Rodent mast cells (MCs) are common experimental tools but are somewhat different from their human counterparts in their responses to certain cytokines and drugs. We examined the expression of more than 10 000 distinct genes in human and mouse cultured MCs using high-density oligonucleotide probe arrays to find molecules similarly regulated and expressed by the 2 MC types. After stimulation via high-affinity Fcε receptor I (FcεRI), the transcriptional levels of several CC chemokines were markedly increased, and I-309 (CCL1), macrophage inflammatory protein-1α (MIP-1α) (CCL3) and MIP-1β (CCL4) were found among the 10 most increased human and mouse transcripts from approximately 12 000 genes (including some expressed sequence tags). In addition, a costimulatory molecule that was originally found on the membrane of activated T cells, 4-1BB (CD137), was found among the 10 most increased transcripts. The FcεRI-induced expression of CC chemokines and 4-1BB was also detected at the protein level in both MC types. The conservation of these responses suggests that MCs play a crucial role in recruitment of various CCR-expressing cells into the tissue in a manner dependent on immunoglobin E, and that FcεRI-mediated induction of several CC chemokines and 4-1BB is highly conserved between human and mouse. Interspecies comparison studies at the whole genome expression level should be useful for the interpretation of experimental data obtained in animal models of human pathobiology.
Introduction
Mast cells (MCs) express the high-affinity immunoglobin E (IgE) receptor (FcεRI) on their surface, and they can be activated to secrete a variety of biologically active mediators by cross-linking of receptor-bound IgE. The release of mediators from MCs is responsible for the IgE-dependent allergic reactions clinically recognized as anaphylactic reactions, acute asthma, and allergic rhinitis.1 However, much information about MC biology has been established by using commonly available rodent counterparts. Recently, it has become clear that human MCs show somewhat distinct responses to antiasthma drugs compared to rodent MCs.2 3Therefore, it is now necessary to determine which observations obtained using rodent MCs are applicable to the human.
A draft reading of all human genome sequences has been completed.4,5 It is expected that in the near future we will resolve previously unanswered questions, such as the probability of development of various diseases, by screening for single nucleotide polymorphisms over the whole genome sequence. Comprehension of the genome has also accelerated our understanding of the transcriptome,6 which is the totality of transcripts present in a cell, and the proteome,7 the proteins present in specific cell. Recently developed techniques—cDNA microarrays8 and oligonucleotide expression probe arrays9—have made systematic analyses of transcriptomes practical. High-density oligonucleotide expression probe array (GeneChip, Affymetrix, Santa Clara, CA) is designed to measure the absolute levels of more than 10 000 transcripts, regardless of the cell type, by using the same set of inner standards on a 1.6-cm2 glass chip. Competition with another cell type is required for cDNA microarray assay, but not with the GeneChip. Thus, we can compare the expression levels of more than 10 000 transcripts even in different cell types by using the high-density oligonucleotide probe array.9-11 In the present study, we used the GeneChip for transcriptomes to analyze the RNA from mouse bone marrow–derived cultured MCs12-14 and human cord blood–derived cultured MCs,15-17 both widely used as experimental tools in MC biology. We found that the mRNA levels of several CC chemokines (ie, a subgroup of small [8-14 kDa], basic, structurally related molecules that regulate cell chemotaxis18) were markedly increased after stimulation via FcεRI in both human and murine MCs.
Materials and methods
Cytokines and antibodies
Recombinant human (rh) interleukin 3 (IL-3) and recombinant mouse (rm) IL-3 were purchased from Intergen (Purchase, NY); rh IL-6 was kindly provided by Kirin Brewery (Maebashi, Japan); and rh IL-4 was purchased from R&D Systems (Minneapolis, MN). Recombinant human and mouse stem cell factors (SCFs) were purchased from PeproTech EC (London, England). Anti-human tryptase monoclonal antibody (mAb) was purchased from Chemicon (Temecula, CA). Anti-human CD137, 4-1BB mAb (clone 4B4-1, mouse IgG1), and its irrelevant antibody (Ab) were purchased from Beckman Coulter Japan (Tokyo, Japan). Anti-mouse CD137, 4-1BB mAb (clone 1AH2), its control Ab, rat IgG1κ, and R-phycoerythrin–conjugated goat anti-mouse and anti-rat Abs were purchased from BD Pharmingen (San Diego, CA).
Purification of human CD34+ cells
All human subjects in this study provided written informed consent, and the study was approved by the ethical review boards at their hospitals. Mononuclear cells were obtained from umbilical cord blood samples derived from healthy nonatopic mothers and separated by density-gradient centrifugation on Lymphocyte Separation Medium (Organon Teknika, Durham, NC). The interface containing mononuclear cells was collected. CD34+ cells were positively selected from cord blood (CB)–derived mononuclear cells by means of a CD34+ cell isolation kit and a magnetic separation column (MACS II, Miltenyi Biotec, Bergisch Gladbach, Germany) used according to the manufacturer's instructions.
Culture of human MCs from CD34+ cells
Human CD34+ cells were suspended in complete Iscove modified Dulbecco medium (IMDM), which consisted of IMDM (Life Technologies, Rockville, MD) supplemented with 1% insulin-transferrin-selenium-A supplement (Life Technologies), 50 μM 2-ME (Life Technologies), 100 units/mL penicillin (Life Technologies), 100 μg/mL streptomycin (Life Technologies), and 0.1% bovine serum albumin (BSA; Sigma, St Louis, MO). CD34+ cells were cultured in the complete IMDM supplemented with 100 ng/mL SCF, 50 ng/mL IL-6, and 2% fetal calf serum (FCS; Cansera, Rexdale, Canada) in 25- or 75-cm2 flasks (Iwaki Glass, Tokyo, Japan) as described elsewhere.10,11,15 After 11 to 14 weeks of culture, the cells (> 99% were tryptase positive) were further treated with IL-3 at 10 ng/mL in addition to the above cytokines for 7 days and then used for transcriptome and cytokine production assay. IL-3 was added after 11 weeks because it stimulates basophil production when added from the beginning of culture19 but prevents the apoptosis of cord blood–derived MCs when added after 10 weeks of culture20and because mouse MCs require mIL-3. In a preliminary study, IL-3 did not promote functional maturation of IgE-dependent histamine release and granulocyte-macrophage colony-stimulating factor (GM-CSF) production.
Activation of human MCs
The human MCs were sensitized with 1 μg/mL human myeloma IgE (a generous gift from Dr Kimishige Ishizaka, La Jolla, CA) at 37°C for 48 hours in the presence of IL-4 plus SCF and IL-6. After washing, the cells were suspended in the complete IMDM with the above cytokines. The cells were then challenged with either 1.5 μg/mL rabbit anti-human IgE Ab (Dako, Glostrup, Denmark) or the culture medium alone at 37°C for 6 hours.
Culture of WEHI-3 cell line
We used conditioned medium from the WEHI-3 cell line (American Type Culture Collection, Rockville, MD) as a source of IL-3. The cells were suspended at 5 × 105 cells/mL and cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% FCS, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; Sigma, and 50 μM 2-ME for 72 hours. The supernatant was then collected after centrifuging at 800g for 15 minutes and stored at −40°C after filtration.
Culture of mouse MCs from bone marrow cells
BALB/c mice were purchased from Japan SLC (Hamamatsu, Japan). NOA mice (Naruto Research Institute Otsuka Atrichia, Naruto, Japan) were of Japanese fancy-mice origin and are reported to have high susceptibility to development of atopic eczemalike dermatitis.21 All animal experiments were performed under the protocol approved by each institutional review board. Cultured mouse MCs were generated from the femoral bone marrow cells of mice as described previously.14 22 Cells were grown in RPMI 1640 medium supplemented with 10% FCS, 50 μM 2-ME, and 20% WEHI3 cell line–conditioned medium as a source of MC growth factors by replacement of half of the medium weekly. After 4 to 5 weeks of culture, more than 98% of the cells were identifiable as MCs by toluidine blue staining. For some of the experiments, the cells were further cultured with rm SCF at 100 ng/mL in addition to the above medium for 7 days.
Activation of mouse MCs
Cultured mouse MCs were sensitized overnight with 2 μg/mL mouse monoclonal antidinitrophenol (anti-DNP) IgE Ab (a generous gift from Dr Kimishige Ishizaka) in the above culture medium. After sensitization, the cells were washed twice and suspended at 1 × 106 cells/mL in the culture medium. The cells were challenged with either an optimal concentration (10 ng/mL) of DNP derivatives of bovine serum albumin (DNP-BSA, containing 35 DNP groups per BSA molecule; Calbiochem, La Jolla, CA) or control solution at 37°C for 6 hours. The spleen was obtained as the control tissue for MC-specific genes.
GeneChip expression analysis
Human genome-wide gene expression was examined by using the Human Genome U95A probe array (GeneChip, Affymetrix), which contains the oligonucleotide probe set for approximately 12 000 full-length genes, according to the manufacturer's protocol (Expression Analysis Technical Manual) and previous reports.10 11 Mouse genome screening was done by using Murine Genome U74A probe array (GeneChip, Affymetrix) containing approximately 6000 full-length genes and 6000 expressed sequence tags (ESTs). Total RNA (3-10 μg) was extracted from approximately 107 cells. Double-stranded cDNA was synthesized by means of a SuperScript Choice system (Life Technologies, Rockville, MD) and a T7-(dT)24 primer (Amersham Pharmacia Biotech, Buckinghamshire, England). The cDNA was subjected to in vitro transcription in the presence of biotinylated nucleoside triphosphates by means of a BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). The biotinylated cRNA was hybridized with a probe array for 16 hours at 45°C. After washing, the hybridized biotinylated cRNA was stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) and then scanned with an HP Gene Array Scanner (Affymetrix). The fluorescence intensity of each probe was quantified with a computer program, GeneChip Analysis Suite 4.0 (Affymetrix). The expression level of a single mRNA was determined as the average fluorescence intensity among the intensities obtained by 6- to 20-paired (perfect-matched and single nucleotide-mismatched) probes consisting of 18- to 25-mer oligonucleotides. If the intensities of mismatched probes were very high, gene expression was judged to be absent even if a high average fluorescence was obtained with the GeneChip Analysis Suite 4.0 program. The level of gene expression was determined as the average difference (AD) using the GeneChip software. The percentages of the specific AD level versus the mean AD level of 6 probe sets for housekeeping genes (β-actin and glyceraldehyde-3-phosphate dehydrogenase) were then calculated.
ELISA for CC-chemokines
Human I-309 (CCL1) was measured by sandwich enzyme-linked immunosorbent assay (ELISA). Ninety-six–well microtitre plates (Nunc-Immuno Module F8 MaxiSorp, Nalge Nunc International, Roskilde, Denmark) were coated with 5 μg/mL of mouse anti-human I-309 mAb (clone no. 35305.11, R&D Systems) in carbonate buffer at 4°C. After overnight incubation the wells were blocked for 2 hours with the blocking solution (Blocking reagent for ELISA, Roche Diagnostics, Mannheim, Germany), and, after washing, 100 μL of samples were allowed to incubate for 18 hours. After incubation, the plates were treated with 100 μL of biotinylated anti-human I-309 Ab (0.3 μg/mL, PeproTech EC) for 3 hours, followed by 100 μL of streptavidin-peroxidase (Life Technologies) for 45 minutes. The plates were developed with the TMB microwell peroxidase substrate system (Kirkegaard & Perry Laboratories, Gaithersburg, MD); the reactions were stopped with 1 M phosphoric acid. Absorbance was measured at 450 nm, and a standard curve was generated by using recombinant I-309 (PeproTech EC). The sensitivity of the assay was 41 pg/mL. Mouse I-309 was also measured by ELISA according to the above method,with some modification. Hamster anti-mouse I-309 (annotated as TCA3) mAb (clone 4B12, BD Pharmingen) and biotinylated anti-mouse I-309 (TCA3) antibody (0.3 μg/mL, R&D Systems) were using the coating and the captured antibodies, respectively. The standard curve was generated by using recombinant I-309 (BD Pharmingen). The sensitivity of the assay was 123 pg/mL. Monocyte chemoattractant protein-1 (MCP-1; CCL2) and macrophage inflammatory protein-1β (MIP-1β; CCL4) were measured by ELISA kits purchased from R&D Systems.
Flow cytometric analyses
MCs were suspended in phosphate-buffered saline (PBS) containing 1% BSA and 0.1% NaN3. The cells were then incubated with each primary Ab or its irrelevant Ab in the presence of human IgG (ICN Biomedicals, Aurora, OH) for 30 minutes. They were then incubated with either fluorescein isothiocyanate (FITC)– or phycoerythrin-conjugated goat anti-mouse IgG Ab or goat anti-rat IgG Ab for 30 minutes at 4°C in the dark. After washing, the cells were analyzed by fluorescence-activated cell sorter (FACS) and Cell Quest software (Becton Dickinson, San Jose, CA). The mean fluorescence intensities (MFIs) of MCs stained with specific Ab and those stained with control Ab were obtained.
Statistical analysis
Differences between 2 paired groups were analyzed by the paired Student t test and were considered significant atP < .05. Values are expressed as the mean ± SEM.
Results
Marked increase in CC chemokine transcripts in activated MCs
The aim of this study was to compare the gene expression profiles of widely used functionally mature MC types derived from humans and mice. Human cord blood–derived cultured MCs require SCF, IL-6, and IL-4, while mouse MCs require IL-3 for their development and functional maturation.14,15 As much as possible, we tried to compensate for differences in the standard culture conditions. We added IL-3 for human MCs and SCF for mouse MCs in this study, although these cytokines did not have a significant effect on cytokine production in a preliminary study. We did not add IL-4 or IL-6 to the mouse culture system, since they induce apoptosis of mouse MCs23 or development of other cell types, such as mouse dendritic cells.24 After stimulation via FcεRI, 4 common molecules were found in the 10 most increased human transcripts and mouse transcripts among approximately 12 000 genes (Table1). Three of the 4 increased transcripts were for CC chemokines: I-309 (CCL1), MIP-1α (CCL3), and MIP-1β (CCL4). The other transcript increased in both human and mouse MCs was for 4-1BB (CD137).
Most increased transcripts in activated human and mouse MCs
| Name of transcript (protein) . | GenBank accession no.* . | Experiment 1†,‡ . | Experiment 2 . | Experiment 3 . | Fold increase1-153 . | |||
|---|---|---|---|---|---|---|---|---|
| Rest . | Act . | Rest . | Act . | Rest . | Act . | |||
| Human MCs | ||||||||
| Class I MHC-restricted T-cell-associated molecule | AF001622 | <0.11-155 | 4.1 | <0.1 | 5.0 | ND | ND | 676.0 |
| CCL1, I-309 | M57506 | 0.1 | 36.7 | 0.1 | 66.9 | ND | ND | 446.0 |
| GM-CSF | M13207 | 0.1 | 2.5 | 0.4 | 52.6 | ND | ND | 108.6 |
| CCL4, MIP-1β | J04130 | 1.6 | 22.5 | 1.5 | 69.1 | ND | ND | 30.1 |
| CD137, 4-1BB | U03397 | <0.1 | 2.6 | 0.2 | 4.9 | ND | ND | 24.2 |
| Gem GTPase | U10550 | 0.4 | 2.3 | 0.4 | 12.4 | ND | ND | 20.2 |
| CXCL8, IL-8 | M28130 | 4.2 | 20.5 | 2.2 | 78.5 | ND | ND | 15.4 |
| CCL3, MIP-1α | D90144 | 3.0 | 7.8 | 2.0 | 65.5 | ND | ND | 14.5 |
| Putative cyclin G1-interacting protein | U61836 | 0.6 | 7.5 | 1.3 | 19.5 | ND | ND | 13.8 |
| IL-3 | M20137 | 0.8 | 2.6 | 0.3 | 12.9 | ND | ND | 13.5 |
| P1 cre recombinase (3′)1-154 | — | 59.9 | 58.6 | 49.0 | 55.4 | ND | ND | |
| P1 cre recombinase (5′) | — | 42.3 | 39.0 | 30.1 | 40.4 | ND | ND | |
| Mean, 6 AD levels of housekeeping genes | 12710 | 15606 | 17850 | 16398 | ND | ND | ||
| Mouse MCs | ||||||||
| CCL1, I-309 | M23501 | <0.11-155 | 17.5 | <0.1 | 4.2 | 0.2 | 25.1 | 208.2 |
| CD137, 4-1BB | AA798611 | 0.2 | 26.4 | 0.5 | 36.4 | 1.0 | 67.1 | 73.9 |
| CCL7, MCP-3 | X70058 | 0.1 | 21.7 | 0.3 | 14.4 | 0.4 | 15.1 | 60.5 |
| Myeloid differentiation primary response gene 118 | X54149 | 0.9 | 18.2 | <0.1 | 2.4 | 0.3 | 5.6 | 21.4 |
| CCL3, MIP-1α | J04491 | 1.2 | 72.9 | 2.2 | 7.2 | 4.0 | 17.5 | 13.2 |
| CCL4, MIP-1β | X62502 | <0.1 | 23.1 | 1.0 | 6.9 | 3.2 | 12.9 | 10.2 |
| Homeodomain interacting protein kinase 2 | AF077659 | 1.4 | 7.9 | 0.5 | 4.9 | <0.1 | 4.9 | 8.9 |
| Asparagine synthetase | U38940 | 0.8 | 6.1 | 4.2 | 38.0 | 6.2 | 48.3 | 8.3 |
| UI-M-BH2.2-aoo-b-05-0-UI.s1 | AW122030 | 1.3 | 7.4 | 3.1 | 26.2 | 4.8 | 34.4 | 7.4 |
| Secreted phosphoprotein 1 | X13986 | 1.1 | 14.6 | 1.1 | 5.6 | 2.4 | 11.2 | 6.9 |
| P1 cre recombinase (3′)1-154 | — | 112.7 | 83.3 | 133.5 | 152.2 | 112.8 | 91.2 | 58.6 |
| P1 cre recombinase (5′) | — | 72.2 | 56.5 | 75.8 | 83.7 | 66.4 | 48.7 | 39.0 |
| Mean, 6 AD levels of housekeeping genes | 13754 | 18553 | 11767 | 11733 | 9223 | 7819 | ||
| Name of transcript (protein) . | GenBank accession no.* . | Experiment 1†,‡ . | Experiment 2 . | Experiment 3 . | Fold increase1-153 . | |||
|---|---|---|---|---|---|---|---|---|
| Rest . | Act . | Rest . | Act . | Rest . | Act . | |||
| Human MCs | ||||||||
| Class I MHC-restricted T-cell-associated molecule | AF001622 | <0.11-155 | 4.1 | <0.1 | 5.0 | ND | ND | 676.0 |
| CCL1, I-309 | M57506 | 0.1 | 36.7 | 0.1 | 66.9 | ND | ND | 446.0 |
| GM-CSF | M13207 | 0.1 | 2.5 | 0.4 | 52.6 | ND | ND | 108.6 |
| CCL4, MIP-1β | J04130 | 1.6 | 22.5 | 1.5 | 69.1 | ND | ND | 30.1 |
| CD137, 4-1BB | U03397 | <0.1 | 2.6 | 0.2 | 4.9 | ND | ND | 24.2 |
| Gem GTPase | U10550 | 0.4 | 2.3 | 0.4 | 12.4 | ND | ND | 20.2 |
| CXCL8, IL-8 | M28130 | 4.2 | 20.5 | 2.2 | 78.5 | ND | ND | 15.4 |
| CCL3, MIP-1α | D90144 | 3.0 | 7.8 | 2.0 | 65.5 | ND | ND | 14.5 |
| Putative cyclin G1-interacting protein | U61836 | 0.6 | 7.5 | 1.3 | 19.5 | ND | ND | 13.8 |
| IL-3 | M20137 | 0.8 | 2.6 | 0.3 | 12.9 | ND | ND | 13.5 |
| P1 cre recombinase (3′)1-154 | — | 59.9 | 58.6 | 49.0 | 55.4 | ND | ND | |
| P1 cre recombinase (5′) | — | 42.3 | 39.0 | 30.1 | 40.4 | ND | ND | |
| Mean, 6 AD levels of housekeeping genes | 12710 | 15606 | 17850 | 16398 | ND | ND | ||
| Mouse MCs | ||||||||
| CCL1, I-309 | M23501 | <0.11-155 | 17.5 | <0.1 | 4.2 | 0.2 | 25.1 | 208.2 |
| CD137, 4-1BB | AA798611 | 0.2 | 26.4 | 0.5 | 36.4 | 1.0 | 67.1 | 73.9 |
| CCL7, MCP-3 | X70058 | 0.1 | 21.7 | 0.3 | 14.4 | 0.4 | 15.1 | 60.5 |
| Myeloid differentiation primary response gene 118 | X54149 | 0.9 | 18.2 | <0.1 | 2.4 | 0.3 | 5.6 | 21.4 |
| CCL3, MIP-1α | J04491 | 1.2 | 72.9 | 2.2 | 7.2 | 4.0 | 17.5 | 13.2 |
| CCL4, MIP-1β | X62502 | <0.1 | 23.1 | 1.0 | 6.9 | 3.2 | 12.9 | 10.2 |
| Homeodomain interacting protein kinase 2 | AF077659 | 1.4 | 7.9 | 0.5 | 4.9 | <0.1 | 4.9 | 8.9 |
| Asparagine synthetase | U38940 | 0.8 | 6.1 | 4.2 | 38.0 | 6.2 | 48.3 | 8.3 |
| UI-M-BH2.2-aoo-b-05-0-UI.s1 | AW122030 | 1.3 | 7.4 | 3.1 | 26.2 | 4.8 | 34.4 | 7.4 |
| Secreted phosphoprotein 1 | X13986 | 1.1 | 14.6 | 1.1 | 5.6 | 2.4 | 11.2 | 6.9 |
| P1 cre recombinase (3′)1-154 | — | 112.7 | 83.3 | 133.5 | 152.2 | 112.8 | 91.2 | 58.6 |
| P1 cre recombinase (5′) | — | 72.2 | 56.5 | 75.8 | 83.7 | 66.4 | 48.7 | 39.0 |
| Mean, 6 AD levels of housekeeping genes | 13754 | 18553 | 11767 | 11733 | 9223 | 7819 | ||
Unreviewed additional data are shown athttp://www.nch.go.jp/imal/English_index.htm.
Rest indicates resting; Act, active; MHC, major histocompatibility complex; GTPase, guanosine triphosphatase; AD, average difference; and ND, not done.
The GenBank is accessible at http://www.ncbi.nlm.nih.gov.
Three different batches of human cord blood-derived cultured MCs at week 14 (experiment 1) and week 11 (experiment 2) were primed with SCF, IL-3, IL-6, and IL-4 (see “Materials and methods”). Sensitized MCs were then challenged with either anti-IgE (activated) or control medium (resting) for 6 hours.
Balb/c mouse bone marrow-derived cultured MCs at week 4 (experiment 1) with IL-3 were further cultured with IL-3 plus SCF for 1 week (experiment 2). NOA mouse bone marrow-derived cultured MCs at week 4 with IL-3 were also cultured with SCF and IL-3 (see “Materials and methods”). Sensitized MCs were then challenged with either DNP-BSA antigen (activated) or control medium (resting) for 6 hours.
Fold increase was obtained by calculating the AD levels of activated MCs vs those of resting MCs.
The percentages of the specific average AD level versus the mean AD level of 6 probe sets for housekeeping genes are shown. Only transcripts that expressed more than 2% of housekeeping gene levels and that were judged to be present in activated cells are shown in this table.
External standards such as P1 cre recombinase protein were added to cRNA of the target sample just prior to the hybridization (see “Materials and methods”) to evaluate the difference between samples. These standards were found at similar levels (an increase of less than 2-fold) in these MC samples after compensation for the housekeeping genes.
Similar chemokine gene expression profiles of human and mouse MCs
Next, we compared gene expression profiles of human MCs and mouse MCs with respect to chemokines, cytokines, and their receptors. As shown in Table 2, remarkable similarities were found in the IgE-dependent transcriptional regulation of CC chemokines between the 2 MC types. Among these similarly regulated chemokines, MCP-1 (CCL2) was highly expressed by activated MCs as well as resting MCs.
Transcripts for chemokines, cytokines, and their receptors expressed by human and mouse MCs
| . | GenBank accession no.* . | Human 1† . | Human 2‡ . | Fold increase2-160 . | GenBank accession no.* . | Mouse 12-153 . | Mouse 22-155 . | Mouse 32-154 . | Fold increase . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rest# . | Act# . | Rest . | Act . | Rest# . | Act# . | Rest . | Act . | Rest . | Act . | |||||
| Chemokines | ||||||||||||||
| CCL1, I-309 | M57506 | 0.1 | 36.7 | 0.1 | 66.9 | 446.0 | M23501 | 0.0 | 17.5 | 0.0 | 4.2 | 0.2 | 25.1 | 208.1 |
| CCL2, MCP-1 | X26683 | 50.0 | 70.7 | 28.3 | 99.3 | 2.2 | M19681 | 19.4 | 35.9 | 30.1 | 39.1 | 29.7 | 43.2 | 1.5 |
| CCL3, MIP-1α | D90144 | 3.0 | 7.8 | 2.0 | 65.5 | 14.5 | J04491 | 1.2 | 72.9 | 2.2 | 7.2 | 4.0 | 17.5 | 13.2 |
| CCL4, MIP-1β | J04130 | 1.6 | 22.5 | 1.5 | 69.1 | 30.1 | X62502 | 0.0 | 23.1 | 1.0 | 6.9 | 3.2 | 12.9 | 10.2 |
| CCL5, RANTES | M21121 | 0.0 | 0.3 | 0.0 | 0.2 | 12.9 | AF065947 | 0.1 | 2.7 | 0.0 | 0.1 | 0.0 | 0.5 | 55.3 |
| CCL7, MCP-3 | X72308 | 3.0 | 4.1 | 0.4 | 2.2 | 1.9 | X70058 | 0.1 | 21.7 | 0.3 | 14.4 | 0.4 | 15.1 | 60.5 |
| CCL8, MCP-2 | Y16645 | 6.9 | 5.1 | 2.2 | 2.8 | 0.9 | AB023418 | 0.2 | 0.0 | 0.3 | 0.2 | 0.3 | 0.8 | 1.3 |
| CCL11, Eotaxin | U46573 | 0.2 | 0.4 | 0.2 | 0.1 | 1.3 | U77462 | 0.1 | 0.1 | 0.2 | 0.3 | 0.4 | 0.3 | 1.2 |
| CCL22, MDC | U83171 | 0.6 | 2.1 | 0.3 | 0.0 | 2.3 | AF052505 | 0.3 | 0.9 | 1.2 | 1.2 | 2.1 | 2.1 | 1.2 |
| CCL25, TECK | U86358 | 1.2 | 1.5 | 0.3 | 1.1 | 1.7 | AJ249480 | 0.0 | 0.1 | 0.2 | 0.2 | 0.3 | 0.2 | 0.9 |
| CX3CL, fractalkine | U84487 | 0.3 | 0.4 | 0.3 | 0.1 | 0.9 | U92565 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 11.8 |
| CXCL4, PF4 | M25897 | 0.1 | 0.3 | 0.3 | 0.5 | 1.5 | AB017491 | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.1 | 9.9 |
| CXCL5, CKA-3 | U81234 | 0.2 | 0.2 | 0.3 | 0.4 | 1.1 | U27267 | 0.0 | 0.1 | 0.0 | 0.1 | 0.3 | 0.2 | 1.2 |
| Cytokines | ||||||||||||||
| G-CSF | X03656 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | M13926 | 1.8 | 1.4 | 3.1 | 1.9 | 4.8 | 2.0 | 0.6 |
| GM-CSF | M13207 | 0.1 | 2.5 | 0.4 | 52.6 | 108.6 | X03020 | 0.0 | 2.3 | 0.0 | 0.0 | 0.0 | 0.7 | 182.7 |
| IFNα | M28585 | 0.0 | 0.1 | 0.0 | 0.0 | 7.7 | M28587 | 0.3 | 0.0 | 0.0 | 0.0 | 0.4 | 0.4 | 0.6 |
| IFNβ1 | V00535 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | V00755 | 0.8 | 2.2 | 0.9 | 2.8 | 5.8 | 6.6 | 1.5 |
| IFNγ | J00219 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | K00083 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.6 |
| IL-1α | M28983 | 0.2 | 0.1 | 0.1 | 0.1 | 0.8 | M14639 | 0.2 | 4.1 | 0.6 | 0.8 | 0.7 | 3.6 | 5.8 |
| IL-1β | M15330 | 12.1 | 13.4 | 1.6 | 4.9 | 1.3 | M15131 | 1.6 | 16.8 | 1.0 | 1.2 | 2.6 | 5.7 | 4.6 |
| IL-2 | M22005 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | K02292 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 |
| IL-5 | X04688 | 0.3 | 0.9 | 0.1 | 5.5 | 16.0 | X06271 | 0.0 | 0.2 | 0.2 | 0.2 | 0.4 | 0.3 | 1.2 |
| IL-6 | X04430 | 0.0 | 0.2 | 0.0 | 0.3 | 9.9 | X54542 | 4.4 | 26.1 | 16.6 | 14.0 | 20.9 | 28.7 | 1.6 |
| IL-7 | J04156 | 0.5 | 0.2 | 0.2 | 0.1 | 0.4 | X07962 | 0.1 | 0.1 | 0.1 | 0.0 | 0.2 | 0.7 | 1.8 |
| IL-10 | U16720 | 0.6 | 0.8 | 3.5 | 4.0 | 1.2 | M37897 | 0.1 | 0.2 | 0.3 | 0.3 | 0.1 | 0.0 | 0.9 |
| IL-12a | M65291 | 0.1 | 0.0 | 0.2 | 0.1 | 0.5 | M86672 | 0.0 | 0.1 | 0.1 | 0.2 | 0.4 | 0.2 | 0.9 |
| IL-12b | M65290 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | M86671 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 |
| IL-13 | U31120 | 0.3 | 0.7 | 0.5 | 2.3 | 3.6 | M23504 | 0.2 | 8.6 | 0.0 | 0.1 | 0.0 | 1.4 | 50.3 |
| IL-16 | M90391 | 0.5 | 0.4 | 0.4 | 0.1 | 0.6 | AF017111 | 0.9 | 0.7 | 3.4 | 2.5 | 6.0 | 3.2 | 0.6 |
| IL-17 | U32659 | 0.0 | 0.0 | 0.0 | 0.0 | 0.3 | L13839 | 0.0 | 0.1 | 0.1 | 0.1 | 0.3 | 0.3 | 1.2 |
| IL-18 | D49950 | 1.2 | 1.5 | 1.0 | 0.7 | 1.0 | D49949 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 |
| M-CSF | M37435 | 5.0 | 6.3 | 4.7 | 41.3 | 4.9 | M21952 | 1.6 | 3.3 | 18.3 | 28.3 | 32.1 | 52.7 | 1.6 |
| TNFα | X02910 | 0.2 | 0.5 | 0.5 | 5.7 | 8.7 | D84196 | 0.2 | 4.3 | 1.0 | 1.5 | 1.1 | 3.9 | 4.1 |
| Chemokine receptors | ||||||||||||||
| CCR1 | D10925 | 0.0 | 0.1 | 0.6 | 1.0 | 1.7 | U29678 | 0.8 | 5.5 | 9.1 | 4.5 | 18.0 | 13.7 | 0.8 |
| CCR2 | U03905 | 0.4 | 0.1 | 0.2 | 0.3 | 0.7 | U56819 | 0.5 | 0.5 | 1.1 | 0.2 | 9.9 | 2.5 | 0.3 |
| CCR3 | U28694 | 0.2 | 0.1 | 0.1 | 0.1 | 1.0 | U29677 | 0.2 | 0.3 | 0.3 | 0.1 | 0.4 | 0.4 | 0.8 |
| CCR6 | U45984 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 | AJ222714 | 0.5 | 0.0 | 1.7 | 0.9 | 1.7 | 2.1 | 0.8 |
| CCR9 | AJ132337 | 0.1 | 0.1 | 0.2 | 0.0 | 0.5 | AJ132336 | 0.1 | 0.0 | 0.1 | 0.5 | 0.2 | 0.0 | 1.6 |
| CX3CR1 | U20350 | 0.0 | 0.0 | 0.1 | 0.0 | 0.7 | AF074912 | 1.3 | 0.5 | 1.2 | 1.5 | 1.4 | 1.3 | 0.8 |
| CXCR3 | X95876 | 0.1 | 0.1 | 0.0 | 0.9 | 15.6 | AF045146 | 0.6 | 0.8 | 1.7 | 1.8 | 2.1 | 2.3 | 1.1 |
| CXCR4 | L06797 | 0.5 | 0.4 | 1.5 | 0.8 | 0.6 | U15208 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 |
| Cytokine receptors | ||||||||||||||
| c-Kit | X06182 | 27.7 | 21.4 | 11.2 | 2.6 | 0.6 | Y00864 | 36.9 | 26.9 | 70.0 | 57.4 | 56.7 | 50.3 | 0.8 |
| G-CSFR | M59818 | 0.2 | 0.0 | 0.3 | 0.5 | 0.9 | U05894 | 1.7 | 2.1 | 2.9 | 4.9 | 4.4 | 1.9 | 1.0 |
| IFNgR | U19247 | 4.5 | 4.9 | 5.2 | 5.0 | 1.0 | M28233 | 0.3 | 0.4 | 1.2 | 0.5 | 1.8 | 1.7 | 0.8 |
| IL-1R | M27492 | 1.0 | 1.5 | 2.1 | 3.0 | 1.4 | M20658 | 0.2 | 0.4 | 0.7 | 0.9 | 0.4 | 1.2 | 1.7 |
| IL-2R | X01057 | 0.1 | 0.6 | 0.1 | 0.4 | 8.0 | M26271 | 0.3 | 0.1 | 0.2 | 0.0 | 0.4 | 0.3 | 0.5 |
| IL-2Rβ | M26062 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | M28052 | 0.1 | 0.1 | 0.1 | 0.6 | 0.3 | 0.0 | 1.6 |
| IL-2Rγ | D11086 | 6.0 | 9.7 | 3.9 | 8.0 | 1.8 | X75337 | 0.8 | 1.0 | 1.2 | 2.0 | 1.1 | 1.7 | 1.5 |
| IL-3Rα | D49410 | 1.0 | 1.4 | 1.2 | 2.8 | 1.9 | X64534 | 1.8 | 0.8 | 2.7 | 2.4 | 6.4 | 5.5 | 0.8 |
| IL-3Rβ1 | AL008637 | 21.4 | 13.6 | 26.8 | 10.4 | 0.5 | M34397 | 36.2 | 30.6 | 63.9 | 68.4 | 34.7 | 30.6 | 1.0 |
| IL-4R | X52425 | 16.8 | 17.4 | 10.5 | 14.8 | 1.2 | M27960 | 13.5 | 6.2 | 34.2 | 20.7 | 46.1 | 21.9 | 0.5 |
| IL-6R | X12830 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | X51975 | 0.2 | 0.0 | 0.3 | 0.9 | 1.2 | 0.9 | 1.1 |
| IL-7R | M29696 | 2.0 | 5.1 | 2.1 | 12.1 | 4.2 | M29697 | 0.2 | 1.1 | 0.6 | 1.3 | 1.5 | 2.5 | 2.0 |
| IL-9R | L39064 | 7.0 | 4.5 | 2.2 | 1.7 | 0.7 | M84746 | 0.0 | 0.0 | 0.2 | 0.9 | 0.0 | 0.4 | 5.4 |
| IL-10R | U00672 | 0.2 | 0.7 | 0.6 | 1.0 | 2.3 | L12120 | 3.8 | 5.9 | 6.0 | 9.5 | 7.5 | 8.9 | 1.4 |
| IL-11R | U32324 | 0.8 | 0.9 | 0.4 | 0.3 | 1.0 | U14412 | 0.6 | 0.4 | 1.2 | 0.0 | 1.9 | 1.2 | 0.4 |
| IL-17R | U58917 | 1.2 | 0.7 | 2.4 | 2.3 | 0.8 | U31993 | 4.5 | 2.4 | 4.3 | 3.7 | 5.8 | 4.6 | 0.7 |
| M-CSFR | X03663 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | X06368 | 0.4 | 0.6 | 0.8 | 0.4 | 0.4 | 1.2 | 1.3 |
| . | GenBank accession no.* . | Human 1† . | Human 2‡ . | Fold increase2-160 . | GenBank accession no.* . | Mouse 12-153 . | Mouse 22-155 . | Mouse 32-154 . | Fold increase . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rest# . | Act# . | Rest . | Act . | Rest# . | Act# . | Rest . | Act . | Rest . | Act . | |||||
| Chemokines | ||||||||||||||
| CCL1, I-309 | M57506 | 0.1 | 36.7 | 0.1 | 66.9 | 446.0 | M23501 | 0.0 | 17.5 | 0.0 | 4.2 | 0.2 | 25.1 | 208.1 |
| CCL2, MCP-1 | X26683 | 50.0 | 70.7 | 28.3 | 99.3 | 2.2 | M19681 | 19.4 | 35.9 | 30.1 | 39.1 | 29.7 | 43.2 | 1.5 |
| CCL3, MIP-1α | D90144 | 3.0 | 7.8 | 2.0 | 65.5 | 14.5 | J04491 | 1.2 | 72.9 | 2.2 | 7.2 | 4.0 | 17.5 | 13.2 |
| CCL4, MIP-1β | J04130 | 1.6 | 22.5 | 1.5 | 69.1 | 30.1 | X62502 | 0.0 | 23.1 | 1.0 | 6.9 | 3.2 | 12.9 | 10.2 |
| CCL5, RANTES | M21121 | 0.0 | 0.3 | 0.0 | 0.2 | 12.9 | AF065947 | 0.1 | 2.7 | 0.0 | 0.1 | 0.0 | 0.5 | 55.3 |
| CCL7, MCP-3 | X72308 | 3.0 | 4.1 | 0.4 | 2.2 | 1.9 | X70058 | 0.1 | 21.7 | 0.3 | 14.4 | 0.4 | 15.1 | 60.5 |
| CCL8, MCP-2 | Y16645 | 6.9 | 5.1 | 2.2 | 2.8 | 0.9 | AB023418 | 0.2 | 0.0 | 0.3 | 0.2 | 0.3 | 0.8 | 1.3 |
| CCL11, Eotaxin | U46573 | 0.2 | 0.4 | 0.2 | 0.1 | 1.3 | U77462 | 0.1 | 0.1 | 0.2 | 0.3 | 0.4 | 0.3 | 1.2 |
| CCL22, MDC | U83171 | 0.6 | 2.1 | 0.3 | 0.0 | 2.3 | AF052505 | 0.3 | 0.9 | 1.2 | 1.2 | 2.1 | 2.1 | 1.2 |
| CCL25, TECK | U86358 | 1.2 | 1.5 | 0.3 | 1.1 | 1.7 | AJ249480 | 0.0 | 0.1 | 0.2 | 0.2 | 0.3 | 0.2 | 0.9 |
| CX3CL, fractalkine | U84487 | 0.3 | 0.4 | 0.3 | 0.1 | 0.9 | U92565 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 11.8 |
| CXCL4, PF4 | M25897 | 0.1 | 0.3 | 0.3 | 0.5 | 1.5 | AB017491 | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.1 | 9.9 |
| CXCL5, CKA-3 | U81234 | 0.2 | 0.2 | 0.3 | 0.4 | 1.1 | U27267 | 0.0 | 0.1 | 0.0 | 0.1 | 0.3 | 0.2 | 1.2 |
| Cytokines | ||||||||||||||
| G-CSF | X03656 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | M13926 | 1.8 | 1.4 | 3.1 | 1.9 | 4.8 | 2.0 | 0.6 |
| GM-CSF | M13207 | 0.1 | 2.5 | 0.4 | 52.6 | 108.6 | X03020 | 0.0 | 2.3 | 0.0 | 0.0 | 0.0 | 0.7 | 182.7 |
| IFNα | M28585 | 0.0 | 0.1 | 0.0 | 0.0 | 7.7 | M28587 | 0.3 | 0.0 | 0.0 | 0.0 | 0.4 | 0.4 | 0.6 |
| IFNβ1 | V00535 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | V00755 | 0.8 | 2.2 | 0.9 | 2.8 | 5.8 | 6.6 | 1.5 |
| IFNγ | J00219 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | K00083 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.6 |
| IL-1α | M28983 | 0.2 | 0.1 | 0.1 | 0.1 | 0.8 | M14639 | 0.2 | 4.1 | 0.6 | 0.8 | 0.7 | 3.6 | 5.8 |
| IL-1β | M15330 | 12.1 | 13.4 | 1.6 | 4.9 | 1.3 | M15131 | 1.6 | 16.8 | 1.0 | 1.2 | 2.6 | 5.7 | 4.6 |
| IL-2 | M22005 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | K02292 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 |
| IL-5 | X04688 | 0.3 | 0.9 | 0.1 | 5.5 | 16.0 | X06271 | 0.0 | 0.2 | 0.2 | 0.2 | 0.4 | 0.3 | 1.2 |
| IL-6 | X04430 | 0.0 | 0.2 | 0.0 | 0.3 | 9.9 | X54542 | 4.4 | 26.1 | 16.6 | 14.0 | 20.9 | 28.7 | 1.6 |
| IL-7 | J04156 | 0.5 | 0.2 | 0.2 | 0.1 | 0.4 | X07962 | 0.1 | 0.1 | 0.1 | 0.0 | 0.2 | 0.7 | 1.8 |
| IL-10 | U16720 | 0.6 | 0.8 | 3.5 | 4.0 | 1.2 | M37897 | 0.1 | 0.2 | 0.3 | 0.3 | 0.1 | 0.0 | 0.9 |
| IL-12a | M65291 | 0.1 | 0.0 | 0.2 | 0.1 | 0.5 | M86672 | 0.0 | 0.1 | 0.1 | 0.2 | 0.4 | 0.2 | 0.9 |
| IL-12b | M65290 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | M86671 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 |
| IL-13 | U31120 | 0.3 | 0.7 | 0.5 | 2.3 | 3.6 | M23504 | 0.2 | 8.6 | 0.0 | 0.1 | 0.0 | 1.4 | 50.3 |
| IL-16 | M90391 | 0.5 | 0.4 | 0.4 | 0.1 | 0.6 | AF017111 | 0.9 | 0.7 | 3.4 | 2.5 | 6.0 | 3.2 | 0.6 |
| IL-17 | U32659 | 0.0 | 0.0 | 0.0 | 0.0 | 0.3 | L13839 | 0.0 | 0.1 | 0.1 | 0.1 | 0.3 | 0.3 | 1.2 |
| IL-18 | D49950 | 1.2 | 1.5 | 1.0 | 0.7 | 1.0 | D49949 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 |
| M-CSF | M37435 | 5.0 | 6.3 | 4.7 | 41.3 | 4.9 | M21952 | 1.6 | 3.3 | 18.3 | 28.3 | 32.1 | 52.7 | 1.6 |
| TNFα | X02910 | 0.2 | 0.5 | 0.5 | 5.7 | 8.7 | D84196 | 0.2 | 4.3 | 1.0 | 1.5 | 1.1 | 3.9 | 4.1 |
| Chemokine receptors | ||||||||||||||
| CCR1 | D10925 | 0.0 | 0.1 | 0.6 | 1.0 | 1.7 | U29678 | 0.8 | 5.5 | 9.1 | 4.5 | 18.0 | 13.7 | 0.8 |
| CCR2 | U03905 | 0.4 | 0.1 | 0.2 | 0.3 | 0.7 | U56819 | 0.5 | 0.5 | 1.1 | 0.2 | 9.9 | 2.5 | 0.3 |
| CCR3 | U28694 | 0.2 | 0.1 | 0.1 | 0.1 | 1.0 | U29677 | 0.2 | 0.3 | 0.3 | 0.1 | 0.4 | 0.4 | 0.8 |
| CCR6 | U45984 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 | AJ222714 | 0.5 | 0.0 | 1.7 | 0.9 | 1.7 | 2.1 | 0.8 |
| CCR9 | AJ132337 | 0.1 | 0.1 | 0.2 | 0.0 | 0.5 | AJ132336 | 0.1 | 0.0 | 0.1 | 0.5 | 0.2 | 0.0 | 1.6 |
| CX3CR1 | U20350 | 0.0 | 0.0 | 0.1 | 0.0 | 0.7 | AF074912 | 1.3 | 0.5 | 1.2 | 1.5 | 1.4 | 1.3 | 0.8 |
| CXCR3 | X95876 | 0.1 | 0.1 | 0.0 | 0.9 | 15.6 | AF045146 | 0.6 | 0.8 | 1.7 | 1.8 | 2.1 | 2.3 | 1.1 |
| CXCR4 | L06797 | 0.5 | 0.4 | 1.5 | 0.8 | 0.6 | U15208 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 |
| Cytokine receptors | ||||||||||||||
| c-Kit | X06182 | 27.7 | 21.4 | 11.2 | 2.6 | 0.6 | Y00864 | 36.9 | 26.9 | 70.0 | 57.4 | 56.7 | 50.3 | 0.8 |
| G-CSFR | M59818 | 0.2 | 0.0 | 0.3 | 0.5 | 0.9 | U05894 | 1.7 | 2.1 | 2.9 | 4.9 | 4.4 | 1.9 | 1.0 |
| IFNgR | U19247 | 4.5 | 4.9 | 5.2 | 5.0 | 1.0 | M28233 | 0.3 | 0.4 | 1.2 | 0.5 | 1.8 | 1.7 | 0.8 |
| IL-1R | M27492 | 1.0 | 1.5 | 2.1 | 3.0 | 1.4 | M20658 | 0.2 | 0.4 | 0.7 | 0.9 | 0.4 | 1.2 | 1.7 |
| IL-2R | X01057 | 0.1 | 0.6 | 0.1 | 0.4 | 8.0 | M26271 | 0.3 | 0.1 | 0.2 | 0.0 | 0.4 | 0.3 | 0.5 |
| IL-2Rβ | M26062 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | M28052 | 0.1 | 0.1 | 0.1 | 0.6 | 0.3 | 0.0 | 1.6 |
| IL-2Rγ | D11086 | 6.0 | 9.7 | 3.9 | 8.0 | 1.8 | X75337 | 0.8 | 1.0 | 1.2 | 2.0 | 1.1 | 1.7 | 1.5 |
| IL-3Rα | D49410 | 1.0 | 1.4 | 1.2 | 2.8 | 1.9 | X64534 | 1.8 | 0.8 | 2.7 | 2.4 | 6.4 | 5.5 | 0.8 |
| IL-3Rβ1 | AL008637 | 21.4 | 13.6 | 26.8 | 10.4 | 0.5 | M34397 | 36.2 | 30.6 | 63.9 | 68.4 | 34.7 | 30.6 | 1.0 |
| IL-4R | X52425 | 16.8 | 17.4 | 10.5 | 14.8 | 1.2 | M27960 | 13.5 | 6.2 | 34.2 | 20.7 | 46.1 | 21.9 | 0.5 |
| IL-6R | X12830 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | X51975 | 0.2 | 0.0 | 0.3 | 0.9 | 1.2 | 0.9 | 1.1 |
| IL-7R | M29696 | 2.0 | 5.1 | 2.1 | 12.1 | 4.2 | M29697 | 0.2 | 1.1 | 0.6 | 1.3 | 1.5 | 2.5 | 2.0 |
| IL-9R | L39064 | 7.0 | 4.5 | 2.2 | 1.7 | 0.7 | M84746 | 0.0 | 0.0 | 0.2 | 0.9 | 0.0 | 0.4 | 5.4 |
| IL-10R | U00672 | 0.2 | 0.7 | 0.6 | 1.0 | 2.3 | L12120 | 3.8 | 5.9 | 6.0 | 9.5 | 7.5 | 8.9 | 1.4 |
| IL-11R | U32324 | 0.8 | 0.9 | 0.4 | 0.3 | 1.0 | U14412 | 0.6 | 0.4 | 1.2 | 0.0 | 1.9 | 1.2 | 0.4 |
| IL-17R | U58917 | 1.2 | 0.7 | 2.4 | 2.3 | 0.8 | U31993 | 4.5 | 2.4 | 4.3 | 3.7 | 5.8 | 4.6 | 0.7 |
| M-CSFR | X03663 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | X06368 | 0.4 | 0.6 | 0.8 | 0.4 | 0.4 | 1.2 | 1.3 |
Numbers in roman indicate “presence” transcripts, which were judged by the GeneChip software; numbers in italics indicate “marginal” or “absence” transcripts.
Rest indicates resting; and Act, active.
The GenBank is accessible at http://www.ncbi.nlm.nih.gov.
Three different batches of human cord blood-derived cultured MCs at week 14 were primed with SCF, IL-3, IL-6, and IL-4 (see “Materials and methods”).
Three different batches of human cord blood-derived cultured MCs at week 11 were primed with SCF, IL-3, IL-6, and IL-4.
Balb/c mouse bone marrow cells were cultured for 4 weeks in the presence of WEHI-3 conditioned medium.
Balb/c mouse bone marrow cells were cultured for 4 weeks in the presence of WEHI-3 conditioned medium and then mrSCF was added to the culture for 1 week.
NOA mouse bone marrow cells were cultured for 4 weeks in the presence of WEHI-3 conditioned medium and then mrSCF was added to the culture for 1 week.
#Sensitized MCs were then challenged with either anti-IgE for human MCs, DNP-BSA for mouse MCs (activated), or control medium (resting) for 6 hours.
Fold increase was obtained by calculating the AD levels of activated MCs vs those of resting MCs.
Protein expression of CC chemokines and 4-1BB (CD137) by human and mouse MCs
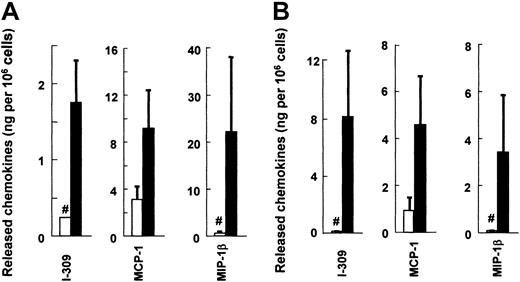
We used ELISA to examine whether these chemokines are also increased at the protein level by IgE-dependent stimulation (Figure1). As expected, the proteins I-309 (CCL1), MCP-1 (CCL2), and MIP-1β (CCL4) were detected in both cultured human MCs and mouse MCs after cross-linking of FcεRI. The protein levels of human MCP-1 and MIP-1β released from activated human MCs were the highest among the cytokines/chemokines we have tested (GM-CSF, IL-5, IL-8, IL-13, and CCL3; MIP-1α).10,15 25 Mouse I-309, MCP-1, and MIP-1β were also produced at high levels. On the other hand, human I-309 proteins were produced at relatively low levels, in spite of abundant expression of their transcripts. We found in a preliminary study that human I-309 was unstable. When we incubated the 2 batches of 106 human MCs with anti-IgE for 6, 24, and 48 hours, I-309 was found at 13.8ng/3.64ng, 1.89ng/0.95ng, and 1.28ng/0.58ng, respectively. Thus, I-309 was rapidly degraded during 6 to 24 hours' incubation with MCs at 37°C. Both human and mouse MCP-1, whose mRNA levels were high in resting MCs, were detected also as proteins before IgE stimulation, whereas the 2 other CC chemokine proteins were not detected in the resting MCs.
FcεRI-dependent release of I-309 (CCL1), MCP-1 (CCL2), and MIP-1β (CCL4) by human and mouse cultured MCs.
Sensitized MCs were challenged with anti-IgE (panel A human, ▪), DNP-BSA (panel B mouse, ▪), or the control medium (■) and incubated at 5 × 105 cells/mL. After 6 hours, the supernatants were collected. Each column and bar represents the mean and SEM of 6 (human) or 3 (mouse) experiments. The FcεRI-mediated production of MCP-1 was judged to be significant. I-309 and MIP-1β were below detectable levels in both supernatants and pellets of the cells incubated with the control medium, and the column represents the detectable level (#).
FcεRI-dependent release of I-309 (CCL1), MCP-1 (CCL2), and MIP-1β (CCL4) by human and mouse cultured MCs.
Sensitized MCs were challenged with anti-IgE (panel A human, ▪), DNP-BSA (panel B mouse, ▪), or the control medium (■) and incubated at 5 × 105 cells/mL. After 6 hours, the supernatants were collected. Each column and bar represents the mean and SEM of 6 (human) or 3 (mouse) experiments. The FcεRI-mediated production of MCP-1 was judged to be significant. I-309 and MIP-1β were below detectable levels in both supernatants and pellets of the cells incubated with the control medium, and the column represents the detectable level (#).
The molecule 4-1BB (CD137), recently found to be an important costimulatory molecule in T cells,26 natural killer (NK) cells,27 monocytes,28 and eosinophils,29 was up-regulated in both human and mouse MCs by FcεRI-mediated stimulation. The surface 4-1BB expression on both human and mouse MCs was up-regulated following FcεRI cross-linking (Figure 2).
Cell surface expression of 4-1BB (CD137).
Cell surface expression of human (A) and mouse (B) 4-1BB is shown as the average values and SEs of the mean fluorescence intensity (MFI) values obtained by 3 independent experiments. (A) Human MCs were reacted with various concentrations of anti-IgE (as indicated) for 6 hours. The net MFI was obtained by subtracting the MFI given by isotype-matched control Ab (4.45-5.35) from the MFI given by Ab against 4-1BB. (B) Mouse MCs were reacted with 10 ng/mL DNP-BSA for 4, 8, and 24 hours. The net MFI was obtained by subtracting the MFI given by isotype-matched control Ab (3.19-5.29) from the MFI given by Ab against 4-1BB.
Cell surface expression of 4-1BB (CD137).
Cell surface expression of human (A) and mouse (B) 4-1BB is shown as the average values and SEs of the mean fluorescence intensity (MFI) values obtained by 3 independent experiments. (A) Human MCs were reacted with various concentrations of anti-IgE (as indicated) for 6 hours. The net MFI was obtained by subtracting the MFI given by isotype-matched control Ab (4.45-5.35) from the MFI given by Ab against 4-1BB. (B) Mouse MCs were reacted with 10 ng/mL DNP-BSA for 4, 8, and 24 hours. The net MFI was obtained by subtracting the MFI given by isotype-matched control Ab (3.19-5.29) from the MFI given by Ab against 4-1BB.
Interspecies comparison of MC-specific transcripts
We used GeneChip to find abundant human and mouse MC-specific transcripts. We measured the 12 000 genes and ESTs by comparing the expression levels in MCs and those in mouse spleen cells or human leukocytes (neutrophils, eosinophils, and mononuclear cells). Then we selected abundant MC-specific transcripts, whose signals were more than 10-fold higher than in these control cell types, by sorting them on the bassis of expression levels (Table 3). As previously reported,11 both human and mouse cultured MCs expressed several proteases, such as tryptase, at the highest levels.
Abundant and cell type–specific transcripts in human and mouse MCs
| Name of transcript (protein) . | GenBank accession no.3-150 . | Expression level, mean ± SD3-151 . |
|---|---|---|
| Human MCs | ||
| Tryptase β | M30038 | 143.81 ± 22.1 |
| Cathepsin G | M16117 | 106.51 ± 14.9 |
| Major basic protein | Z26248 | 97.21 ± 33.6 |
| Tissue inhibitor of metalloproteinases | D11139 | 88.41 ± 21.3 |
| Galectin-1 | AI535946 | 79.81 ± 19.6 |
| Clusterin | M25915 | 66.11 ± 16.1 |
| CCL2, MCP-1 | M26683 | 62.11 ± 30.3 |
| Nc61c12.r1 | AA420624 | 56.61 ± 13.0 |
| Heat shock protein 90 | J04988 | 56.31 ± 20.0 |
| PGD2 synthase | AF150241 | 54.71 ± 10.3 |
| Carboxypeptidase A | M73720 | 53.01 ± 17.7 |
| L-histidine decarboxylase | D16583 | 45.01 ± 15.4 |
| 15-hydroxyprostaglandin dehydrogenase | L76465 | 44.71 ± 19.5 |
| Macrophage capping protein (G-actin) | M94345 | 44.71 ± 19.3 |
| Galectin-3 | AB006780 | 44.71 ± 19.6 |
| Mouse MCs | ||
| MMCP5 Chymase1 | M68898 | 176.91 ± 30.0 |
| MMCP6 Tryptase β | M57626 | 156.21 ± 68.9 |
| Galectin | X15986 | 127.41 ± 77.4 |
| IL-3Rβ2 | M29855 | 125.31 ± 43.2 |
| Carboxypeptidase A | J05118 | 118.11 ± 31.9 |
| Tryptophan hydroxylase | J04758 | 108.51 ± 29.8 |
| ERATO Doi 411 (Cytochrome P450) | AW121619 | 103.51 ± 31.8 |
| CD63 | D16432 | 94.91 ± 32.4 |
| UI-M-BH2.3-anx-g-02-0-U.sl | AW120614 | 91.11 ± 29.5 |
| Insulinlike growth factor binding protein 5 | L12447 | 90.61 ± 61.7 |
| Gly96 | X67644 | 88.41 ± 37.2 |
| Aldo-keto reductase | AB027125 | 87.41 ± 53.7 |
| MMCP7 Tryptase | L00653 | 83.31 ± 63.5 |
| Lymphocyte antigen 84 | D13695 | 81.51 ± 16.3 |
| FcεRIα | J05018 | 81.21 ± 19.6 |
| Name of transcript (protein) . | GenBank accession no.3-150 . | Expression level, mean ± SD3-151 . |
|---|---|---|
| Human MCs | ||
| Tryptase β | M30038 | 143.81 ± 22.1 |
| Cathepsin G | M16117 | 106.51 ± 14.9 |
| Major basic protein | Z26248 | 97.21 ± 33.6 |
| Tissue inhibitor of metalloproteinases | D11139 | 88.41 ± 21.3 |
| Galectin-1 | AI535946 | 79.81 ± 19.6 |
| Clusterin | M25915 | 66.11 ± 16.1 |
| CCL2, MCP-1 | M26683 | 62.11 ± 30.3 |
| Nc61c12.r1 | AA420624 | 56.61 ± 13.0 |
| Heat shock protein 90 | J04988 | 56.31 ± 20.0 |
| PGD2 synthase | AF150241 | 54.71 ± 10.3 |
| Carboxypeptidase A | M73720 | 53.01 ± 17.7 |
| L-histidine decarboxylase | D16583 | 45.01 ± 15.4 |
| 15-hydroxyprostaglandin dehydrogenase | L76465 | 44.71 ± 19.5 |
| Macrophage capping protein (G-actin) | M94345 | 44.71 ± 19.3 |
| Galectin-3 | AB006780 | 44.71 ± 19.6 |
| Mouse MCs | ||
| MMCP5 Chymase1 | M68898 | 176.91 ± 30.0 |
| MMCP6 Tryptase β | M57626 | 156.21 ± 68.9 |
| Galectin | X15986 | 127.41 ± 77.4 |
| IL-3Rβ2 | M29855 | 125.31 ± 43.2 |
| Carboxypeptidase A | J05118 | 118.11 ± 31.9 |
| Tryptophan hydroxylase | J04758 | 108.51 ± 29.8 |
| ERATO Doi 411 (Cytochrome P450) | AW121619 | 103.51 ± 31.8 |
| CD63 | D16432 | 94.91 ± 32.4 |
| UI-M-BH2.3-anx-g-02-0-U.sl | AW120614 | 91.11 ± 29.5 |
| Insulinlike growth factor binding protein 5 | L12447 | 90.61 ± 61.7 |
| Gly96 | X67644 | 88.41 ± 37.2 |
| Aldo-keto reductase | AB027125 | 87.41 ± 53.7 |
| MMCP7 Tryptase | L00653 | 83.31 ± 63.5 |
| Lymphocyte antigen 84 | D13695 | 81.51 ± 16.3 |
| FcεRIα | J05018 | 81.21 ± 19.6 |
Unreviewed additional data are shown athttp://www.nch.go.jp/imal/English_index.htm.
The GenBank is accessible at http://www/ncbi.nlm.nih.gov.
Expression level is the percentage of the specific AD level vs the mean AD level of 6 probe sets for housekeeping genes; shown are means for 4 (human) or 6 (mouse) experiments.
We selected orthologous genes (homologous genes in different species evolving from the same common ancestral gene)30 of cytokines, chemokines, their receptors, CD molecules, housekeeping, mouse MC-specific, and human MC-specific molecules from the 12 000 distinct genes. The pairs of orthologs were selected primarily on the basis of perfectly coincident annotation. If the annotation was partially matched, we examined the homology between the 2 MC transcripts by consulting the UniGene Web site (http://www.ncbi.nlm.nih.gov/UniGene/) and the Human-Mouse Homology Map (http://www.ncbi.nlm.nih.gov/Homology/; this map became available during preparation of this paper).31 Finally, we selected 287 pairs of orthologous genes, as shown in Figure3. We confirmed that the gene expression of several CC chemokines, such as I-309 (CCL1), was regulated in a very similar manner. The names and expression levels of these 287 genes are shown as unreviewed additional material at our Web site (http://www.nch.go.jp/imal/English_index.htm).
Interspecies comparison of orthologous genes expressed by human and mouse MCs.
The expression levels of 287 pairs of orthologous genes were compared. The horizontal direction stands for the expression levels of human genes and the vertical direction represents those of mouse genes. Each point represents the average of 2 (human) or 3 (mouse) independent experiments shown in Tables 1 and 2. The expression levels of I-309 (CCL1), MCP-1 (CCL2), MIP-1α (CCL3), MIP-1β (CCL4), and 4-1BB (CD137) genes in resting MCs (panel A) were increased in activated MCs (panel B), while those of tryptase and major basic protein (MBP) remained at similar levels. The expression levels were normalized into a percentage of the average of 6 housekeeping genes (♦). The oblique broken lines indicate a 10-fold difference.
Interspecies comparison of orthologous genes expressed by human and mouse MCs.
The expression levels of 287 pairs of orthologous genes were compared. The horizontal direction stands for the expression levels of human genes and the vertical direction represents those of mouse genes. Each point represents the average of 2 (human) or 3 (mouse) independent experiments shown in Tables 1 and 2. The expression levels of I-309 (CCL1), MCP-1 (CCL2), MIP-1α (CCL3), MIP-1β (CCL4), and 4-1BB (CD137) genes in resting MCs (panel A) were increased in activated MCs (panel B), while those of tryptase and major basic protein (MBP) remained at similar levels. The expression levels were normalized into a percentage of the average of 6 housekeeping genes (♦). The oblique broken lines indicate a 10-fold difference.
It should be noted that several MC-specific transcripts could not be compared. Human cells are known to lack a β2 subunit of the IL-3 receptor, and the homology between human IL-3 and mouse IL-3 proteins is less than 30%.32 Mouse mast cell protease (MMCP)7 was found as a pseudogene in human genome.33 IL-8 (CXCL8) is also not found in the mouse genome.18 Interestingly, mouse MCs did not express eosinophil granule major basic protein (MBP), which has recently been found to be abundantly present in all human MC types, both in vitro and in vivo.11
Discussion
The aim of this study was to elucidate which molecules are commonly expressed in both SCF- and IL-6–dependent cultured human cord blood–derived MCs and IL-3–dependent cultured mouse bone marrow–derived MCs. Owing to the differences in cytokine dependency, we did not strongly expect to find many molecules expressed in both human and mouse MCs. However, following IgE-dependent activation, 3 CC chemokines and 4-1BB (CD137) were found in the 10 most up-regulated transcripts among approximately 12 000 molecules in both cultured human MCs and cultured mouse MCs. Another CC chemokine, MCP-1 (CCL2), was also highly expressed in both human and mouse MCs in a resting state as well as in an activated state.
Mouse MCs have already been reported by many investigators34-38 to produce a variety of cytokine/chemokine proteins, including I-309 (CCL1), MCP-1 (CCL2), MIP-1α (CCL3), and MIP-1β (CCL4) among the transcripts listed in Table 2. However, we demonstrated for the first time that these CC chemokines were expressed at the highest levels of the transcriptome. We had previously reported10,25 that in activated human MCs MIP-1α, IL-8 (CXCL8), IL-13, and GM-CSF were up-regulated at the protein levels. Thus, we chose to measure the protein levels of I-309, MCP-1, MIP-1β, and 4-1BB (CD137) in this study. We demonstrated the FcεRI-induced protein production of I-309, MCP-1, and MIP-1β. MCP-1–deficient mice are known to lack Th2 cell development.39 Mouse MCs are reported to produce MIP-1β in the antigen-induced late skin reaction characterized by T-cell recruitment.40 In the present study, the MCP-1 and MIP-1β proteins released from activated human and mouse MCs were noted to be at the highest concentrations among the cytokines produced by these MCs,10,15,25 as were the corresponding transcript levels. Mouse I-309 protein was also produced at the highest levels, whereas human I-309 protein was detected at relatively low levels, probably owing to its instability. I-309 seems to be of particular importance, since it is a chemokine that can recruit CCR8-positive Th2 cells18,41-44 into inflammatory human lung in the late-phase reaction after allergen challenge.45Furthermore, the deletion of CCR8 genes markedly reduced airway hyperresponsiveness in a mouse model of asthma, whereas the other CC chemokine receptor deletion failed to do so.46 Since the expression of CCR8 on the Th2 cells is transient during late-phase allergic reaction,45 its ligand I-309 need not be very stable. Production of I-309 by mouse MCs38 and a human mast cell leukemia cell line, HMC-1,47 has been reported. We demonstrated for the first time that human MCs could newly produce I-309 after IgE stimulation, and that the expression level was the highest among human and mouse MC transcripts, suggesting that IgE may play an important role in the recruitment of activated Th2 cells through MC activation. Thus, both human and mouse MCs may play a crucial role in recruiting CCR-expressing T cells.
The molecule 4-1BB (CD137), recently found to be an important costimulatory molecule in various immune cell types, 26-29was up-regulated at transcriptional and protein levels in both human and mouse MCs by FcεRI-mediated stimulation. Functionally, this molecule is not fully characterized, and contradictory findings have been reported. Whereas some investigators have observed cell proliferation by 4-1BB activation,26,48,49 Langstein et al50 reported the induction of apoptosis by its activation. Future studies are needed to clarify whether 4-1BB primarily activates or deactivates MC functions.
Animal models, especially mice, are common surrogates for studying human diseases. However, clinical trials sometimes fail owing to the fact that the results obtained in animal studies cannot be reproduced in humans. For instance, anti–IL-5 antibody completely blocked the airway hypersensitivity in experimental animal models of asthma,51 while the therapeutic application of humanized anti–IL-5 antibody did not improve the bronchial hypersensitivity of asthmatics.52 Recently, many human and mouse orthologous genes have become available at genome-wide level in electronic format (http://www.ncbi.nlm.nih.gov/Homology/), which facilitates interspecies comparisons.31 However, it has not been shown that these structure-based orthologs are similarly regulated. We compared for the first time the expression levels of these orthologous genes by selecting 287 gene pairs. Among the ortholog pairs, the regulation pattern of I-309 (CCL1) turned out to be highly conserved between human and mouse. Thus the targeting of I-309 is an attractive approach for potential clinical applications, since investigation of I-309 in mouse models may be more predictive of the human responses. For other orthologous genes, we found that mRNA levels are regulated differently in mouse and human MCs. Therefore, studies on the function of molecules highly expressed only in mouse cells have to be carefully interpreted with regard to their potential function in humans. Interspecies comparison studies of whole genome expression should be useful for interpretation of experimental data from animal models of human pathogenesis.
We thank Dr Kiyoshi Kawashima, Dr Shigenobu Shoda, and the staff of the Department of Obstetrics, Gyoda Chuo Hospital, for generously providing umbilical cord blood. We also thank Dr Florian Gantner at Bayer Yakuhin for proofreading the manuscript; Dr Shigeru Okumura for discussion; and Mr Hisashi Tomita, Mr Keisuke Yuki, Ms Noriko Hashimoto, and Ms Futaba Sekiya at National Research Institute for Child Health and Development and Ms Atsuko Ikeda at National Sagamihara Hospital for skillful technical assistance.
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2002-02-0602.
Supported in part by a grant from the Organization for Pharmaceutical Safety and Research and the Ministry of Health, Labour and Welfare (the Millennium Genome Project, MPJ-5) and by a grant from RIKEN Research Center for Allergy and Immunology.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hirohisa Saito, Department of Allergy and Immunology, National Research Institute for Child Health and Development, 3-35-31 Taishido, Setagaya-ku, Tokyo 154-8567, Japan; e-mail: hsaito@nch.go.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal