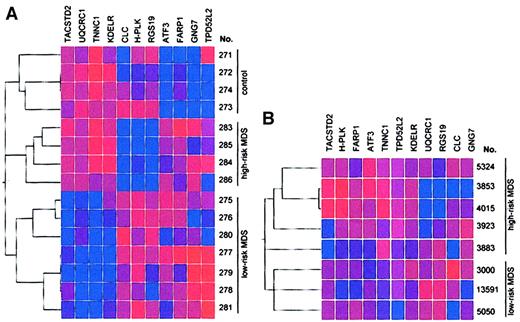
Using the first data set (Table 1, samples 275-286), we identified, by class membership prediction, 11 genes (Table5) whose expression can be used to differentiate with high accuracy between patients with low-risk MDS, those with high-risk MDS, and healthy controls. To verify the power of the genes selected by class membership prediction, we used the 11 genes for hierarchical clustering with Spearman confidence correlation (Figure 2A). We generated 3 clusters. All controls were in one cluster (with a maximum of 2 subclusters). Furthermore, patients with high-risk MDS and low-risk MDS were separated into different clusters, with a maximum of 3 subclusters. As a control experiment, unsupervised hierarchical cluster analysis using expression data from 1000 randomly selected genes did not result in any specific subclusters (data not shown) underlining the importance of the 11 selected genes for the prediction of MDS risk groups.
Genes selected by class membership prediction analysis to be predictive in the distinction between control, low-risk MDS, and high-risk MDS groups
| Gene . | Accession . | Map . | Description . |
|---|---|---|---|
| TACSTD2 | J04152 | 1p32 | Tumor-associated calcium signal transducer 2 |
| UQCRC1 | L16842 | 3p21.3 | Ubiquinol-cytochrome C reductase core protein 1 |
| TNNC1 | M37984 | 3p21.3 | Troponin C, slow |
| KDELR | M88458 | 7p | Endoplasmic reticulum protein retention receptor |
| CLC | L01664 | 19q13.1 | Charcot-Leyden crystal |
| H-PLK | M55422 | 7 | Krueppel-related zinc finger protein |
| RGS19 | X91809 | — | Regulator of G-protein signaling 19 |
| ATF3 | L19871 | 1 | Activating transcription factor 3 |
| FARP1 | AI701049 | — | FERM, RhoGEF, and pleckstrin domain protein 1 |
| GNG7 | AW051450 | — | Guanine nucleotide-binding protein gamma 7 |
| TPD52L2 | U44429 | 6q22-q23 | Tumor protein D52-like 2 |
| Gene . | Accession . | Map . | Description . |
|---|---|---|---|
| TACSTD2 | J04152 | 1p32 | Tumor-associated calcium signal transducer 2 |
| UQCRC1 | L16842 | 3p21.3 | Ubiquinol-cytochrome C reductase core protein 1 |
| TNNC1 | M37984 | 3p21.3 | Troponin C, slow |
| KDELR | M88458 | 7p | Endoplasmic reticulum protein retention receptor |
| CLC | L01664 | 19q13.1 | Charcot-Leyden crystal |
| H-PLK | M55422 | 7 | Krueppel-related zinc finger protein |
| RGS19 | X91809 | — | Regulator of G-protein signaling 19 |
| ATF3 | L19871 | 1 | Activating transcription factor 3 |
| FARP1 | AI701049 | — | FERM, RhoGEF, and pleckstrin domain protein 1 |
| GNG7 | AW051450 | — | Guanine nucleotide-binding protein gamma 7 |
| TPD52L2 | U44429 | 6q22-q23 | Tumor protein D52-like 2 |
Samples from patients with high-risk and low-risk MDS and from healthy controls could be correctly classified by examining the CD34+ marrow cells for gene expression profile of these 11 genes by class membership prediction.
Accession indicates GenBank accession number.
Identification of genes expressed in CD34+ marrow cells that can distinguish between patients with low-risk MDS, those with high-risk MDS, and healthy controls.
(A) Results represent analysis by hierarchical clustering with Spearman confidence correlation of 15 samples of CD34+ bone marrow cells (training set). Eleven genes were selected to predict the class membership of each of the samples. Vertical list is each of the samples. Horizontal list displays the 11 genes. (B) Validation of the 11 predictive genes used to distinguish between high-risk and low-risk MDS by gene expression profiling. The 11 predictive genes were used for clustering analysis in a second data set obtained from CD34+ cells from patients with low-risk and high-risk MDS (test set). Two clusters corresponding to the IPSS classifications high-risk and low-risk were found. No misclassification of any of the samples of the test set occurred, demonstrating the power of the selected genes for risk group prediction. One high-risk sample, however, was classified in a separate subcluster (sample 3883). Blue indicates low expression; red, high expression. Intensity of the color reflects the reliability of the expression data.
Identification of genes expressed in CD34+ marrow cells that can distinguish between patients with low-risk MDS, those with high-risk MDS, and healthy controls.
(A) Results represent analysis by hierarchical clustering with Spearman confidence correlation of 15 samples of CD34+ bone marrow cells (training set). Eleven genes were selected to predict the class membership of each of the samples. Vertical list is each of the samples. Horizontal list displays the 11 genes. (B) Validation of the 11 predictive genes used to distinguish between high-risk and low-risk MDS by gene expression profiling. The 11 predictive genes were used for clustering analysis in a second data set obtained from CD34+ cells from patients with low-risk and high-risk MDS (test set). Two clusters corresponding to the IPSS classifications high-risk and low-risk were found. No misclassification of any of the samples of the test set occurred, demonstrating the power of the selected genes for risk group prediction. One high-risk sample, however, was classified in a separate subcluster (sample 3883). Blue indicates low expression; red, high expression. Intensity of the color reflects the reliability of the expression data.
We used the 11 predictive genes to perform clustering analysis in a second data set obtained from CD34+ cells of patients with low-risk and high-risk MDS (Table 1, samples 3000-13 591). The aim of these additional experiments was to evaluate independently the predictive power of the 11 selected genes using a new series of patient samples. As shown in Figure 2B, we found 2 clusters corresponding to the IPSS classifications high-risk and low-risk. No misclassification of any of the samples of the test set occurred, demonstrating the power of the selected genes for risk-group prediction. One high-risk sample was, however, classified in a separate subcluster (sample 3883).
Gene expression analysis by real-time PCR
We tested 14 genes that were differentially expressed in CD34+ cells from patients with low-risk MDS compared to CD34+ cells from healthy controls (data not shown). Real-time PCR analysis was performed in 2 steps. First, we evaluated the expression of genes selected by a raw data restriction of 1000. Of 7 genes—IEX1,RAI3, FNTB, ATF3, TM4SF1,STIP1, and DNM—we confirmed the expression of 3 (43%) of them. Therefore, we increased our stringency level from a raw data cutoff of 1000 to 2500. The second set of genes for confirmation included BTUB, KDELR, STRAT,TXNR, OCT3, CARD2, andANXA2. In this cohort of more stringently selected genes, the differential expression measured by oligonucleotide microarray was confirmed by real-time PCR in 6 of 7 genes (86% confirmation rate).
Discussion
The lack of suitable experimental models for MDS (eg, cell lines or animal systems) hampers progress in understanding the biology of this disease. Therefore, new approaches to elucidate the underlying abnormalities are needed. We and others2 14 have shown that to begin to characterize the cellular and molecular defects of clinical samples from patients with MDS requires the analysis of freshly isolated hematopoietic stem cells. Even those highly purified cells may demonstrate broad biologic variability, especially in low-risk MDS because the CD34+ cells in this subgroup of MDS may not always be monoclonal. In addition, the selection procedure of stem cells from whole bone marrow cells might influence biologic behavior and gene expression in these selected cells. We did focus on optimizing the cell selection procedure and the use of small amounts of RNA.
The nanogram scale assay used in this work enabled amplification of small amounts of total RNA available for the analysis of the gene expression profile of CD34+ cells from patients with low-risk and high-risk MDS in comparison to those from healthy, unstimulated CD34+ cells. We were able to correlate the gene expression patterns in CD34+ cells with the clinical course of MDS. In addition, by analyzing the expression data of 9670 different genes, we identified key genes whose expression was aberrant in hematopoietic stem cells from patients with low-risk and high-risk MDS.
Class membership prediction analysis and hierarchical clustering based on expression data of 11 selected genes enabled us to discriminate between patients with low-risk and high-risk MDS and to discriminate between each of these subgroups and healthy controls according to their gene expression profiles (Figure 2A). No misclassification of any of the samples occurred. The low number of subclusters substantiates the importance of the selected genes to discriminate between the 3 different groups. Analysis of a paired sample from a patient with low-risk MDS at the time of initial diagnosis and after transformation to high-risk MDS (samples 277 and 284) demonstrates that the power of the 11 selected genes to classify disease status is stronger than the similarity of gene expression within paired samples from the same patient (Figure 2A).
A groundbreaking study showed that global gene expression analysis can distinguish with high accuracy between tumor samples from patients with AML or with acute lymphoblastic leukemia (ALL).9Expression data from 51 selected genes were used for classification of AML versus ALL. These initial results were extended by class membership prediction analysis of breast cancers carrying mutations of 2 different genes (BRCA1 and BRCA2).10 The expression data of 9 and 11 genes, respectively, were successfully used to categorize the tumor samples to the correct group having either a BRCA1 or a BRCA2 mutation.
Recently, we and others11,15-21 have shown that microarray analysis can provide sufficient data to detect genes or gene patterns associated with alterations of specific cellular pathways or signal cascades in tumor cells. For example, several new genes regulated by p53 have been identified that are important in p53-induced cell cycle arrest.22 Likewise, the regulation of gene expression in response to ionizing radiation was carefully dissected in human cells.23 In addition, the knowledge of the genes involved in specific pathways facilitates the discovery of those coding for interactive proteins, giving insight into their functional importance. Several redox and mitochondrial elements that control either resistance or sensitivity to apoptosis have been identified by microarray.7 Recent studies in genome-wide identification of putative DNA-binding sites for transcription factors have been accomplished using the microarray approach.24
We showed earlier11 that the application of restrictions (raw data values and -fold changes) to the microarray data resulted in the selection of genes whose expression could be confirmed by 2 independent techniques (real-time PCR and immunohistochemistry) in approximately 90% of genes. This may be true when using standard in vitro transcription to generate cRNA for hybridization with oligonucleotide microarrays. This report is the first to describe the double in vitro transcription for microarray analysis of small amounts of RNA from isolated CD34+ cell samples. Our results show that genes selected from the microarray data should have raw data of at least 2500. We focused on genes associated with cell growth and survival, apoptosis, and transcriptional factors because their dysregulation is a likely target of MDS.
In low-risk MDS, enhanced intramedullary apoptosis may contribute to ineffective hematopoiesis.25-27 We found that the expression of the IEX1 gene was decreased in low-risk MDS.IEX1 was initially identified in human squamous carcinoma cells, and its expression was induced by radiation.28Tumor necrosis factor alpha (TNF-α), which is overexpressed in bone marrow cells from patients with MDS, also induced the expression ofIEX1,29 which controls apoptosis in healthy cells.30 In addition, several of the other down-regulated genes in low-risk MDS (MIG2, STIP1) code for proteins that protect cells from stresses caused by mutagens or other factors. In summary, our data suggest that CD34+ cells from patients with low-risk MDS lack certain defensive proteins that may result in their increased susceptibility to cell damage.
Overexpression of DLK1 was described previously in patients with high-risk MDS, and it was implicated as being involved in MDS.14 The function of this gene in hematopoiesis is unknown, but it is genomically imprinted and expressed only from the paternal allele.31 The mechanism of imprinting of this gene follows the same pattern known from the reciprocally imprinted genes IGF2 (insulinlike growth factor 2) and H19. We found that this gene was markedly up-regulated in our series of patients with low-risk MDS compared with healthy CD34+marrow cells. We propose that this gene is associated with cellular proliferation and that possible loss of imprinting of this gene in MDS enhances the expression of DLK1, giving the abnormal clone a growth advantage.
This study provides evidence that microarray analysis can detect gene expression profiles strongly associated with different risk groups in MDS. Therefore, we believe that prognoses for patients with this disease may be predicted by using the gene expression of purified CD34+ cells. We have shown that microarray analysis can be used with small amounts of RNA, which can be obtained from cells during routine diagnostic bone marrow aspirate. This method may facilitate therapeutic decisions for patients in whom diagnosis or risk evaluation is not possible through morphologic and classical cytogenic techniques. Lastly, such analysis of global gene expression provides new insights into alterations of cellular pathways in early hematopoietic stem cells in low-risk MDS.
We thank Caroline Zander and Sandra Wagner for technical help and Dietrich Henne for advice in several computer techniques. We appreciate the expertise of the VASDHS Gene Chip Core Laboratory and the University of California at San Diego (UCSD) Cancer Center Microarray Facility for technical assistance with the microarray assays.
Supported by the National Institutes of Health (H.P.K.), the C. and H. Koeffler Fund, Parker Hughes Trust, Fredrich Begell Foundation, and the Joseph Troy Leukemia Fund. W.K.H. is a recipient of a scholarship from the Deutsche Forschungsgemeinschaft (HO2207/1-1). S.d.V. was supported by the University of California at Los Angeles (UCLA) Specialty Training and Research (STAR) Program as a Hematology/Oncology fellow and as an Advanced Research fellow. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars Sinai Medical Center/UCLA School of Medicine.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wolf-K. Hofmann, Department of Hematology, University Hospital Theodor-Stern-Kai 7, 60596 Frankfurt/Main, Germany; e-mail: w.k.hofmann@em.uni-frankfurt.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal