Adaptive immunity necessitates the proliferation of lymphocytes. In the mouse, we have previously shown that antigen-experienced T cells that have lost their proliferative potential express the killer cell lectinlike receptor G1 (KLRG1). By using a newly generated monoclonal antibody specific for human KLRG1, we now demonstrate that expression of KLRG1 also identifies T cells in humans that are capable of secreting cytokines but that fail to proliferate after stimulation. Furthermore, our data show that proliferative incapacity of CD8 T cells correlates better with KLRG1 expression than with absence of the CD28 marker. In peripheral blood lymphocytes (PBLs) from healthy adult donors, KLRG1 was expressed on 44% ± 14% of CD8 and 18% ± 10% of CD4 T cells. KLRG1 expression was restricted to antigen-experienced T cells. Here, KLRG1+ cells were preferentially found in the CCR7− effector T-cell pool. Besides T cells, a significant portion (approximately 50%) of human natural killer (NK) cells expressed KLRG1. Interestingly, these KLRG1+ NK cells were found exclusively in the CD56dim NK-cell subset. Thus, the expression of KLRG1 identifies a subset of NK cells and antigen-experienced T cells in humans that lack proliferative capacity.
Introduction
T cells have been shown to express inhibitory natural killer (NK) receptors that can down-regulate T-cell receptor (TCR)–mediated T-cell functions.1-5 These inhibitory receptors can be divided into 2 general structural types.6-8 One type, termed killer immunoglobulinlike receptor (KIR), represents type 1 integral membrane proteins that belong to the immunoglobulin (Ig) superfamily. The other type, termed killer cell lectinlike receptor (KLR), is represented by the mouse Ly49 family and by CD94/NKG2 receptors that are type 2 integral membrane proteins belonging to the C-type lectin family. The killer cell lectinlike receptor G1 (KLRG1) was first identified on the rat mucosal-type mast cell line RBL-2H3 and was originally named mast cell function–associated antigen (MAFA).9,10 In the mouse, KLRG1 expression was found on NK cells and on a subset of effector/memory CD8 T cells but not on mast cells.11-14Most strikingly, the expression of KLRG1 identified effector and memory CD8 T cells in mice that were fully capable of performing effector cell functions but were severely impaired in their ability to proliferate after antigen stimulation.15 In humans, KLRG1 mRNA has been found in lymphoid organs and in NK and basophilic cell lines,16 but to date reports on KLRG1 protein expression, cell subset distribution, and functional studies are lacking. To address these issues, a monoclonal antibody (mAb) specific for human KLRG1 was generated. The present study represents the first analysis of KLRG1 protein expression in humans. In addition, it demonstrates that human T cells lacking proliferative capacities can be identified on the basis of KLRG1 expression.
Materials and methods
Generation of mAb specific for human KLRG1
KLRG1 cDNA from human peripheral blood lymphocytes (PBLs) was amplified with KLRG1-specific primers (HMAF 38: 5′-TGACAGTGTTATTTATTCCATGTTAGAG-3′; HMAF 658: 5′-CCTCCTTTTAGGGATACATG-3′) and was cloned into pcDNA3.1/NT-GFP-TOPO (Invitrogen, Groningen, The Netherlands) to generate a chimeric molecule consisting of KLRG1 fused to green fluorescence protein (GFP). BALB/c mice were first DNA-immunized at 2- to 3-week intervals by 3 intramuscular injections of KLRG1-GFP plasmid (100 μg per mouse), followed by 3 intraperitoneal injections of 107live COS7 cells transiently transfected with KLRG1. Three days after the last immunization, spleen cells were fused with SP2/0 myeloma cells. Hybridoma supernatants were screened by flow cytometry using COS7 cells transiently transfected with KLRG1-GFP. Eight of 720 clones stained positive, and one clone (13A2) was chosen for further analysis. The 13A2 mAb was purified from hybridoma supernatants by affinity chromatography over protein G–agarose and was biotinylated using standard procedures.
Flow cytometry
PBLs isolated by Ficoll-Paque (Uppsala, Sweden) gradient centrifugation were resuspended in phosphate-buffered saline (PBS) containing 2% (fetal calf serum) FCS and 0.1% NaN3at a concentration of 106 to 107 cells/mL, followed by incubation at 4°C for 20 minutes with 100 μL of appropriately diluted mAb. For direct staining of whole blood cells, 10 U/mL heparin was added to the staining buffer. The following mAbs (all from BD PharMingen, San Diego, CA, except anti-CD11a, which was from Immunotech, Marseille, France) were used: anti-CD3 (clone UCHT1), anti-CD4 (clone RPA-T4), anti-CD8 (clone RPA-T8), anti-CD16 (clone 3G8), anti-CD19 (clone HIB19), anti-CD28 (clone CD28.2), anti-CD45RA (clone HI100), anti-CD45RO (clone UCHL1), anti-CD56 (clone B159), anti-CD62L (clone Dreg 56), anti-CD11a (clone 25.3), anti-CCR4 (clone 1G1.1), and anti-CXCR3 (clone 1C6). The mAbs were directly labeled with fluorescein isothiocyanate (FITC) or phycoerythrin (PE), or they were biotinylated. For the latter, allophycocyanin (APC)–coupled or PE-coupled streptavidin (BD PharMingen) was used as a secondary reagent for detection. To induce cytokine secretion, PBLs (3 × 106/mL) were activated for 4 hours with phorbol 12-myristate 13-acetate (PMA; 10−8 M) and ionomycin (5 μg/mL) in the presence of brefeldin A (15 μg/mL; Sigma Chemical, Taufkirchen, Germany). Activated cells were first surface stained, fixed, and permeabilized using Cytofix-Cytoperm solution (BD PharMingen); this was followed by staining with anti–interferon-γ (anti–IFN-γ, clone 4S.B3; BD PharMingen) or anti–interleukin (anti–IL-4, clone 8D4-8; BD PharMingen) mAb. CCR7 was stained with purified mAb (clone 2H4; BD PharMingen), followed by FITC-labeled goat anti-mouse IgM (Southern Biotechnology, Birmingham, AL). Basophils from peripheral human blood were identified using anti-CD203c (clone 97A6; Beckman-Coulter, Krefeld, Germany). The mAbs 2F113 and G639 specific for mouse and rat KLRG1, respectively, were kindly provided by D. Raulet (University of California-Berkeley, San Francisco) and I. Pecht (The Weizmann Institute of Science, Rehovot, Israel). Binding of these mAbs was detected by staining with PE-labeled goat–anti-hamster IgG (for 2F1) and goat–anti-mouse IgG (for G63; both Caltag, Burlingame, CA). Cells were analyzed on a FACSort flow cytometer using CELLQuest II software (both from Becton Dickinson). Before analysis of PBLs, red blood cells were lysed using FACS-Lysing Solution (BD PharMingen). Cell sorting was performed on a MoFlo cell sorter (Cytomation, Fort Collins, CO).
Cell culture
T-cell subpopulations were purified to more than 98% by cell sorting using either anti-CD8 in combination with anti-KLRG1 and anti-CD28 or anti-CD4 in combination with anti-KLRG1 and anti-CD45RA mAbs. Purified cells were labeled with 5- (and 6-)carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Leiden, The Netherlands) as described17 and were cultured in flat-bottomed microtiter plates at 105 cells per well, together with 105 unlabeled autologous PBL as feeder cells previously treated with mitomycin C (3 μg/mL) for 30 minutes at 37°C. Cells were stimulated with phytohemagglutinin (PHA, 2.5 μg/mL; Sigma) in Iscove modified Dulbecco medium (IMDM) culture medium containing 10% FCS, penicillin/streptomycin, 0.001 M β-mercaptoethanol, and 40 U/mL recombinant human IL-2 (BD PharMingen). Cells were analyzed on day 4 or 5 of culture. Unsorted CFSE-labeled PBLs were stimulated for 3 days with PHA and IL-2 or for 5 days with PHA, IL-2, and recombinant human IL-7 and IL-15 (both at 25 ng/mL; RDI, Flanders, NJ).
Immunoprecipitation
PBLs were surface-iodinated using precoated iodination tubes (Pierce, Rockford, IL) and were lysed in lysis buffer containing 20 mM Tris-HCl, pH 8, 137 mM NaCl, 2 mM EDTA (ethylenediaminetetraacetic acid), 10% glycerol, and 1% NP40. Lysates, obtained by microcentrifugation, were precleared (3 × 3 hours at 4°C) with bovine serum albumin (BSA)–coupled Sepharose 4B beads (Amersham Pharmacia Biotech, Uppsala, Sweden) and then were immunoprecipitated overnight at 4°C with Sepharose 4B precoupled with purified 13A2 mAbs. Antibodies were coupled to cyanogen bromide–activated Sepharose 4B at 5 mg antibody/1 mL gel, according to the manufacturer's instructions. Immunoprecipitates were washed 3 times in lysis buffer and were subjected to 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Autoradiographs were developed using a PhosphorImager and the BAS2000 system (Fujix, Tokyo, Japan).
Results
Generation of a mAb specific for human KLRG1
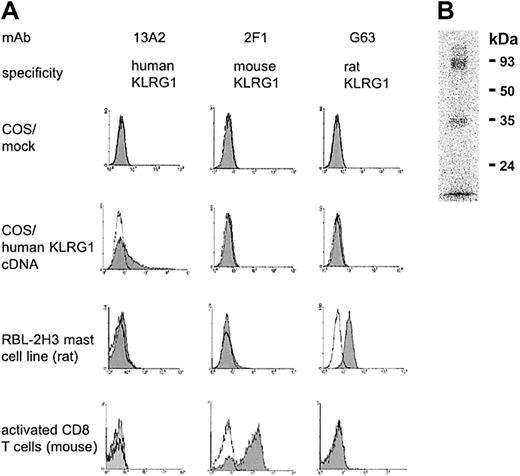
To analyze cell surface expression of human KLRG1, a mAb (13A2) was generated. This mAb reacted with COS7 cells transiently transfected with human KLRG1 cDNA but not with mock-transfected cells (Figure1). The primary sequence of human KLRG1 is more closely related to its homologues from rat and mouse (70% and 71% similarity, respectively) than to any other known human C-type lectin. Therefore, cross-reactivity of the 13A2 mAb to other C-type lectins was tested using KLRG1+ RBL-2H3 mast cells from rat and KLRG1+ CD8 T cells from mice infected with lymphocytic choriomeningitis virus (LCMV). These cells could be stained with the corresponding mAbs G63 and 2F1 specific for the rat or mouse homologue of KLRG1, respectively, but not with the 13A2 mAb (Figure 1A).
The 13A2 mAb is specific for human KLRG1.
(A) COS7 cells were transiently transfected with empty expression vector (mock) or with expression vector containing human KLRG1 cDNA. Afterward, cells were stained with the indicated mAb (shaded histograms) and analyzed by flow cytometry. Open histograms represent negative controls. To determine cross-reactivity of the 13A2 mAb, KLRG1+ RBL-2H3 cells from rat and KLRG1+ CD8 T cells from mice were examined in a similar fashion, including mAb G63 and 2F1 specific for rat and mouse KLRG1, respectively, as positive controls. (B) Immunoprecipitation of KLRG1. Detergent extracts of surface iodinated human PBL were incubated with beads coupled with mAb 13A2, and bound material was analyzed in 12.5% SDS-PAGE under nonreducing conditions.
The 13A2 mAb is specific for human KLRG1.
(A) COS7 cells were transiently transfected with empty expression vector (mock) or with expression vector containing human KLRG1 cDNA. Afterward, cells were stained with the indicated mAb (shaded histograms) and analyzed by flow cytometry. Open histograms represent negative controls. To determine cross-reactivity of the 13A2 mAb, KLRG1+ RBL-2H3 cells from rat and KLRG1+ CD8 T cells from mice were examined in a similar fashion, including mAb G63 and 2F1 specific for rat and mouse KLRG1, respectively, as positive controls. (B) Immunoprecipitation of KLRG1. Detergent extracts of surface iodinated human PBL were incubated with beads coupled with mAb 13A2, and bound material was analyzed in 12.5% SDS-PAGE under nonreducing conditions.
The antigen recognized by 13A2 mAb was immunoprecipitated from lysates prepared from human PBLs that had been subjected to surface125iodination. As shown in Figure 1B, SDS-PAGE revealed 2 bands with molecular weights of 30 to 40 and 80 to 90 kDa. These bands corresponded to the monomeric and homodimeric forms of KLRG1, similar to those found with rat and mouse KLRG1.9 13 Taken together, these results indicate that the 13A2 mAb is specific for human KLRG1.
KLRG1 is expressed on memory T cells and NK cells
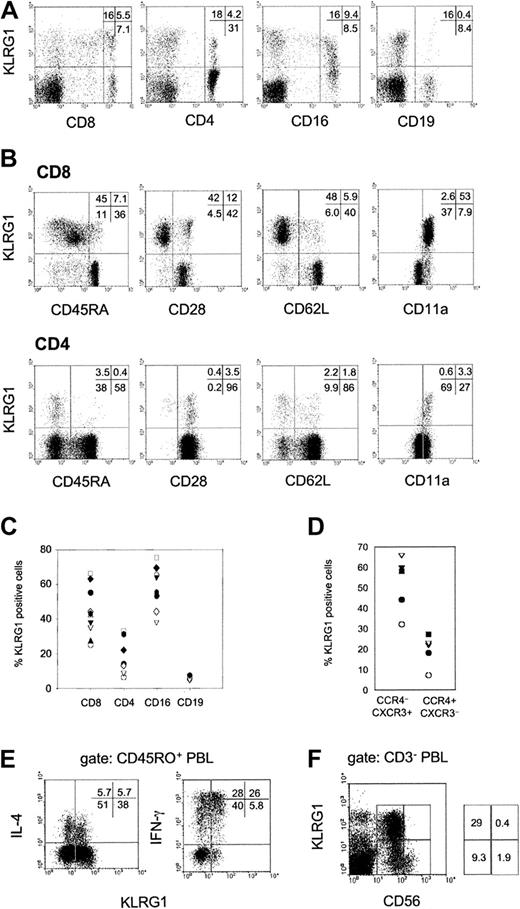
Flow cytometric analysis of human PBLs from healthy donors showed that KLRG1 was expressed on distinct subsets of CD8 and CD4 T cells and NK cells (CD16+) but not on B cells (CD19+; Figure 2A). Among different donors, the percentage of KLRG1 expression varied considerably. On average, 44% ± 14% of CD8, 18% ± 10% of CD4, and 58% ± 13% of CD16+ cells expressed KLRG1 on the cell surface (Figure2C). Additional staining using mAbs specific for CD45RA, CD28, CD62L, and CD11a further revealed that KLRG1 was predominantly found on T cells with an effector/memory phenotype (Figure 2B). Memory T cells are known to express distinct patterns of chemokine receptors.18 19 KLRG1 was expressed at a 2- to 3-fold higher frequency among T-helper 1 (TH1)–biased CXCR3+/CCR4− cells when compared with the TH2-biased CXCR3−/CCR4+ population (Figure 2D). Intracellular cytokine staining of PMA/ionomycin-activated PBLs further revealed that IFN-γ–secreting cells were more frequent in the KLRG1+ memory T-cell population (CD45RO+) than in the KLRG1− memory cell population. In contrast, IL-4–secreting cells were present in KLRG1+ and KLRG1− populations with similar frequencies (Figure 2E).
Expression of KLRG1 in human PBL.
(A) PBLs from a representative healthy donor were doubly stained with mAb specific for KLRG1 and CD8, CD4, CD16, or CD19. (B) Coexpression of KLRG1 versus CD45RA, CD28, CD62L, and CD11a gated on CD8 and CD4 T cells. (C) Percentages of KLRG1+ cells in the indicated cell subsets of healthy adult donors (20 to 40 years of age). Each symbol represents the value from one donor. (D) CD4 T cells were analyzed for coexpression of KLRG1, CCR4 and CXCR3. The plot shows percentages of KLRG1+ cells in the Th1-biased CCR4 CXCR3+ and the Th2-biased (CCR4+CXCR3) cell populations. Each symbol represents the value from one donor. (E) KLRG1 versus intracellular IL-4 and IFN-γ staining gated on CD45RO+ memory T cells. PBLs were activated by PMA and ionomycin for 4 hours before cytokine analysis. (F) KLRG1 expression on NK cells. PBLs were triple-stained with mAbs specific for CD3, CD56, and KLRG1. The dot plot displays KLRG1 versus CD56 expression gated on CD3− PBLs.
Expression of KLRG1 in human PBL.
(A) PBLs from a representative healthy donor were doubly stained with mAb specific for KLRG1 and CD8, CD4, CD16, or CD19. (B) Coexpression of KLRG1 versus CD45RA, CD28, CD62L, and CD11a gated on CD8 and CD4 T cells. (C) Percentages of KLRG1+ cells in the indicated cell subsets of healthy adult donors (20 to 40 years of age). Each symbol represents the value from one donor. (D) CD4 T cells were analyzed for coexpression of KLRG1, CCR4 and CXCR3. The plot shows percentages of KLRG1+ cells in the Th1-biased CCR4 CXCR3+ and the Th2-biased (CCR4+CXCR3) cell populations. Each symbol represents the value from one donor. (E) KLRG1 versus intracellular IL-4 and IFN-γ staining gated on CD45RO+ memory T cells. PBLs were activated by PMA and ionomycin for 4 hours before cytokine analysis. (F) KLRG1 expression on NK cells. PBLs were triple-stained with mAbs specific for CD3, CD56, and KLRG1. The dot plot displays KLRG1 versus CD56 expression gated on CD3− PBLs.
Human NK cells can be divided into 2 subsets based on their CD56 cell surface expression. CD56bright NK cells have the capacity to proliferate and to produce large amounts of cytokines, whereas CD56dim cells exhibit a high cytotoxic potential.20-23 Interestingly, KLRG1+ cells were found exclusively in the CD56dim NK cell subset (Figure 2F).
Originally, KLRG1 was identified on the rat mast cell line RBL-2H39, and human KLRG1 expression at the RNA level has also been detected in the basophilic cell line KU-812 using reverse transcription–polymerase chain reaction (RT-PCR).16However, we failed to detect KLRG1 protein expression by flow cytometry on KU-812 cells, CD203c+ basophilic granulocytes, and CD14+ monocytes from human blood (data not shown).
Predominant expression of KLRG1 on CCR7− effector memory T cells
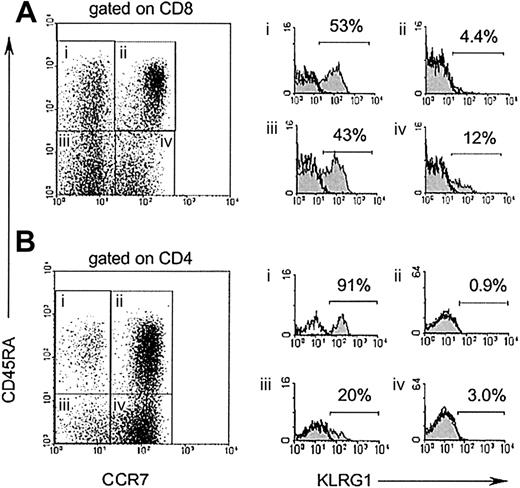
Memory T cells have been recently classified into 2 subsets, based on differential expression of the chemokine receptor CCR7.24 Central memory T cells express CCR7 and are thought to migrate to lymphoid tissues, whereas effector memory T cells lack CCR7 expression and are capable of immediate effector cell functions. To define KLRG1 expression on these 2 subsets of memory T cells, 4-color staining using antibodies specific for CD8/CD4, CD45RA, CCR7, and KLRG1 were performed. As illustrated in Figure3, KLRG1+ cells were present at low frequencies (0.9%-4.4%) among naive T cells defined by high expression of CD45RA and CCR7 (gate ii). Among CCR7+CD45RA− central memory T cells (gate iv), a slight increase in the percentage (3%-12%) of KLRG1+ cells was found, whereas 20% to 43% of CCR7−CD45RA− effector memory T cells (gate iii) expressed KLRG1. The highest percentages of KLRG1+cells (53%-90%) were observed in the CD45RA+CCR7− cell subset (gate i), which has been proposed to represent terminally differentiated effector T cells.25 26
KLRG1 is preferentially expressed on CCR7 effector T cells.
CD8 (A) and CD4 (B) T cells from PBL of a healthy adult donor were stained with mAbs to CD45RA, CCR7, and KLRG1. Filled histograms (right) display KLRG1 expression electronically gated on the 4 subsets indicated in the dot blot (left). Open histograms display isotype-staining controls of the same populations.
KLRG1 is preferentially expressed on CCR7 effector T cells.
CD8 (A) and CD4 (B) T cells from PBL of a healthy adult donor were stained with mAbs to CD45RA, CCR7, and KLRG1. Filled histograms (right) display KLRG1 expression electronically gated on the 4 subsets indicated in the dot blot (left). Open histograms display isotype-staining controls of the same populations.
KLRG1+ T cells lack proliferative capacity
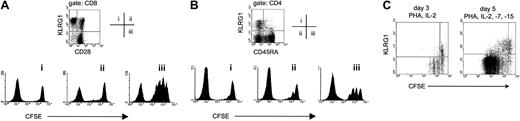
We have recently demonstrated that murine CD8 T cells expressing KLRG1 could efficiently lyse target cells and secrete cytokines but were severely impaired in their ability to proliferate after antigen stimulation.15 In humans, it is well known that CD8 T cells lacking CD28 expression exhibit impaired proliferative potential in vitro.25,27,28 Because almost all CD28− T cells from human PBLs expressed KLRG1 (Figure 2B), it was important to determine whether the replicative incapacity primarily correlated with lack of CD28 or with KLRG1 expression. Therefore, the proliferative response of cell-sorted CD28−KLRG1+, CD28+KLRG1+, and CD28+KLRG1−CD8 T cells to PHA stimulation was compared using the CFSE dilution method to track cell division.17 Unlabeled autologous feeder PBLs and IL-2 were added to the cultures to provide optimal cell growth conditions. On day 5 after stimulation, CD28+KLRG1− cells (gate iii) had divided extensively, whereas CD28−KLRG1+ (gate i) and CD28+KLRG1+ (gate ii) populations failed to proliferate (Figure 4A). This result indicates that there is a better correlation of proliferative incapacity of CD8 T cells with KLRG1 expression than with lack of the CD28 marker.
KLRG1+ T cells lack proliferative capacity.
(A) The indicated 3 CD8 T-cell populations from PBLs of a healthy adult donor were isolated by cell sorting, according to their expression of KLRG1 and CD28. Afterward, cells were labeled with CFSE and were stimulated with PHA/IL-2 in the presence of unlabeled autologous feeder cells. Cell division was analyzed by flow cytometry after 4 days. Histograms display CFSE profiles of the populations indicated. The left peak in each histogram represents unlabeled autologous feeder cells. (B) The indicated 3 CD4 T-cell populations from PBLs were isolated by cell sorting according to their expression of KLRG1 and CD45RA. Afterward, CFSE-labeled cells were stimulated with PHA/IL-2 in the presence of unlabeled autologous feeder cells, and cell division was analyzed after 5 days. The left peak in each histogram represents unlabeled autologous feeder cells. (C) Total PBLs were labeled with CFSE and were stimulated with PHA and the indicated cytokines in vitro. Three and 5 days later, cultures were stained with mAbs to CD8 and KLRG1. Dot plots shown are gated on CD8 T cells.
KLRG1+ T cells lack proliferative capacity.
(A) The indicated 3 CD8 T-cell populations from PBLs of a healthy adult donor were isolated by cell sorting, according to their expression of KLRG1 and CD28. Afterward, cells were labeled with CFSE and were stimulated with PHA/IL-2 in the presence of unlabeled autologous feeder cells. Cell division was analyzed by flow cytometry after 4 days. Histograms display CFSE profiles of the populations indicated. The left peak in each histogram represents unlabeled autologous feeder cells. (B) The indicated 3 CD4 T-cell populations from PBLs were isolated by cell sorting according to their expression of KLRG1 and CD45RA. Afterward, CFSE-labeled cells were stimulated with PHA/IL-2 in the presence of unlabeled autologous feeder cells, and cell division was analyzed after 5 days. The left peak in each histogram represents unlabeled autologous feeder cells. (C) Total PBLs were labeled with CFSE and were stimulated with PHA and the indicated cytokines in vitro. Three and 5 days later, cultures were stained with mAbs to CD8 and KLRG1. Dot plots shown are gated on CD8 T cells.
To extend these results to the CD4 subset, the rate of cell division after PHA stimulation of naive (CD45RA+KLRG1−) and of KLRG1+ and KLRG1−memory CD4 T cells (CD45RA−) was determined. As shown in Figure 4B, naive CD4 T cells (gate iii) proliferated extensively after PHA stimulation, whereas the proliferative response of CD45RA− memory cells (gates i and ii) was less pronounced. Among them, however, cell division was observed in the KLRG1− (gate ii) but not in the KLRG1+ (gate i) population.
To exclude that the lack of cell division of cell-sorted human KLRG1+ T cells was caused by inhibitory signals generated by antibody-mediated cross-linking of KLRG1, unsorted CFSE-labeled human PBLs were also analyzed after PHA stimulation in vitro. As depicted in Figure 4C (left), KLRG1− CD8 T cells underwent 3 cell cycles within 3 days after PHA/IL-2 stimulation, whereas CD8 T cells expressing KLRG1 did not divide. Similar results were obtained when plate-bound anti-CD3 mAb was used for stimulation (data not shown). IL-7 and IL-15 are important cytokines for homeostatic proliferation of CD8 T cells.29 Nonetheless, addition of these cytokines to the culture medium failed to restore the proliferative capacity of PHA-stimulated KLRG1+ CD8 T cells (Figure 4C, right). Moreover, this experiment showed that IL-15 did not induce the expression of KLRG1 on activated T cells in vitro, in contrast to its ability to induce expression of other related C-type lectins such as CD94 and NKG2A.30 Taken together, these results indicate that the expression of KLRG1 identifies effector and memory T cells in humans that lack replicative potential in the CD4 and the CD8 T-cell compartment.
Discussion
Induction of KLRG1 expression in murine T cells requires a large number (more than 10) of cell divisions after antigen challenge.15 In human PBLs from healthy adult donors, KLRG1 was expressed on approximately 40% of CD8 and 20% of CD4 T cells. These values are significantly higher than those found in adult mice that have not undergone deliberate immunization, in which only 2% to 10% of CD8 and 1% to 2% of CD4 T cells express KLRG1. However, viral infections are known to induce extensive proliferation of lymphocytes and to dramatically increase the number of KLRG1+ T cells (30%-60% of CD8) in mice.15Therefore, the increased frequency of KLRG1+ T cells in humans compared with laboratory mice is most likely attributed to the multiple infections experienced by humans during their longer lifetimes. In both mice and humans, higher percentages of KLRG1+ cells were found in the CD8 subset than in the CD4 subset. This result fits in well with the postulated cell division–dependent expression of KLRG1 because CD8 T cells are known to proliferate more vigorously after antigen challenge in vivo than CD4 T cells.31-33 Also consistent with this notion is the higher frequency of KLRG1+ cells among CCR7−effector memory T cells that have undergone more cell divisions than CCR7+ central memory cells.24
It is unknown whether KLRG1+ memory T cells can revert to KLRG1− cells. In mice, purified KLRG1+ CD8 T cells that had been adoptively transferred into recipient mice did not revert to KLRG1− cells and could still be detected in PBLs of the recipients 6 months after transfer (D.V., unpublished observations, June 2001). Given the striking similarity of KLRG1+ expression in the lymphocytes of mice and humans, one may hypothesize that human T cells also show a linear, irreversible differentiation from KLRG1− to KLRG1+ cells. Unfortunately, this important issue cannot easily be addressed by in vitro experiments because KLRG1 expression is slowly down-regulated in long-term T-cell cultures (data not shown).
Numerous studies have shown that the lack of CD28 expression on T cells correlates with immediate effector T-cell function,25,34reduced proliferative capacity,28,35,36 and shortened telomers.37 The presence of KLRG1 on almost all T cells lacking CD28 further supports the view that KLRG1 expression on human T cells also requires a large number of cell divisions. The present work identified an additional T-cell subset in humans (KLRG1+CD28+) that exhibits a proliferative incapacity similar to CD28− T cells. This implies that the proliferative incapacity of T cells correlates better with KLRG1 expression than with loss of CD28.
It is unclear whether the inhibition of cell-cycle progression of KLRG1+ T cells is due to inhibitory signals mediated by KLRG1. An inhibitory function of KLRG1 on FcεRI-mediated degranulation has been demonstrated in the rat RBL-2H3 mast cell line.9 Furthermore, antibody-mediated cross-linking of KLRG1 on T cells from transgenic mice overexpressing KLRG1 has recently been shown to result in a small inhibition of Ca++mobilization and of cell-mediated cytotoxicity.14 The immunoreceptor tyrosine-based inhibitory motif (ITIM), present in the cytoplasmic tail of the KLRG1 protein from mice, rats and humans, provides a structural basis for the inhibitory potential of this molecule. However, our attempts to demonstrate an inhibitory function of KLRG1 on T cells from healthy mice and humans have failed so far. The lack of cell division of cell-sorted KLRG1+ T cells was not caused by antibody-mediated cross-linking of KLRG1 because KLRG1+ T cells from unseparated total PBL cultures also failed to proliferate after PHA stimulation (Figure 4C). Nonetheless, inhibition of cell division under these conditions could still be mediated by KLRG1 through its unknown physiological ligand(s) present on neighboring cells in the culture.
In addition to T cells, a significant portion (approximately 50%) of human NK cells expressed KLRG1. Strikingly, KLRG1+ cells were found exclusively in the CD56dim NK cell subset. CD56dim NK cells have potent cytolytic activity but proliferate poorly in response to mitogenic cytokines, whereas CD56bright NK cells have proliferative capacity and produce large amounts of cytokines.20-23 In mice, 30% to 40% of NK cells express KLRG113 and a recent study has shown that KLRG1 expression on these cells correlated inversely with their ability to produce IFN-γ.38 Because NK cells in mice do not express a murine homologue of CD56, it is unknown whether KLRG1+ NK cells in the mouse are restricted to a particular subset analogous to the CD56dim population in humans. In mice lacking major histocompatibility complex (MHC) class 1 molecules, the number of KLRG1+ NK cells is reduced substantially.13 Similarly, patients with low MHC class 1 expression because of a deficiency in the peptide transporter associated with antigen processing (TAP) have recently been shown to contain lower numbers of CD56dim NK cells.39Thus, it will be interesting to analyze KLRG1 expression on NK cells from these patients.
In conclusion, we have generated a mAb specific for human KLRG1. This novel reagent allows identification of a subset of NK cells and antigen-experienced T cells in humans that lack proliferative capacity. These cells can now be monitored in patients with infections, tumors, immune deficiencies, and autoimmune diseases.
We thank Drs. S. Batsford and S. Ehl for comments on the manuscript.
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2002-02-0657.
Supported by the State of Baden-Württemberg (Zentrum für Klinische Forschung I/ Teilprojekt B5; Universitätsklinikum Freiburg) and by the Deutsche Forschungsmeinschaft (SFB 620 Teilprojekt B2).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hanspeter Pircher, Institute for Medical Microbiology and Hygiene, Department of Immunology, Hermann-Herder-Str. 11, University of Freiburg, D-79104 Freiburg, Germany; e-mail: pircher@UKL.uni-freiburg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal