The CDM (ced-5 of Caenorhabditis elegans,DOCK180 [downstream of Crkwith molecular weight of 180 kDa] of humans, andmyoblast city of Drosophila melanogaster) family of proteins has been shown to play a pivotal role in the integrin-mediated signaling pathway under the regulation of an adaptor moleculec-CT10–related kinase II (c–Crk-II) in adherent cells. Recently, hematopoietic cell–specific CDM protein DOCK2 has been shown to be indispensable for lymphocyte migration. However, the regulatory mechanism for DOCK2 is still unknown because DOCK2 lacks a c–Crk-II binding consensus motif. In this study, we demonstrated that DOCK2 bound to CrkL, which is present exclusively in hematopoietic cells both in vivo and in vitro, and we also found that 2 separate regions of DOCK2 contributed to its binding to Src homology 3 (SH3) domain of CrkL. Colocalization of DOCK2 with Crk-like (CrkL) and F-actin was shown by immunocytochemical analysis with the use of Jurkat cells. We also found that CrkL-induced activation of small guanine triphosphatase (GTPase) Rac1 was significantly inhibited by the DOCK2-dCS mutant in 293T cells. Furthermore, the association of DOCK2 and Vav, the guanine-nucleotide exchanging factor (GEF) for Rac1, was demonstrated in Jurkat cells. Finally, the stable expression of DOCK2-dCS mutant in Jurkat cells was shown to reduce cell attachment. These data suggest the presence of a novel protein complex of CrkL, DOCK2, and Vav to regulate Rac1 in leukemia cell lines.
Introduction
The CDM family of proteins is a newly emerging class of molecules that includes ced-5 ofCaenorhabditis elegans,1DOCK180 (downstream ofCrk with molecular weight of 180kDa) of humans,2 and myoblast city ofDrosophila melanogaster,3 and has been shown to play a pivotal role in the integrin signaling pathway in the nonhematopoietic lineages of cells. The prototype of the CDM protein, DOCK180, was isolated as one of the effector molecules of the signaling adaptor proteinc-CT10–related kinase II (c–Crk-II),2 and has been shown to regulate cytoskeletal movement through small guanine triphosphatase (GTPase) Rac, leading to various cellular responses, such as phagocytosis, cell migration, and cytoplasmic spreading.4-6 As DOCK180 was not present in hematopoietic cells,2 the role of CDM proteins in hematopoietic cells was still under investigation.
DOCK2, found exclusively in hematopoietic cells, belongs to the CDM family of proteins, and was identified as a homolog of DOCK180 with the use of a cDNA library of the leukemia cell line.7,8 DOCK2 has been reported to activate Rac,8 and recently, analysis of DOCK2 knockout mice clearly demonstrated that DOCK2 plays an essential role in lymphocyte migration toward chemokines.9 However, the regulatory mechanism of DOCK2 was still unknown because DOCK2 lacks a binding consensus sequence for the Src homology 3 (SH3) domain of c–Crk-II.8
Crk-like (CrkL), which is composed of an SH2 domain and two SH3 domains, was isolated as a homolog of c–Crk-II, and its expression was reported to be dominant in hematopoietic cells.10,11 CrkL has been shown to be tyrosine phosphorylated in leukemia cell lines and, in association withCrk SH3 domain binding GEF (C3G), c-Abl, and DOCK180, is involved in cell adhesion and migration.12-14 However, a hematopoietic cell–specific role for CrkL has not yet been established.
The Rho family of small GTPases, including Rho, Rac, and Cdc42 subfamilies, has been shown to regulate the reorganization of the actin cytoskeleton.15-18 The guanine-nucleotide exchanging factors (GEFs) of the Rho family of proteins share 2 conserved regions, the Dbl homology (DH) domain19 and the pleckstrin homology (PH) domain.20 The expression of Vav, which is one of the GEFs for Rac, was observed exclusively in hematopoietic cells,21 and has been shown to mediate the integrin signaling pathway by its association with tyrosine kinase Syk.22 23
In this study, to analyze the role of the CDM family of proteins in hematopoietic cells, we examined the regulatory mechanism of DOCK2 and found a novel protein complex of CrkL, DOCK2, and Vav, which leads to the regulation of Rac1 activity and cell adhesion.
Materials and methods
Plasmids
The pCXN2-Flag-DOCK2, pCXN2-Flag-DOCK2-dCS, pCXN2-Flag-DOCK2-dN, and pCXN2-Flag-Vav were described previously.8 The pCXN2-Flag-DOCK2-dBE, which encodes amino acids (aa) 926 to 1478; pCXN2-Flag-DOCK2–carboxyl-terminal region (pCXN2-Flag-DOCK2-CT) with aa 1640 to 1830; pCXN2-Flag-DOCK2–internal domain 1 (pCXN2-Flag-DOCK2-INT1) with aa 386 to 593; and pCXN2-Flag-DOCK2-INT2 with aa 586 to 883 were constructed. Human Rac1 cDNA was a gift from Dr Silivo J. Gutkind (National Institutes of Health, Bethesda, MD) and was subcloned into pCXN2 designated pCXN2-Flag-Rac1. CrkL cDNA was a gift from Dr John Groffen (Children's Hospital of Los Angeles, CA). The pGEX-CrkL-SH3(N), from Dr Brian J. Druker (Oregon Health Sciences University, Portland), was described previously.24 The pGEX-CrkIISH3(N), pGEX-CrkIISH3(C), pGEX-CrkIISH3(N)W169L, pGEX-Grb2SH3, pGEX-cAblSH3, pGEX-vSrcSH3, pGEX-p85SH3, pGEX-MLKSH3, and pGEX-CrkII (wild type) were described previously.25 A cDNA fragment encoding the Rac-binding domain (RBD) of human p21-activated kinase 2 (PAK2) encompassing amino acids 66 through 147 was obtained with the use of human T-cell line Jurkat cells by reverse-transcriptase–polymerase chain reaction (RT-PCR), and the PCR product was subcloned into pGEX-4T (Amersham Pharmacia Biotech, Piscataway, NJ) designated pGEX-PAK2-RBD.
Antibodies, reagent, and cells
Anti-DOCK2 antibody (Ab) and anti-DOCK180 Ab have been described previously.2 8 Anti-Flag tag M2 monoclonal Ab, anti-CrkL Ab (C19), and anti-Vav Ab were purchased from Sigma (St Louis, MO), Santa Cruz Biotechnology (Santa Cruz, CA), and B&D Transduction Laboratories (Lexington, KY), respectively. Phalloidin conjugated with Alexa 594, antimouse immunoglobulin Ab conjugated with Alexa 488/594, and antirabbit immunoglobulin Ab with Alexa-488 were purchased from Molecular Probes (Eugene, OR).
The 293T human embryonal kidney cells with simian virus40 (SV40) large T antigen were maintained in Dulbecco modified minimum essential medium (DMEM) containing 10% fetal bovine serum (FBS). Jurkat and MOLT4 human T cells, Raji human B cells, and K562 human myeloblast cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% FBS. For establishment of Jurkat cell lines stably expressing DOCK2 and its mutants, cells were transfected with either pCXN2-Flag-DOCK2 wild type (WT) or pCXN2-Flag-DOCK2-dCS by electroporation and selected by 400 μg/mL neomycin for 3 weeks. Several independent colonies were isolated, and protein expression levels were examined by immunoblotting by means of anti-Flag Ab. For establishment of K562 cell lines stably expressing DOCK2, cells were cotransfected with pBabe-Puro and either pCXN2-Flag-DOCK2 WT or pCXN2-Flag-DOCK2-dCS by electroporation and selected by 1 μg/mL puromycin for 1 week.
Preparation of human platelets
For this study, approval was obtained from the Institutional Review Board of Hokkaido University School of Medicine, and informed consent was provided according to the Declaration of Helsinki. Blood from healthy volunteers was drawn by venipuncture, after informed consent was obtained, and placed into 1:10 volume of 3.8% (wt/vol) trisodium citrate and gently mixed. Platelet-rich plasma (PRP) was prepared by centrifugation of whole blood at 200g for 20 minutes. PRP was aspirated and incubated with 2 μM aspirin for 30 minutes at room temperature. After the addition of prostaglandin E1 at a final concentration of 1 μM, the PRP was spun at 800g to form a soft platelet pellet. The pellet was suspended in 1 mL modified Hepes (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)–Tyrode buffer containing 129 mM NaCl, 8.9 mM NaHCO3, 0.8 mM KH2PO4, 0.8 mM MgCl2, 5.6 mM dextrose, and 10 mM Hepes (pH 7.4) supplement with apyrase 0.6 U/mL, and washed twice. Platelets (3 × 108 cells per milliliter) were suspended for experiments in modified Hepes-Tyrode buffer supplemented with apyrase (0.6 U/mL) and RGDS peptide (200 μg/mL).
Immunoprecipitation and pull-down assay using GST fusion protein
Cells were washed with phosphate-buffered saline (PBS) and lysed by Lysis buffer containing 1% Triton-X 100, 10 mM Tris-HCl (pH 7.4), 5 mM EDTA (ethylenediaminetetraacetic acid), 150 mM NaCl, 0.5 mM Na3VO4, 10 mM NaF, 1 μg/mL aprotinin, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and centrifuged at 12 000 rpm at 4°C for 15 minutes. The supernatant was incubated with primary Abs such as anti-CrkL Ab and anti-DOCK2 Ab for 1 hour, and then with a mixture of Protein A–Sepharose 4A and Protein G–Sepharose 4A and anti-CrkL Ab at 4°C for 1 hour with rotation. After washing, the immunocomplexes were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by immunoblotting with anti-CrkL, anti-Vav, and anti-DOCK2 Abs.
Pull-down assay using glutathione S-transferase (GST) fusion proteins was performed as described.26 27 Briefly, GST fusion proteins were isolated from bacterial lysates by means of glutathione Sepharose beads (Amersham Pharmacia Biotech). After washing with lysis buffer, the beads were incubated with lysates of either cell lines or platelets for 1 hour. Proteins bound to GST fusion proteins were separated by SDS-PAGE, and analyzed by immunoblotting with anti-DOCK2 and anti-GST Abs.
Pull-down assay for active form of Rac1
The detection of GTP-bound Rac1 was performed as described previously.28 29 The 293T cells were transfected with expression plasmids with the use of FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) and incubated for 24 hours. For Jurkat and K562 cells, plasmids were transfected by electroporation, and drug-selected cells were used. Cells were lysed with lysis buffer composed of 1% Nonidet P-40, 25 mM HEPES (pH 7.4), 150 mM NaCl, 10% glycerol, 1 mM EDTA, 10 mM MgCl2, 1 μg/mL aprotinin, and 1 mM PMSF. Lysates were centrifuged at 12 000 rpm at 4°C for 1 minute. The supernatant was incubated with 10 μg purified GST-PAK2-RBD and glutathione-Sepharose 4B beads. The beads were washed 3 times with lysis buffer. The precipitates were analyzed by immunoblotting with anti-Flag M2 Ab, and positive signals were visualized by ECL chemiluminescence reaction (Amersham Pharmacia Biotech), and quantified with the use of LAS-1000 plus image analyzer (Fuji Film, Tokyo, Japan).
Immunostaining
Jurkat cells were transfected with expression plasmids with the use of X-tremeGene Q2 transfection reagent (Roche Diagnostics), and after 24 hours, the cells were plated on fibronectin-coated chamber slides and incubated for 24 hours. Thereafter, cells were fixed with 4% paraformaldehyde in PBS with 0.33 M sucrose at room temperature for 15 minutes, permeabilized with 0.2% Triton X-100 in PBS for 5 minutes, preincubated with 0.1% goat serum in PBS for 10 minutes, and incubated with indicated antibodies or Alexa-594–conjugated phalloidin for 1 hour. For those cases using anti-Flag and anti-CrkL Ab stain, cells were further incubated with antimouse immunoglobulin Ab conjugated with Alexa 488 or Alexa 594, and antirabbit immunoglobulin Ab with Alexa 488. The cells were observed by confocal laser scanning microscopy (Olympus, FV300, Tokyo, Japan).
Assessment of cell attachment
Jurkat cells were plated on collage type I–coated dishes, incubated at 37°C for 20 minutes, and fixed with 4% paraformaldehyde for 10 minutes at room temperature. Morphology of attached cells was observed by phase contrast microscopy (Olympus).
Results
Association of CrkL with DOCK2
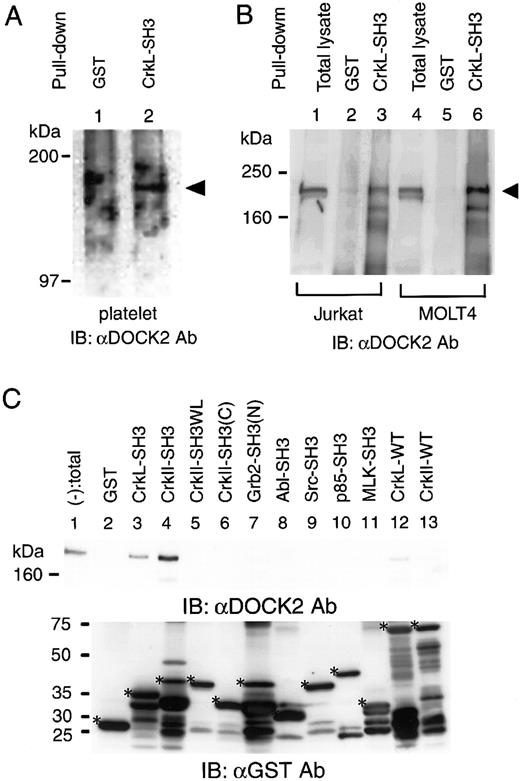
Because the expression of both CrkL and DOCK2 was observed exclusively in hematopoietic cells, we hypothesized that CrkL could be associated with DOCK2 as the analogy of the binding of c–Crk-II and DOCK180 found in nonhematopoietic lineages of cell lines. By pull-down assay, we found that the GST-fusion form of the N-terminal SH3 domain of CrkL could precipitate endogenous DOCK2 in human normal platelets (Figure 1A). We could also observe that DOCK2 was precipitated with GST-CrkL-SH3 in human T-cell lines Jurkat and MOLT4. In this assay, DOCK2 could not be precipitated in human B-cell line Raji or myeloblast cell line K562 (Figure 1B). Among several SH3 domains, including those of Grb2, the p85 subunit of phosphatidyl inositol 3 kinase (PI-3K), v-Src, c-Abl, and mixed lineage kinase 3 (MLK3), we confirmed that none of them bound to DOCK2 (Figure 1C). We also observed that the GST fusion form of the SH3 domain of c–Crk-II precipitated DOCK2, and that introducing point mutation into the essential part of the SH3 domain of c–Crk-II, designated CrkIISH3W169L, failed to maintain the binding to DOCK2 (Figure 1C, lane 5). The full-length form of the wild-type CrkL provided weak signals of DOCK2, whereas the wild-type c–Crk-II did not (Figure 1C).
The association of DOCK2 and GST-CrkL-SH3(N).
(A) Lysates of human normal platelets were subjected to pull-down assay by GST (lane 1) or GST-CrkL-SH3(N) (lane 2). The precipitates were analyzed by immunoblotting with anti-DOCK2 (αDOCK2) Ab. Arrowhead indicates the size of DOCK2. (B) Lysates of Jurkat and MOLT4 were subjected to pull-down assay by GST (lanes 2 and 5) or GST-CrkL-SH3(N) (lanes 3 and 6). Arrowhead indicates the size of DOCK2. (C) Analysis of the binding of DOCK2 and various SH3 domains (indicated at the top) by pull-down assay in Jurkat cells lysates. The precipitates were analyzed by immunoblotting by means of anti-DOCK2 Ab (top panel) and anti-GST Ab (bottom panel). Lane 1, pull-down (−) indicates total cell lysates. In the bottom panel, asterisks indicate the expected size of each SH3 domain–containing proteins.
The association of DOCK2 and GST-CrkL-SH3(N).
(A) Lysates of human normal platelets were subjected to pull-down assay by GST (lane 1) or GST-CrkL-SH3(N) (lane 2). The precipitates were analyzed by immunoblotting with anti-DOCK2 (αDOCK2) Ab. Arrowhead indicates the size of DOCK2. (B) Lysates of Jurkat and MOLT4 were subjected to pull-down assay by GST (lanes 2 and 5) or GST-CrkL-SH3(N) (lanes 3 and 6). Arrowhead indicates the size of DOCK2. (C) Analysis of the binding of DOCK2 and various SH3 domains (indicated at the top) by pull-down assay in Jurkat cells lysates. The precipitates were analyzed by immunoblotting by means of anti-DOCK2 Ab (top panel) and anti-GST Ab (bottom panel). Lane 1, pull-down (−) indicates total cell lysates. In the bottom panel, asterisks indicate the expected size of each SH3 domain–containing proteins.
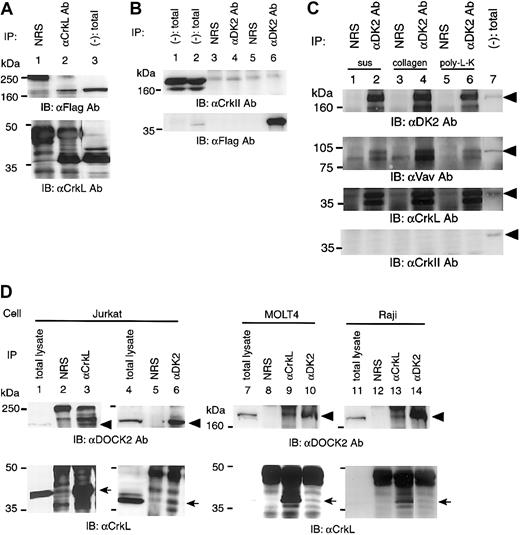
To confirm the binding of CrkL and DOCK2, the expression plasmids for CrkL and DOCK2 were transiently transfected in 293T cells, and immunoprecipitation analysis was performed. Anti-CrkL Ab could precipitate DOCK2 (Figure 2A), anti-DOCK2 Ab was also found to precipitate CrkL (data not shown). In this transient protein expression system, we could not find any coprecipitation of c–Crk-II with DOCK2 (Figure 2B). Furthermore, we analyzed the binding of endogenous DOCK2 to CrkL in Jurkat, MOLT4, and Raji cells and found that anti-CrkL Ab and anti-DOCK2 Ab could precipitate DOCK2 and CrkL, respectively (Figure 2D). Stimulation of Jurkat cells by collagen type I or poly-L-lysine did not alter their binding (Figure 2C). We could not find any binding of DOCK2 to c–Crk-II regardless of the stimulation (Figure 2C).
The association of DOCK2 and CrkL in vivo.
(A) Analysis of the association of CrkL with DOCK2 in 293T cells. The pCAGGS-CrkL and pCXN2-Flag-DOCK2 were transiently transfected, and an immunoprecipitation assay was performed by anti-CrkL Ab (lane 2) or normal rabbit serum (NRS; lane 1). Immunoblotting was performed with the use of anti-Flag Ab (top panel) and anti-CrkL Ab (bottom panel). Total cell lysates appear in lane 3. (B) Analysis of the association of c–Crk-II with DOCK2 in 293T cells. The sets of cotransfected plasmid were as follows: pCAGGS–c–Crk-II and pCXN2 vector (lanes 1, 3, and 4); pCAGGS–c–Crk-II and pCXN2-Flag-DOCK2 (lanes 2, 5, and 6). Cell lysates were analyzed by immunoprecipitation with the use of anti-DOCK2 (αDK2) Ab or NRS (lanes 3-6). Total cell lysates are shown in lanes 1 and 2. (C) Association of CrkL and DOCK2 in Jurkat cells with stimulation. Cells were in suspension (sus; lanes 1 and 2), plated on collagen type I–coated dishes (collagen; lanes 3 and 4), or plated on poly-L-lysine–coated dish (poly-L-K; lanes 5 and 6). Cells were incubated for 15 minutes at 37°C, and cell lysates were immunoprecipitated with anti-DOCK2 (αDK2) Ab (lanes 2, 4, and 6) or NRS (lanes 1, 3, and 5), and precipitates were analyzed by immunoblotting with antibodies, indicated below each panel. Total cell lysates appear in lane 7. The arrowheads indicate DOCK2 in the top panel, Vav in the second panel, CrkL in the third panel, and c–Crk-II in the bottom panel. (D) Association of endogenous CrkL and DOCK2 in Jurkat, MOLT4, and Raji cells. Cell lysates were immunoprecipitated by anti-CrkL Ab (αCrkL; lane 3, 9, and 13) or anti-DOCK2 Ab (αDK2; lane 6, 10, and 14) and probed with anti-DOCK2 Ab (top panels) and with anti-CrkL Ab (bottom panels).
The association of DOCK2 and CrkL in vivo.
(A) Analysis of the association of CrkL with DOCK2 in 293T cells. The pCAGGS-CrkL and pCXN2-Flag-DOCK2 were transiently transfected, and an immunoprecipitation assay was performed by anti-CrkL Ab (lane 2) or normal rabbit serum (NRS; lane 1). Immunoblotting was performed with the use of anti-Flag Ab (top panel) and anti-CrkL Ab (bottom panel). Total cell lysates appear in lane 3. (B) Analysis of the association of c–Crk-II with DOCK2 in 293T cells. The sets of cotransfected plasmid were as follows: pCAGGS–c–Crk-II and pCXN2 vector (lanes 1, 3, and 4); pCAGGS–c–Crk-II and pCXN2-Flag-DOCK2 (lanes 2, 5, and 6). Cell lysates were analyzed by immunoprecipitation with the use of anti-DOCK2 (αDK2) Ab or NRS (lanes 3-6). Total cell lysates are shown in lanes 1 and 2. (C) Association of CrkL and DOCK2 in Jurkat cells with stimulation. Cells were in suspension (sus; lanes 1 and 2), plated on collagen type I–coated dishes (collagen; lanes 3 and 4), or plated on poly-L-lysine–coated dish (poly-L-K; lanes 5 and 6). Cells were incubated for 15 minutes at 37°C, and cell lysates were immunoprecipitated with anti-DOCK2 (αDK2) Ab (lanes 2, 4, and 6) or NRS (lanes 1, 3, and 5), and precipitates were analyzed by immunoblotting with antibodies, indicated below each panel. Total cell lysates appear in lane 7. The arrowheads indicate DOCK2 in the top panel, Vav in the second panel, CrkL in the third panel, and c–Crk-II in the bottom panel. (D) Association of endogenous CrkL and DOCK2 in Jurkat, MOLT4, and Raji cells. Cell lysates were immunoprecipitated by anti-CrkL Ab (αCrkL; lane 3, 9, and 13) or anti-DOCK2 Ab (αDK2; lane 6, 10, and 14) and probed with anti-DOCK2 Ab (top panels) and with anti-CrkL Ab (bottom panels).
Among the representative GEFs for Rac, the expression of Vav is observed exclusively in hematopoietic cells; we thus examined whether Vav was involved in DOCK2-dependent activation of Rac. In Jurkat cells, Vav was found to be coprecipitated with DOCK2 (Figure 2C).
Analysis of the CrkL-binding region of DOCK2
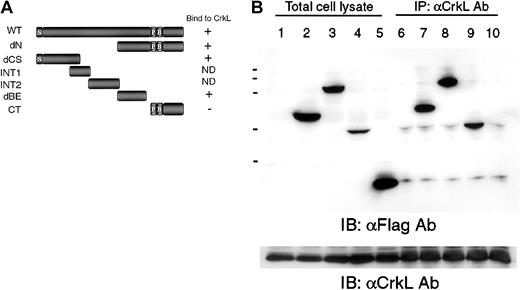
To determine the CrkL-binding regions of DOCK2, we generated truncated mutants of DOCK2 (Figure 3A) and transiently transfected them into 293T cells with the CrkL expression plasmid. As shown in Figure 3B, anti-CrkL antibody precipitated both the dCS mutant composed of 515 amino acids (aa 1-515) of the N-terminus region of DOCK2 (DOCK2-dCS) and the dBE mutant with 538 amino acids (aa 939-1476) (DOCK2-dBE). The CT mutant, which includes a proline-rich region, was not coprecipitated with CrkL. We did not obtain satisfactory expression levels of mutants of INT1 or INT2 (data not shown).
Analysis of the association of CrkL with DOCK2 truncated mutants.
(A) Schematic structure of wild-type DOCK2 and the mutants designated dN (aa 939 to 1830), dCS (aa 1 to 515), INT1 (aa 386 to 593), INT2 (aa 586 to 883), dBE (aa 926 to 1478), and CT (aa 1640 to 1830). ND indicates not determined; P, proline-rich sequence; B, basic domain; and S, SH3 domain. (B) Association of CrkL and DOCK2 mutants were analyzed by immunoprecipitation. The 293T cells were transfected with pCAGGS-CrkL and the DOCK2 expression vector, and cell lysates were immunoprecipitated with anti-CrkL Ab and were analyzed by immunoblotting with anti-Flag monoclonal Ab (Ab; lanes 6-10). Protein expressions of DOCK2 mutants and CrkL were examined by anti-Flag mAb (top panel, lanes 1-5) and anti-CrkL Ab (bottom panel). Transfected plasmids were as follows: lanes 1 and 6, pCAGGS; lanes 2 and 7, pCXN2-Flag-DOCK2-dCS; lanes 3 and 8, pCXN2-Flag-DOCK2-dN; lanes 4 and 9, pCXN2-Flag-DOCK2-dBE; and lanes 5 and 10, pCXN2-Flag-DOCK2-CT.
Analysis of the association of CrkL with DOCK2 truncated mutants.
(A) Schematic structure of wild-type DOCK2 and the mutants designated dN (aa 939 to 1830), dCS (aa 1 to 515), INT1 (aa 386 to 593), INT2 (aa 586 to 883), dBE (aa 926 to 1478), and CT (aa 1640 to 1830). ND indicates not determined; P, proline-rich sequence; B, basic domain; and S, SH3 domain. (B) Association of CrkL and DOCK2 mutants were analyzed by immunoprecipitation. The 293T cells were transfected with pCAGGS-CrkL and the DOCK2 expression vector, and cell lysates were immunoprecipitated with anti-CrkL Ab and were analyzed by immunoblotting with anti-Flag monoclonal Ab (Ab; lanes 6-10). Protein expressions of DOCK2 mutants and CrkL were examined by anti-Flag mAb (top panel, lanes 1-5) and anti-CrkL Ab (bottom panel). Transfected plasmids were as follows: lanes 1 and 6, pCAGGS; lanes 2 and 7, pCXN2-Flag-DOCK2-dCS; lanes 3 and 8, pCXN2-Flag-DOCK2-dN; lanes 4 and 9, pCXN2-Flag-DOCK2-dBE; and lanes 5 and 10, pCXN2-Flag-DOCK2-CT.
Activation of Rac1 by CrkL and DOCK2 in 293T cells
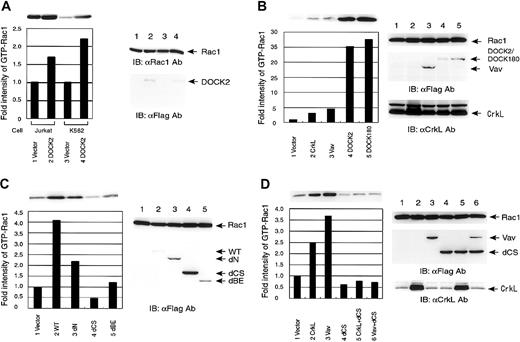
To examine the mechanism of the DOCK2-induced Rac1 activation, we employed a pull-down assay using PAK2-RBD (Rac-binding domain) to measure the amount of the active form of Rac1. First we established the Jurkat and K562 cell lines stably expressing DOCK2, and examined the levels of GTP form of endogenous Rac1. In spite of a polyclonal state, the levels of GTP-Rac1 in the cells stably expressing DOCK2 were elevated about 2-fold compared with vector-control cells in both cell lines (Figure 4A). In 293T cells, we found that CrkL activated Rac1 2- to 3-fold over the vector control, and confirmed significant activation of Rac1 by DOCK2, similar to the DOCK180 as previously reported8 (Figure 4B). We also confirmed that Vav activated Rac1 2- to 3-fold over the control in 293T cells (Figure 4B).
Analysis of the activation of Rac1 by pull-down assay using PAK2-RBD.
(A) Activation of endogenous Rac1 in Jurkat and K562 cells. Jurkat and K562 cells were transfected with either empty vector or pCXN2-Flag-DOCK2, and drug-resistant cells were lysed and incubated with GST-PAK2-RBD and glutathione-Sepharose beads. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-Rac1 mAb (top and right panels). Expression of DOCK2 proteins was examined by immunoblotting by anti-Flag mAb (right panels). The intensities of precipitated Rac1 were measured and are depicted as a bar graph comparing each with the value of vector control as 1.0. (B-D) The 293T cells were transfected with pCXN2-Flag-Rac1. Expression vectors for the proteins are indicated at the bottom. Cells were lysed and incubated with GST-PAK2-RBD and glutathione-Sepharose beads. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-Flag mAb (top panel and bottom graph). Expression of transfected proteins was examined by immunoblotting by anti-Flag mAb for Vav, DOCK2, and DOCK180, and by anti-CrkL Ab (right panels). The intensities of precipitated Rac1 were measured and are depicted as a bar graph comparing each with the value of vector control as 1.0. Panel B shows activation of Rac1 by CrkL, Vav, DOCK2, DOCK180; panel C, activation of Rac1 by DOCK2 and its truncated mutants; and panel D, suppression of CrkL- and Vav-induced Rac1 activation by the DOCK2 dCS mutant.
Analysis of the activation of Rac1 by pull-down assay using PAK2-RBD.
(A) Activation of endogenous Rac1 in Jurkat and K562 cells. Jurkat and K562 cells were transfected with either empty vector or pCXN2-Flag-DOCK2, and drug-resistant cells were lysed and incubated with GST-PAK2-RBD and glutathione-Sepharose beads. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-Rac1 mAb (top and right panels). Expression of DOCK2 proteins was examined by immunoblotting by anti-Flag mAb (right panels). The intensities of precipitated Rac1 were measured and are depicted as a bar graph comparing each with the value of vector control as 1.0. (B-D) The 293T cells were transfected with pCXN2-Flag-Rac1. Expression vectors for the proteins are indicated at the bottom. Cells were lysed and incubated with GST-PAK2-RBD and glutathione-Sepharose beads. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-Flag mAb (top panel and bottom graph). Expression of transfected proteins was examined by immunoblotting by anti-Flag mAb for Vav, DOCK2, and DOCK180, and by anti-CrkL Ab (right panels). The intensities of precipitated Rac1 were measured and are depicted as a bar graph comparing each with the value of vector control as 1.0. Panel B shows activation of Rac1 by CrkL, Vav, DOCK2, DOCK180; panel C, activation of Rac1 by DOCK2 and its truncated mutants; and panel D, suppression of CrkL- and Vav-induced Rac1 activation by the DOCK2 dCS mutant.
To determine the region of DOCK2 responsible for the activation of Rac1, the truncated mutants of DOCK2 were cotransfected with Rac1 into 293T cells and subjected to pull-down assay. The DOCK2 dN mutant induced a 2-fold activation of Rac1, while the DOCK2-dBE did not activate Rac1. The levels of activated Rac1 were below the basal level of using vector alone when we introduced DOCK2-dCS (Figure 4C). Because DOCK2-dCS seemed to decrease the levels of the active form of Rac1, we examined whether DOCK2-dCS functions as a dominant-negative molecule against Rac1 activation. In the pull-down assay, dCS suppressed the CrkL-induced Rac1 activation (Figure 4D).
As DOCK2 does not contain a homologous region as GEF for Rac1, we examined the possible interaction of GEF with DOCK2, and found that Vav was coprecipitated with DOCK2 in Jurkat cells (Figure 2C). The pull-down assay for Rac1 showed that Vav was found to activate Rac1 (Figure 4B), and this was suppressed by DOCK2-dCS (Figure 4D).
Colocalization of DOCK2 with CrkL and F-actin in Jurkat cells
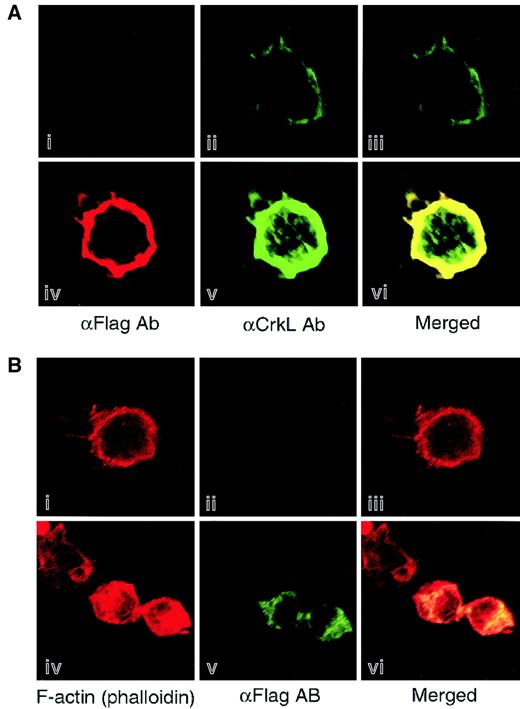
To confirm the association of CrkL and DOCK in hematopoietic cells, the subcellular localization of these 2 proteins was analyzed by immunofluorescent staining. As we found by immunoprecipitation analysis, DOCK2 was colocalized with CrkL to the cell membrane of Jurkat cells that were transiently transfected with the expression vectors of CrkL and DOCK2 (Figure 5A). Furthermore, we observed that the colocalization of DOCK2 and F-actin appears in the cortical ring of Jurkat cells (Figure 5B).
Colocalization of DOCK2 with CrkL and F-actin in the hematopoietic cell line.
Jurkat cells were transfected with expression vectors of both CrkL and DOCK2 (panels Aiv-v), of DOCK2 (panels Biv-v), or vector alone (panels Ai-ii,Biv-v), and at 24 hours after transfection, cells were plated on fibronectin-coated slides and fixed with paraformaldehyde. Localization of CrkL and DOCK were analyzed by anti-CrkL (panels Aii,v) and anti-Flag Abs (panels Ai,iv,Bii,v), respectively. F-actin was visualized by phalloidin conjugated with Alexa-594 (panels Bi,iv). Cells were observed with laser scanning confocal microscopy, and merged images are displayed (panels Aiii,vi,Biii,vi). Original magnification × 400.
Colocalization of DOCK2 with CrkL and F-actin in the hematopoietic cell line.
Jurkat cells were transfected with expression vectors of both CrkL and DOCK2 (panels Aiv-v), of DOCK2 (panels Biv-v), or vector alone (panels Ai-ii,Biv-v), and at 24 hours after transfection, cells were plated on fibronectin-coated slides and fixed with paraformaldehyde. Localization of CrkL and DOCK were analyzed by anti-CrkL (panels Aii,v) and anti-Flag Abs (panels Ai,iv,Bii,v), respectively. F-actin was visualized by phalloidin conjugated with Alexa-594 (panels Bi,iv). Cells were observed with laser scanning confocal microscopy, and merged images are displayed (panels Aiii,vi,Biii,vi). Original magnification × 400.
Suppression of cell attachment in Jurkat cells by stable expression of DOCK2-dCS
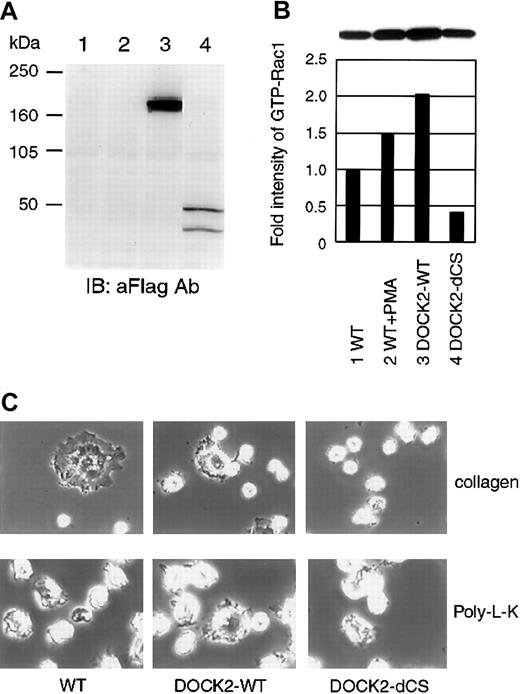
To examine the functional interaction of CrkL and DOCK2 in hematopoietic cells, we established Jurkat cell lines expressing the DOCK2-WT and its mutant, DOCK2-dCS. The expressed proteins were analyzed by immunoblotting using anti-Flag tag Ab (Figure6A), and we confirmed that DOCK2-WT induced Rac1 activation and that the dCS mutant suppressed this activation (Figure 6B). Correlating this with the suppression of Rac1 activity by the dCS mutant, Jurkat cells expressing dCS did not exhibit significant attachment of the cells onto collagen-coated dishes (Figure6C). However, Jurkat-dCS cells attached onto poly-L-lysine–coated dishes in a manner similar to parental cells or those with DOCK2-WT (Figure 6C). We did not observe a significant difference in cell adhesion of Jurkat cells with DOCK2-WT, when compared with their parental Jurkat cells, even on collagen-coated dishes (Figure 6C).
Establishment of Jurkat cell line stably expressing DOCK2 and DOCK2-dCS.
(A) Expression levels of DOCK2 of proteins were examined by immunoblotting with anti-Flag mAb. Lane 1, parental Jurkat cells; lane 2, Jurkat cells with vector control; lane 3, Jurkat cells with DOCK2-WT; lane 4, Jurkat cells with DOCK2-dCS. (B) Measurement of active form of Rac1 using established cell lines. For the positive control, Jurkat cells were preincubated with phorbol 12-myristate 13-acetate (PMA; Sigma) (1 μM) for 10 minutes. Cells were lysed and incubated with GST-PAK2-RBD and glutathione-Sepharose beads. The bound proteins were separated by SDS-PAGE, and endogenous Rac1 was detected by immunoblotting with anti-Rac1 mAb (B&D Transduction Laboratories). The intensities of precipitated Rac1 were measured and are depicted as a bar graph comparing each with the value of nontransfected control as 1.0. (C) Assessment of cell adhesion. Cells were plated on collage type I–coated (top panels) or poly-L-lysine–coated (bottom panels) dishes, and incubated at 37°C for 20 minutes. Cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature and observed by phase contrast microscopy. Original magnification × 400.
Establishment of Jurkat cell line stably expressing DOCK2 and DOCK2-dCS.
(A) Expression levels of DOCK2 of proteins were examined by immunoblotting with anti-Flag mAb. Lane 1, parental Jurkat cells; lane 2, Jurkat cells with vector control; lane 3, Jurkat cells with DOCK2-WT; lane 4, Jurkat cells with DOCK2-dCS. (B) Measurement of active form of Rac1 using established cell lines. For the positive control, Jurkat cells were preincubated with phorbol 12-myristate 13-acetate (PMA; Sigma) (1 μM) for 10 minutes. Cells were lysed and incubated with GST-PAK2-RBD and glutathione-Sepharose beads. The bound proteins were separated by SDS-PAGE, and endogenous Rac1 was detected by immunoblotting with anti-Rac1 mAb (B&D Transduction Laboratories). The intensities of precipitated Rac1 were measured and are depicted as a bar graph comparing each with the value of nontransfected control as 1.0. (C) Assessment of cell adhesion. Cells were plated on collage type I–coated (top panels) or poly-L-lysine–coated (bottom panels) dishes, and incubated at 37°C for 20 minutes. Cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature and observed by phase contrast microscopy. Original magnification × 400.
Discussion
Recently, one of the roles of the hematopoietic cell–specific CDM protein DOCK2 has been demonstrated through the analysis of DOCK2 knockout mice in which DOCK2 was shown to play an essential role for cell migration of both T and B lymphocytes toward chemokines.9 Although the study of DOCK2 knockout mice clearly demonstrated the novel function of DOCK2, the regulatory mechanism of DOCK2 remains unknown because DOCK2 lacks the SH3 domain–binding consensus motif of c–Crk-II, such as PPXLPXK.30 Considering that CrkL shares high sequence homology with c–Crk-II and that its expression is dominant in hematopoietic cells, CrkL seemed to function as the upstream regulatory protein of DOCK2, and we have proven the association of CrkL and DOCK2 both in vivo and in vitro. The colocalization of CrkL and DOCK2 was observed by immunocytochemical analysis. Furthermore, the suppression of CrkL-induced Rac1 activation by DOCK2-dCS mutant also suggested the association of CrkL and DOCK2.
As c–Crk-II did not associate with DOCK2, the complex formation of CrkL and DOCK2 suggests the differential binding specificity of SH3 domains of highly homologous proteins such as CrkL and c–Crk-II. Although we did not detect the binding between c–Crk-II and DOCK2, the SH3 domain of c–Crk-II alone, such as GST–c–Crk-II–SH3(N), was unexpectedly found to precipitate DOCK2 in Jurkat cell lysates. The steric hindrance of the full-length form of c–Crk-II masking its SH3(N) domain may explain the differential binding ability of c–Crk-II and its deletion mutant c–Crk-II–SH3(N). As the SH2 domain of CrkL has been reported to bind to CD34 while c–Crk-II has not,31 CrkL and c–Crk-II may have a unique SH2 domain–binding upstream regulator and an SH3 domain–binding downstream effector.
The analysis with DOCK2 mutants showed that CrkL bound to 2 separate regions of DOCK2, such as dCS and dBE. However, these regions do not contain any proline-rich sequence which would possibly bind to SH3 domains. The CT mutant contains proline-rich sequences but did not bind to CrkL. Therefore, a possible proline-independent fashion of SH3 binding seems to be involved. Recently, the SH3 domain of another adaptor molecule, Grb2, has been shown to interact with the SH3 domain of Vav in a proline-independent manner. Three-dimensional structural analysis revealed that the SH3 domain of Vav formed a structure mimicking the binding surface of the proline-rich consensus motif for association with the Grb2 SH3 domain.32 One of these CrkL-binding regions of DOCK2, dCS region, has an SH3 domain; the novel interaction found in the complex of the SH3 domain of Grb2 and Vav may be involved in the binding between CrkL and DOCK2.
As DOCK2 does not contain a homologous region for GEF for small G proteins, we hypothesized that some GEFs may form a complex with DOCK2. Abundant expression of Vav in hematopoietic cells prompted us to examine whether DOCK2 binds to Vav, and we found the complex formation in Jurkat cells. However, the association of DOCK2 and Vav may be indirect because we did not observe direct interaction of overexpressed forms of these 2 proteins in 293T cells (data not shown). As DOCK2 activated Rac1 at a significantly higher level than did Vav, DOCK2 is considered to use other GEFs, in addition to Vav, for Rac1. In fact, introduction of the dominant-negative form of Vav (VavΔDH) did not suppress DOCK2-induced Rac1 activation (data not shown).
For further analysis of the binding of CrkL and DOCK2, we established a Jurkat cell line expressing the dCS mutant of DOCK2. In cell attachment assays, this Jurkat-DOCK2-dCS cell line reduced its binding ability onto collagen-coated dishes. As CrkL has been shown to regulate cell adhesion through C3G, which is GEF for Rap1 and R-Ras,12,14,33 34 the DOCK2 dCS mutant may affect the interaction of CrkL and C3G, in addition to that of CrkL and DOCK2 itself. The overexpression of DOCK2-dCS mutant theoretically dissociates both CrkL-DOCK2 and CrkL-C3G complexes, because DOCK2 and C3G share the same binding site, the CrkL-SH3 domain. Therefore, we conclude that the overexpression of DOCK2-dCS interacts with wild-type CrkL in vivo with a certain affinity resulting in the alteration of biological phenotype shown, for example, in cell attachment onto collagen-coated dishes. Thus, this functional interference of the binding of CrkL and its effectors by dCS firmly demonstrated the binding of CrkL and DOCK2 in Jurkat cells.
In this study, we demonstrated the association between CrkL and DOCK2 and its regulation of Rac1 in leukemia cell lines. Considering the recent report describing that the lymphocytes of DOCK2 knockout mice decreased chemotaxis,9 the complex of CrkL and DOCK2 may regulate cell motility through Rac, in addition to regulating cell attachment by the CrkL/C3G/Rap1–mediated signaling mechanism. It is worth describing that DOCK2-overexpressed Jurkat cells did not exhibit enhanced cell migration by chemokines in a transwell assay, and the current interpretation is that migration ability is maxmum in the wild-type cells (data not shown). We also tried to demonstrate DOCK2 Rac1 activation in primary B or T lymphocytes and measured the alteration of chemotactic ability to ensure the physiological relevance of the CrkL/DOCK2 signaling pathway. However, lower transfection efficiency hindered the analysis, and future experiments should use an improved transfection technique such as the adenovirus system.
As Rac has been reported to induce actin polymerization by T-cell antigen receptor (TCR) stimulation,35 DOCK2 may play a role in the TCR-mediated signaling pathway through Rac activation, leading to various cellular responses. In fact, DOCK2 was found to activate the IL-2 promotor, and we are currently analyzing that mechanism (data not shown). As CrkL has been shown to enhance cell migration by integrin stimulation36 in leukemia cell line, and as a recent study of double-transgenic mice with CrkL and Brc/Abl demonstrated enhanced leukemogenesis,37 the CrkL/DOCK2 complex may be involved in the fundamental mechanism for leukemogenesis. Furthermore, inhibition of the function of the CrkL-SH3 domain by peptide has been shown to suppress proliferation of primary blast cells from CML patients,38 and the association of CrkL and DOCK2 is thought to be one of the therapeutic targets for leukemia.
We thank Dr Michiyuki Matsuda (Osaka University, Japan) for cDNA of DOCK180 and DOCK2 and its mutants; Dr John Groffen (Children's Hospital of Los Angeles) for CrkL cDNA; Dr Silvio J. Gutkind (National Institutes of Health) for Rac1 cDNA; Junichi Miyazaki (Osaka University, Japan) for pCXN2 vector; Dr Brian J. Druker (Oregon Health Sciences University) for pGEX-CrkL-SH3(N); Dr J. M. Adams (Institute of Medical Research, Melbourne, Australia) for c-Vav cDNA; and Sumie Oikawa for technical assistance.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2001-11-0032.
Supported in part by Grants-in-Aid from The Ministry of Education, Science, Culture, and Sports in Japan; Human Frontier Science Program (A.O.); Mishima Kaiun Memorial Foundation (A.O.); Mitsui Insurance Welfare Foundation (A.O.); and Welfide Medicinal Research Foundation (A.O.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shinya Tanaka, Laboratory of Molecular & Cellular Pathology, Hokkaido University School of Medicine, N 15, W 7, Kita-ku, Sapporo, 060-8638, Japan; e-mail:sitanaka@patho2.med.hokudai.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal