HIV-1–derived lentivectors are promising for gene transfer into hematopoietic stem cells but require preclinical in vivo evaluation relevant to specific human diseases. Nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice accept human hematopoietic stem cell grafts, providing a unique opportunity for in vivo evaluation of therapies targeting human hematopoietic diseases. We demonstrate for the first time that hematopoietic stem cells from patients with X-linked chronic granulomatous disease (X-CGD) give rise to X-CGD–phenotype neutrophils in the NOD/SCID model that can be corrected using VSV-G–pseudotyped, 3rd-generation, self-inactivating (SIN) lentivector encoding gp91phox. We transduced X-CGD patient-mobilized CD34+ peripheral blood stem cells (CD34+PBSCs) with lentivector–gp91phox or amphotropic oncoretrovirus MFGS–gp91phox and evaluated correction ex vivo and in vivo in NOD/SCID mice. Only lentivector transduced CD34+PBSCs under ex vivo conditions nonpermissive for cell division, but both vectors performed best under conditions permissive for proliferation (multiple growth factors). Under the latter conditions, lentivector and MFGS achieved significant ex vivo correction of X-CGD CD34+PBSCs (18% and 54% of cells expressing gp91phox, associated with 53% and 163% of normal superoxide production, respectively). However, lentivector, but not MFGS, achieved significant correction of human X-CGD neutrophils arising in vivo in NOD/SCID mice that underwent transplantation (20% and 2.4%, respectively). Thus, 3rd-generation SIN lentivector–gp91phox performs well as assessed in human X-CGD neutrophils differentiating in vivo, and our studies suggest that the NOD/SCID model is generally applicable for in vivo study of therapies evaluated in human blood cells expressing a specific disease phenotype.
Introduction
X-linked chronic granulomatous disease (X-CGD) is an inherited defect in phagocyte oxidase resulting from a deficiency of gp91phox.1-5 Female X-CGD carriers exhibit mosaicism of oxidase activity in neutrophils. Carriers with more than 5% oxidase normal neutrophils generally have a normal phenotype, indicating a gene therapy goal of more than 5% corrected neutrophils.6 Even this modest goal for gene marking has not been achieved in myeloid cells in human subjects in vivo after ex vivo stem cell gene therapy.7 One obstacle is the inability of oncoretrovirus vectors to transduce nondividing G0 or G1 hematopoietic totipotent stem cells.8-11 Although vectors derived from lentiviruses have the potential to overcome this obstacle,12-17 lentivectors considered for clinical application must be modified to eliminate pathogenicity and potential to regain replication function yet retain the desirable feature of transducing totipotent hematopoietic stem cells. The replication-incompetent 3rd-generation lentivector used in this study is self-inactivating, stripped of all HIV accessory proteins, lacks regulatory Tat protein, and is strictly dependent on complementation of Rev protein in trans; thus, it appears to satisfy these requirements.18 19
Most work with lentivectors has been performed with nontherapeutic marker genes such as eGFP.13,20-25 It is important to demonstrate that 3rd-generation SIN lentivector efficiently transduces primitive hematopoietic stem cells with each therapeutic gene of interest, such as the gp91phox required for correction of X-CGD. The gp91phox is a 570-amino acid, highly glycosylated transmembrane protein that must form a heterodimer with p22phox and interact with other oxidase subunits for functional activity.4 In this study we demonstrate that 3rd-generation SIN lentivector encoding gp91phox achieves ex vivo transduction of nondividing, mobilized CD34+PBSCs from patients with X-CGD; corrects the functional oxidase defect in myeloid cells derived in vitro from these transduced CD34+PBSCs; and efficiently transduces NOD/SCID mouse-repopulating X-CGD CD34+PBSCs, in which it achieves persistent expression of gp91phox in human granulocytes differentiated in vivo in these chimeras.
Materials and methods
Source of CD34+PBSCs or engineered K562–X-CGD cell line
Granulocyte–colony-stimulating factor (G-CSF; 10 μg/kg per day for 5 days) mobilized CD34+PBSCs were obtained from healthy adult volunteers and 2 adult patients with X-CGD after informed consent was obtained (National Institutes of Health institutional review board–approved protocols 94-I-0073 and 95-I-0134).26 The 2 patients with X-CGD lack gp91phox-protein and oxidase activity in neutrophils; and gp91phox gene mutation analysis shows an open-reading frame del1356GT in one patient and a change from gtagg to gcaag at the start of intron 2 leading to loss of exon 2 from the mRNA in the other patient. Ten-liter apheresis PBSC collection (CS3000 Plus; Baxter Healthcare, Fenwal Division, Deerfield, IL) was performed at peak mobilization. CD34+PBSCs were purified to 70% to 80% from the apheresis product (ISOLEX300i; Nexell Therapeutics, Irvine, CA).7
We also used a human K562 cell model of X-CGD (K562–X-CGD) that we engineered to contain p47phox and p67phox (and that naturally expresses p22phox mRNA).27 Only when K562–X-CGD are transduced to also produce gp91phox do these cells become capable of generating superoxide in response to phorbol 12-myristate 13-acetate (PMA) stimulation. Procedures for transduction and for analysis of gp91phoxexpression and oxidase activity in K562–X-CGD were similar to those outlined below for CD34+PBSCs.
Construction of vectors encoding gp91phox and eGFP
Human gp91phox or eGFP cDNA was inserted in theNcoI-BamHI cloning site of MFGS oncoretrovirus vector plasmid.7,28-31 MFGS–gp91phox or MFGS–eGFP plasmid was transfected into amphotropic packaging line 293-SPA, and vector-producing clones were selected.8-10Construction of SIN lentivector transfer plasmid pRRLsin.hCMVGFPpre.2 containing the eGFP transgene has been described.18 19 Cutting withBamHI-SalI removed the eGFP sequence allowing insertion of gp91phox or eCFP (cyan fluorescence protein). For some experiments the cytomegalovirus (CMV) promoter in lentivector–gp91phox was replaced with human phosphoglycerate kinase (PGK) or elongation-factor-1-alpha (EF1α) promoter. However, all studies in vivo in NOD/SCID mice were performed with lentivector–CMV promoter constructs.
Vector production
Amphotropic MFGS–gp91phox or MFGS–eGFP vector was collected over 12 hours from confluent cultures of producer lines, at titers of 2 × 107 or 1 × 106 infectious particles/mL, respectively. Supernate was filtered, stored at −70°C, and used at 90% of neat supernatant for transductions.
VSV-G–pseudotyped lentivector–gp91phox, –eGFP, or –eCFP particles were generated by transient cotransfection of the specific transfer vector plasmid with the 3 packaging plasmids (pMDLg/pRRE, the gag-pol plasmid; pRSV-Rev, a Rev expressing plasmid; and pMD.G, a VSV-G envelope expressing plasmid) into 293T cells as described previously.18 Lentivector supernatant was filtered, concentrated by ultracentrifugation (43 000g for 3 hours), and stored at −70°C. All lentivectors were adjusted to a titer of 4 × 106 infectious particles/mL at a final dilution for transductions.
CD34+PBSC culture and transduction procedures
Cultures (37°C, 7% CO2) of CD34+PBSCs were initiated at 2 × 105 cells/mL (5 mL per well in 6-well plates) in complete growth medium (X-VIVO 10 [BioWhittaker, Walkersville, MD] containing 1% human serum albumin [HSA] plus stem cell factor [SCF] at 50 ng/mL [R&D Systems, Minneapolis, MN], FLT3-ligand at 100 ng/mL [FLT3-L, a gift from Immunex, Seattle, WA], thrombopoietin (TPO) at 20 ng/mL [R&D Systems], Pixykine [PIXY321; interleukin-3/granulocyte-macrophage–colony-stimulating factor fusion protein; a gift from Immunex] at 20 ng/mL, and G-CSF at 10 ng/mL). Optimum transduction conditions for MFGS or lentivirus vectors were achieved with 5 growth factors, protamine at 6 μg/mL, fibronectin fragment, CH-296 (RetroNectin TaKaRa Shuzo, Otsu, Japan) coating of plates, and spinoculation (centrifugation in plates at 1200g at 32°C) for 20 minutes at the start of each 7-hour transduction.31,32 Transductions were performed on culture days 1, 2, and 3, after which cells were either maintained in complete growth medium for further culture ex vivo or were washed and resuspended in phosphate-buffered saline (PBS) containing 0.1% HSA for intravenous injection into NOD/SCID mice. Cultures of nontransduced X-CGD or normal CD34+PBSCs served as negative and positive controls, respectively, for assays of oxidase activity and gp91phox expression. Some experiments were performed in which CD34+PBSCs were cultured initially for 3 days with FLT3-L at 50 ng/mL as the only growth factor to maintain cell viability while avoiding stimulation of cell division (proliferation-limiting conditions). In those experiments, with transductions performed under proliferation-limiting conditions, the CD34+PBSCs were maintained overnight with FLT3-L and then were subjected to a single 7-hour transduction with either lentivector–gp91phox or MFGS–gp91phox. Vector medium was replaced with fresh medium containing only FLT3-L. Twenty-four hours later, cells were switched to complete medium containing 5 growth factors. For other experiments, CD34+PBSCs remaining in the G0phase of the cell cycle at 18 hours of culture in complete growth medium were sorted by labeling with Hoechst 33342 (DNA dye) and PyroninY (RNA dye).33
Analysis of transduction and correction of oxidase activity
CD34+ cells were maintained in liquid culture for up to 28 days and were analyzed for PMA-stimulated superoxide production (chemiluminescence light units [LU]). For analyses of human hematopoietic cells engrafted in the marrow of NOD/SCID mice, marrow cells were analyzed by flow cytometry to determine the expression of human CD45 (all leukocytes) and human CD13 (myeloid cells). Human gp91phox was detected using fluorescein isothiocyanate (FITC)–conjugated murine monoclonal antibody 7D5, which does not bind to mouse gp91phox.34
Vector copy number in transduced human cells was determined by real-time quantitative TaqMan polymerase chain reaction (PCR) (PE Applied Biosystems, Foster City, CA). Endogenous oncoretroviruses exist in the genome of NOD/SCID mice. Therefore, we designed primer sets that amplified only vector inserts from respective MFGS vectors. This design used a common forward primer for all MFGS vectors located just upstream of the transgene, a labeled probe that overlapped the start of transgene sequence, and a reverse primer located within the 5′ coding region of each transgene. MFGS–gp91phox: forward primer, GTGAAGGCTGCCGACCC; 6FAM-labeled probe, TGGACCATCCTCTAGACTGCCATG; reverse primer, CCAAACCAGAATGACAAAAATGG; MFGS–eGFP: same forward primer; 6FAM-labeled probe, TGGACCATCCTCTAGACTGCCATGGC; reverse primer, CTCGCCCTTGCTCACCAT.
For the lentivectors we designed primers and probe completely within the extended LTR lentivector sequence that could be used regardless of the transgene (gp91phox or eGFP). Lentivector: forward primer, TGAAAGCGAAAGGGAAACCA; 6FAM-labeled probe, AGCTCTCTCGACGCAGGACTC; reverse primer, CCGTGCGCGCTTCAG.
For studies comparing vector copy and mRNA transcript numbers in MFGS- versus lentivector–gp91phox-transduced K562-X-CGD, we designed primers and probe spanning cDNA exon 3 and exon 4 regions that amplified only the gp91phox transgene insert and not the native gp91phox genomic sequence, regardless of which type of vector was used. gp91phox cDNA: forward primer, GTCGAAATCTGCTGTCCTTCCT; 6FAM-labeled probe, TTCCAGTGCGTGCTGCTCAACAAGA; reverse primer, TTCGAAGACAACTGGACAGGAAT.
Quantitative real-time TaqMan PCR analysis of percentage human cell engraftment in NOD/SCID mice was found to correlate closely with human CD45+ cells by flow cytometry. A primer set and fluorescent probe were devised to detect the human housekeeping gene, phenol sulfotransferase gene (STP), within chimeric bone marrow.35 36 Human STP: forward primer, GGTGCCCTTCCTTGAGTTCA; 6FAM-labeled probe, CCCCAGGGATTCCCTCAGGTGTGT; reverse primer, CCCCTTGCACCCAGGAC.
Genomic DNA from mixtures of mouse and human blood cells were used as standards. For TaqMan PCR analysis, the following incubation periods were applied for all primer sets: 2 minutes at 50°C, 10 minutes at 95°C, 40 cycles of 15 seconds at 95°C, and 60 seconds at 60°C. Standard curves for the TaqMan PCR analyses were obtained by using vector single-copy clones of K562 cells transduced with the respective vectors.
Transplantation of human CD34+PBSCs into NOD/SCID mice
Cultured CD34+PBSCs (107/mouse) were transplanted by tail-vein injection into sublethally (325 cGy) irradiated 7- to 9-week-old NOD/SCID mice (Jackson Laboratory, Bar Harbor, ME). For all lentivector studies and most MFGS vector studies, mice were killed after 3.5 months, and bone marrow was harvested from femurs and tibias.20 For one set of experiments with MFGS–gp91phox transduced X-CGD CD34+PBSCs, the NOD/SCID/β2m−/− mouse model was used and analyzed at 2 months after transplantation. Although total engraftment of human cells was higher in the latter model, the percentage of transduced human cells appeared to be the same with MFGS vectors in both mouse models when analyzed after 2 months or longer.
Results
Lentivector–gp91phox transduction of nondividing X-CGD CD34+PBSCs corrects oxidase function
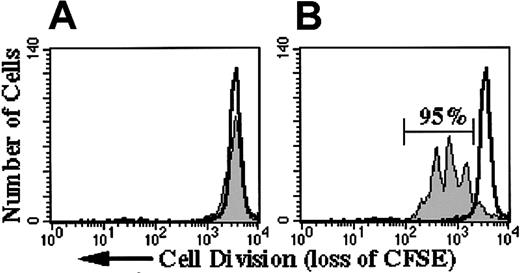
Initial studies focused on determining the potential of our lentivectors to transduce nondividing, human, mobilized CD34+PBSCs. In preliminary studies, we demonstrated that CD34+PBSCs cultured with 50 ng/mL FLT3-L alone were viable for 3 days but did not proliferate. This was confirmed by pulse labeling of CD34+PBSCs with PKH26 or CFSE (carboxyfluorescein diacetate succinimidyl ester).37-39 CFSE fluorescence of CD34+PBSCs remained constant until day 3 in medium containing FLT3-L only (Figure 1A), whereas 95% of CD34+PBSCs cultured in medium containing 5 growth factors had divided 1 to 4 times (Figure 1B).
Effect of growth factors on proliferation of CD34+PBSCs in 3-day culture.
(A) CD34+PBSCs cultured in medium with only FLT3-L did not proliferate (30% viable). (B) CD34+PBSCs cultured in FLT3-L, SCF, G-CSF, TPO, and Pixykine resulted in 95% of cells demonstrating 1 to 4 divisions (more than 90% viable). Colchicine (100 ng/mL)-treated CD34+PBSCs served as a control for nondivided cells (open unshaded histogram in panels A and B).
Effect of growth factors on proliferation of CD34+PBSCs in 3-day culture.
(A) CD34+PBSCs cultured in medium with only FLT3-L did not proliferate (30% viable). (B) CD34+PBSCs cultured in FLT3-L, SCF, G-CSF, TPO, and Pixykine resulted in 95% of cells demonstrating 1 to 4 divisions (more than 90% viable). Colchicine (100 ng/mL)-treated CD34+PBSCs served as a control for nondivided cells (open unshaded histogram in panels A and B).
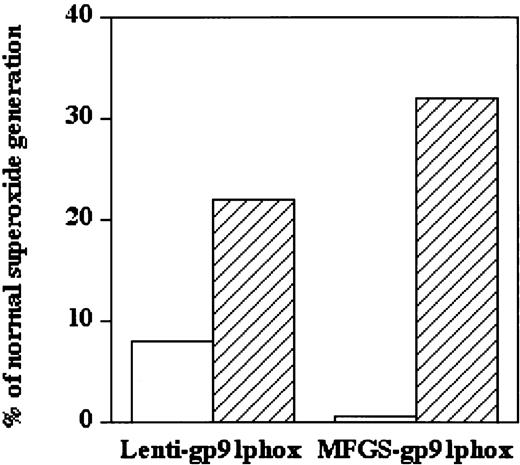
As X-CGD CD34+PBSCs differentiated in culture, endogenous oxidase subunits (excluding the deficient gp91phox) began to appear by day 7. Thus, the appearance of oxidase activity is correlated with gp91phox transgene expression. Figure2 shows the appearance of oxidase activity (% of normal) in cultures of X-CGD CD34+PBSCs that had been transduced once with 3rd-generation SIN lentivector–gp91phox (left pair of bars) or MFGS–gp91phox (right pair of bars) under proliferation-limiting (open bars) or proliferation-permissive (hatched bars) conditions (conditions corresponding to those shown in Figure 1A-B).
Effect of growth factor stimulation on lentivector- or oncoretrovirus vector–mediated ex vivo functional correction of oxidase activity in X-CGD CD34+PBSCs.
Shown is oxidase activity at 17 days of culture following a single 7-hour transduction of nondividing or proliferating X-CGD CD34+PBSCs using either third-generation SIN lentivector–gp91phox or MFGS–gp91phox (average of 2 experiments). For the first 2 days of culture, including transduction on day 1, PBSCs were maintained in FLT3-L (50 ng/mL) alone (proliferation nonpermissive conditions, ■) or in the 5-cytokine combination (proliferation-permissive conditions, ▨). Twenty-four hours after transduction, all cells were switched to fresh medium containing the 5-cytokine combination. PMA-stimulated superoxide generation (chemiluminescence assay) is expressed as percentage of the stimulated oxidase activity of a similar 17-day culture of normal CD34+PBSCs. Negative control: nontransduced X-CGD CD34+PBSCs at 17 days of culture demonstrated PMA-stimulated superoxide generation that was 0.3% of the normal control (not shown).
Effect of growth factor stimulation on lentivector- or oncoretrovirus vector–mediated ex vivo functional correction of oxidase activity in X-CGD CD34+PBSCs.
Shown is oxidase activity at 17 days of culture following a single 7-hour transduction of nondividing or proliferating X-CGD CD34+PBSCs using either third-generation SIN lentivector–gp91phox or MFGS–gp91phox (average of 2 experiments). For the first 2 days of culture, including transduction on day 1, PBSCs were maintained in FLT3-L (50 ng/mL) alone (proliferation nonpermissive conditions, ■) or in the 5-cytokine combination (proliferation-permissive conditions, ▨). Twenty-four hours after transduction, all cells were switched to fresh medium containing the 5-cytokine combination. PMA-stimulated superoxide generation (chemiluminescence assay) is expressed as percentage of the stimulated oxidase activity of a similar 17-day culture of normal CD34+PBSCs. Negative control: nontransduced X-CGD CD34+PBSCs at 17 days of culture demonstrated PMA-stimulated superoxide generation that was 0.3% of the normal control (not shown).
These studies confirm that 3rd-generation lentivector gp91phox retained the ability to transduce nonproliferating X-CGD CD34+PBSCs and that MFGS–gp91phox lacked any significant capacity to transduce nondividing CD34+PBSCs. However, the activation of CD34+PBSCs with multiple cytokines did significantly enhance the capacity of 3rd-generation SIN lentivector to transduce human CD34+PBSCs. Most published studies of lentivector transduction of unstimulated hematopoietic stem cells have been performed with either cord blood or bone marrow, each of which appears to transduce with lentivector efficiently, even without cytokine prestimulation. Mobilized CD34+PBSCs in the current study required stimulation for optimum transduction with 3rd-generation SIN lentivectors. Therefore, for our studies of transplantation of human CD34+PBSCs into NOD/SCID mice, all transductions were performed under conditions of optimum proliferation (multiple growth factors).
Results of 3 daily 7-hour transductions of X-CGD CD34+PBSCs under optimum proliferation conditions
X-CGD CD34+PBSCs were transduced ex vivo with lentivector gp91phox or MFGS–gp91phox on 3 consecutive days under optimum transduction conditions, as noted in “Materials and methods,” and were analyzed for the expression of gp91phox transgene on culture day 17 (Figure3). Similar cultures of nontransduced X-CGD CD34+PBSCs and normal CD34+PBSCs were used as negative and positive controls, respectively, for gp91phox expression. By day 17 of ex vivo culture, CD34+PBSCs had differentiated such that 24% of nontransduced normal CD34+PBSCs appeared to be granulocytes (neutrophils, band forms, eosinophils) that expressed high levels of native gp91phox (Figure 3, open bar). Similar analyses of differentiating cultures of naive, nontransduced X-CGD CD34+PBSCs indicated similar numbers of granulocytes by visual light microscopy inspection but demonstrated no detectable labeling with anti-gp91phox antibody (not shown). X-CGD CD34+PBSCs transduced with lentivector–gp91phox or MFGS–gp91phox under optimum conditions demonstrated gp91phox transgene expression in 18% and 54% of cells, respectively, at day 17 of culture (Figure 3, stippled and hatched bars).
Expression of gp91phox in cultured X-CGD CD34+PBSCs under optimum conditions for ex vivo transduction with lentivector or oncoretrovirus vector.
Shown is the percentage of 17-day cultured CD34+PBSCs expressing gp91phox comparing nontransduced normal CD34+PBSCs (open bar) with X-CGD CD34+PBSCs that had been subjected to 3 daily 7-hour transductions under growth-stimulating conditions (5 cytokines) using either 3rd-generation SIN lentivector–gp91phox (dotted bar) or MFGS–gp91phox (hatched bar). Cultured CD34+PBSCs were labeled with FITC-conjugated anti-gp91phox and analyzed by flow cytometry, and the data were expressed as the percentage of cells that exhibit an expression of gp91phox that is higher than the 95thpercentile of isotype antibody control. By this criterion nontransduced X-CGD CD34+PBSCs do not give rise to any cells that are positive for the expression of gp91phox (not shown). Normal control CD34+PBSCs differentiating in culture first gave rise to differentiated myeloid cells that expressed native gp91phox by approximately day 8 of culture, and this increased to a steady state maximum by approximately day 15, which, in the example shown at day 17 of culture, demonstrated approximately 24% of cells positive for the expression of gp91phox (open bar). Results are representative of 2 experiments.
Expression of gp91phox in cultured X-CGD CD34+PBSCs under optimum conditions for ex vivo transduction with lentivector or oncoretrovirus vector.
Shown is the percentage of 17-day cultured CD34+PBSCs expressing gp91phox comparing nontransduced normal CD34+PBSCs (open bar) with X-CGD CD34+PBSCs that had been subjected to 3 daily 7-hour transductions under growth-stimulating conditions (5 cytokines) using either 3rd-generation SIN lentivector–gp91phox (dotted bar) or MFGS–gp91phox (hatched bar). Cultured CD34+PBSCs were labeled with FITC-conjugated anti-gp91phox and analyzed by flow cytometry, and the data were expressed as the percentage of cells that exhibit an expression of gp91phox that is higher than the 95thpercentile of isotype antibody control. By this criterion nontransduced X-CGD CD34+PBSCs do not give rise to any cells that are positive for the expression of gp91phox (not shown). Normal control CD34+PBSCs differentiating in culture first gave rise to differentiated myeloid cells that expressed native gp91phox by approximately day 8 of culture, and this increased to a steady state maximum by approximately day 15, which, in the example shown at day 17 of culture, demonstrated approximately 24% of cells positive for the expression of gp91phox (open bar). Results are representative of 2 experiments.
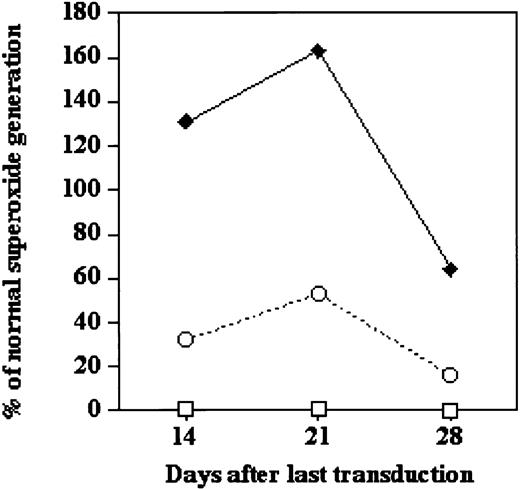
Figure 4 shows reconstitution of oxidase activity over 4 weeks of culture after transduction, with the results expressed as percentage of oxidase activity appearing in differentiating cultures of normal CD34+PBSCs. There was little detectable oxidase activity in the cultures of naive, nontransduced X-CGD CD34+PBSCs (Figure 4, open squares). With both groups of transduced X-CGD CD34+PBSCs, the apparent correction peaked at 3 weeks in culture, at which time X-CGD cells transduced with the lentivector–gp91phoxdemonstrated approximately 53% of normal activity, whereas MFGS–gp91phox-transduced X-CGD cells generated 163% of normal activity. The supranormal levels of oxidase activity in the MFGS–gp91phox-transduced population likely resulted from high expression of gp91phox transgene in differentiating myeloid cells that had begun to express low levels of the complementary oxidase subunits. We have previously shown that transduction-mediated high expression of any one of the oxidase subunits, even in normal early myeloid cells expressing low levels of oxidase factor subunits, results in higher levels of oxidase activity.31 This is probably a simple chemical mass-action effect. Early in differentiation, when all the subunits are present in limiting amounts, increasing the concentration of any one of the multiple subunits enhances assembly to form the active oxidase.
Functional correction of oxidase activity in cultured X-CGD CD34+PBSCs under optimum conditions for ex vivo transduction with lentivector or oncoretrovirus vector.
Shown is the correction of oxidase activity over 28 days (after last transduction) of culture of X-CGD CD34+PBSCs under growth-stimulating conditions (5 cytokines) comparing nontransduced control X-CGD CD34+PBSCs (■) with the same cells that had been subjected to 3 daily 7-hour transductions using third-generation SIN lentivector–gp91phox (○) or MFGS–gp91phox (♦). PMA-stimulated superoxide generation, as measured in a chemiluminescence assay, is expressed as the percentage of the stimulated superoxide generation by a similar culture of normal CD34+PBSCs. Results are representative of 2 experiments.
Functional correction of oxidase activity in cultured X-CGD CD34+PBSCs under optimum conditions for ex vivo transduction with lentivector or oncoretrovirus vector.
Shown is the correction of oxidase activity over 28 days (after last transduction) of culture of X-CGD CD34+PBSCs under growth-stimulating conditions (5 cytokines) comparing nontransduced control X-CGD CD34+PBSCs (■) with the same cells that had been subjected to 3 daily 7-hour transductions using third-generation SIN lentivector–gp91phox (○) or MFGS–gp91phox (♦). PMA-stimulated superoxide generation, as measured in a chemiluminescence assay, is expressed as the percentage of the stimulated superoxide generation by a similar culture of normal CD34+PBSCs. Results are representative of 2 experiments.
A similar comparison of lentivector–eGFP and MFGS–eGFP transduction of normal CD34+PBSCs on 3 successive days in the optimum growth conditions is shown in Figure 5. Again, the oncoretrovirus vector achieved higher rates of transduction of the overall population of differentiating CD34+PBSCs, as assessed over 28 days of ex vivo culture, than lentivector–eGFP.
Expression of eGFP in cultured normal CD34+PBSCs under optimum conditions for ex vivo transduction with lentivector or oncoretrovirus vector.
Shown is analysis over 28 days of culture of the percentage of normal CD34+PBSC expressing eGFP following 3 daily 7-hour transductions using 3rd-generation SIN lentivector–eGFP (○) or MFGS–eGFP (♦) under conditions stimulating proliferation (5 cytokines). Cells were analyzed by flow cytometry, and data were expressed as the percentage of cells that show greater fluorescence than the 99th percentile of nontransduced cells in similar cultures. Results are representative of 6 experiments.
Expression of eGFP in cultured normal CD34+PBSCs under optimum conditions for ex vivo transduction with lentivector or oncoretrovirus vector.
Shown is analysis over 28 days of culture of the percentage of normal CD34+PBSC expressing eGFP following 3 daily 7-hour transductions using 3rd-generation SIN lentivector–eGFP (○) or MFGS–eGFP (♦) under conditions stimulating proliferation (5 cytokines). Cells were analyzed by flow cytometry, and data were expressed as the percentage of cells that show greater fluorescence than the 99th percentile of nontransduced cells in similar cultures. Results are representative of 6 experiments.
It is not the transduction of CD34+PBSCs, as assessed ex vivo in long-term culture, but the transduction of the most primitive long-term marrow repopulating cells that determines the clinical efficacy of a vector system. Determination of transduction efficiency in those human CD34+PBSCs capable of engrafting the NOD/SCID mouse likely is more predictive of the clinical usefulness of a vector system, and this assessment was performed next.
Persistent expression of gp91phox by gene-corrected human X-CGD neutrophils in NOD/SCID mouse chimeras
Lentivector–gp91phox- or MFGS–gp91phox-transduced X-CGD CD34+PBSCs and lentivector–eGFP- or MFGS–eGFP-transduced normal CD34+PBSCs described in the previous section were injected intravenously into NOD/SCID mice (approximately 107cultured CD34+PBSCs per mouse, representing approximately a 3.5-fold expansion during 4 days of ex vivo culture). Some control mice received nontransduced X-CGD or normal CD34+PBSCs cultured similarly to transduced cells. Mouse–human chimeric bone marrow was harvested at 3.5 months (2 months in one MFGS–gp91phoxexperiment).
NOD/SCID chimeric bone marrow was analyzed by flow cytometry to assess engraftment of human cells and expression of either gp91phox or eGFP in human cells. For the data derived by flow cytometry and shown in Figure 6, analysis was gated on the region with forward and side scatter characteristic of human and murine neutrophils and on the human CD45+ population. This region also contained all human CD13+ (myeloid) cells. The range of human cell engraftment was 15% to 60%, and there was close correlation between the CD45+ flow cytometry analysis and the STP gene PCR assessment used to verify the extent of human cell engraftment. Figure 6 shows summary data for the in vivo experiments. It is important to recall that for gp91phox and eGFP vector comparisons, MFGS oncoretrovirus vectors achieved higher transduction as measured ex vivo (Figures 4-5).
Expression of the gp91phox therapeutic gene or eGFP marker gene in human cells in vivo in NOD/SCID mice that underwent transplantation with lentivector or oncoretrovirus transduced CD34+PBSC.
Shown is the percentage of the high side scatter CD45+human cells (granulocytes) detected in engrafted NOD/SCID mouse–human chimeric marrow that express transgene (human gp91phox for transplantation with transduced X-CGD CD34+PBSCs or eGFP for transplantation with transduced normal CD34+PBSCs [3 experiments; N = total number of mice]). Approximately 107 CD34+PBSCs were transplanted by tail-vein injection into irradiated (325 cGy) NOD/SCID mice. Marrow was harvested at 2 to 3.5 months after transplantation. All mice analyzed for this study demonstrated 15% to 60% human cell chimerism. Experiments in which CD34+PBSCs were transduced with lentivector, ■; those transduced with MFGS, ▪. NOD/SCID mice also underwent transplantation with similarly cultured, but nontransduced X-CGD CD34+PBSCs or normal CD34+PBSCs (not shown here, but see Figures 7-9). There was significant human cell chimerism in the marrow of these latter groups of animals (more than 15%), but no human cells expressing gp91phox were detected in mice that received transplants of nontransduced X-CGD CD34+PBSCs and no human cells expressing eGFP detected in mice that received transplants of nontransduced normal CD34+PBSCs. In mice engrafted with normal CD34+PBSCs, 70% to 85% of the high side scatter CD45+ human cells expressed native gp91phox.
Expression of the gp91phox therapeutic gene or eGFP marker gene in human cells in vivo in NOD/SCID mice that underwent transplantation with lentivector or oncoretrovirus transduced CD34+PBSC.
Shown is the percentage of the high side scatter CD45+human cells (granulocytes) detected in engrafted NOD/SCID mouse–human chimeric marrow that express transgene (human gp91phox for transplantation with transduced X-CGD CD34+PBSCs or eGFP for transplantation with transduced normal CD34+PBSCs [3 experiments; N = total number of mice]). Approximately 107 CD34+PBSCs were transplanted by tail-vein injection into irradiated (325 cGy) NOD/SCID mice. Marrow was harvested at 2 to 3.5 months after transplantation. All mice analyzed for this study demonstrated 15% to 60% human cell chimerism. Experiments in which CD34+PBSCs were transduced with lentivector, ■; those transduced with MFGS, ▪. NOD/SCID mice also underwent transplantation with similarly cultured, but nontransduced X-CGD CD34+PBSCs or normal CD34+PBSCs (not shown here, but see Figures 7-9). There was significant human cell chimerism in the marrow of these latter groups of animals (more than 15%), but no human cells expressing gp91phox were detected in mice that received transplants of nontransduced X-CGD CD34+PBSCs and no human cells expressing eGFP detected in mice that received transplants of nontransduced normal CD34+PBSCs. In mice engrafted with normal CD34+PBSCs, 70% to 85% of the high side scatter CD45+ human cells expressed native gp91phox.
We used TaqMan PCR to determine the copy number calculated per transgene-expressing human cell of lentivector or MFGS vector transgene in genomic DNA from NOD/SCID chimeric bone marrow. Average lentivector insert copy numbers per transgene-expressing engrafted human cell in these experiments was 2.5 and 2.2 for the lentivector–gp91phox (8 mice analyzed) and lentivector–eGFP (6 mice analyzed), respectively. Average insert copy numbers per transgene-expressing cells in the same cells ex vivo before transplantation (2-3 preparations each) were 5.7 and 10, respectively. This is consistent with the notion that committed progenitors, which do not engraft, are more easily transduced to higher copy numbers. With MFGS–gp91phox and MFGS–eGFP, insert copy numbers ex vivo per transgene-expressing cells were 11.9 and 6.0, respectively. The very low percentage of human cells expressing transgene in vivo in the MFGS experiments (see next paragraph) made it difficult to reliably calculate the in vivo copy number per transgene-expressing cell (approximate range, 1-4 copies). Additional ex vivo transduction experiments were performed in which vector copy number was determined in CD34+PBSCs that were sorted by flow cytometry into populations that did or did not express transgene. Transgene expression-negative cells had a vector copy number of less than 0.01, whereas vector copy numbers in transgene-expressing cells were consistent with those indicated above for the experiments in which ex vivo–transduced cells were transplanted into NOD/SCID mice. It is important to note that in the lentivector and the MFGS vector studies in the NOD/SCID mice, silencing of transgene expression might have occurred and would have increased the calculated copy insert number per transgene-expressing cell. Particularly with MFGS, vector silencing might have contributed significantly to the low expression in vivo.
Despite higher transduction measured ex vivo with MFGS–gp91phox or MFGS–eGFP, the percentages of human NOD/SCID repopulating cells that expressed transgene in vivo with these vectors averaged only 2.4% and 0.3%, respectively. This emphasizes the fact that the apparent transduction of the total CD34+PBSC culture population as measured ex vivo was not predictive of the targeting of the primitive stem cells capable of engrafting in NOD/SCID mice.
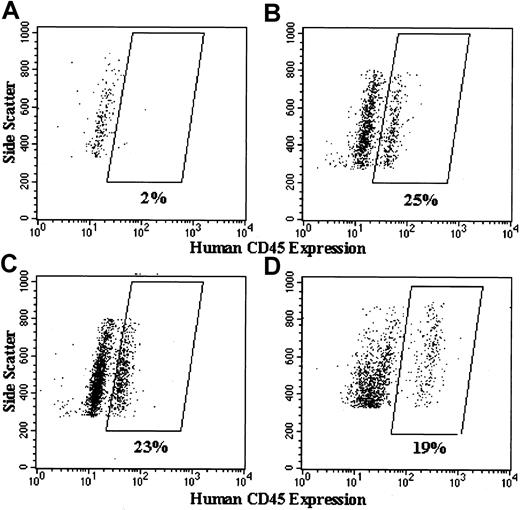
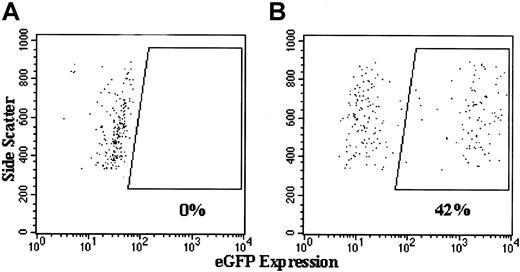
Figures 7,8, and 9show examples of dot plots of flow cytometry analyses used to generate the summary data shown in Figure 6. The 4 representative dot plots in Figure 7 include cells with forward and side scatter parameters characteristic of granulocytes, and they indicate how we gated on the human CD45+ cells to assess the percentage of human cell engraftment. Figure 8 shows examples of dot plot analyses of eGFP transgene expression in the human CD45+ gated population from NOD/SCID chimeric marrow, with or without transduction with lentivector–eGFP. In the example from the lentivector–eGFP transduction experiment shown in Figure 8B, 42% of the lentivector–eGFP-transduced human normal CD34+PBSCs engrafted in this NOD/SCID mouse bone marrow expressed high levels of eGFP. The control for this experiment (engraftment of nontransduced normal human cells) is shown in Figure 8A, where no human cells express eGFP. We do not show the analysis of an example from the MFGS–eGFP transduction experiments because the percentages of human cells from the NOD/SCID chimeras that appeared to express eGFP were lower than 0.4% in every case.
Flow cytometric analyses of high side scatter human CD45+ cells in the bone marrow of NOD/SCID mice that underwent transplantation with human CD34+PBSCs.
Shown are representative dot plots of analysis of bone marrow cells from (A) a NOD/SCID mouse that had not received any transplant and from NOD/SCID mice that received transplants of (B) 4-day cultured but nontransduced X-CGD CD34+PBSCs, (C) lentivector–gp91phox-transduced X-CGD CD34+PBSCs, and (D) lentivector–eGFP-transduced normal CD34+PBSCs. Side scatter is plotted on the vertical axis, and labeling with Per-CP–conjugated antihuman CD45 antibody is plotted on the horizontal axis. Because some of the plots are from experiments analyzed on different days, it demonstrates the analysis variability seen in the apparent separation of the CD45+ cells from the negative cells, in which the boxed areas enclosed the CD45+cells. All NOD/SCID mice that underwent transplantation and were analyzed for this study (Figures 6, 8, 9) had at least 15% CD45+ cells in the chimeric bone marrow as determined by the type of analysis shown in this figure.
Flow cytometric analyses of high side scatter human CD45+ cells in the bone marrow of NOD/SCID mice that underwent transplantation with human CD34+PBSCs.
Shown are representative dot plots of analysis of bone marrow cells from (A) a NOD/SCID mouse that had not received any transplant and from NOD/SCID mice that received transplants of (B) 4-day cultured but nontransduced X-CGD CD34+PBSCs, (C) lentivector–gp91phox-transduced X-CGD CD34+PBSCs, and (D) lentivector–eGFP-transduced normal CD34+PBSCs. Side scatter is plotted on the vertical axis, and labeling with Per-CP–conjugated antihuman CD45 antibody is plotted on the horizontal axis. Because some of the plots are from experiments analyzed on different days, it demonstrates the analysis variability seen in the apparent separation of the CD45+ cells from the negative cells, in which the boxed areas enclosed the CD45+cells. All NOD/SCID mice that underwent transplantation and were analyzed for this study (Figures 6, 8, 9) had at least 15% CD45+ cells in the chimeric bone marrow as determined by the type of analysis shown in this figure.
Flow cytometric analyses of the expression of eGFP by high side scatter human CD45+ cells in the bone marrow of NOD/SCID mice that underwent transplantation with normal human CD34+PBSCs transduced with lentivector–eGFP.
Shown are representative dot plots of analyses of eGFP expression in human (CD45+) cells engrafted in chimeric bone marrow from NOD/SCID mice that underwent transplantation with (A) 4-day cultured but nontransduced normal CD34+PBSCs and (B) lentivector–eGFP-transduced normal CD34+PBSCs. Side scatter is plotted on the vertical axis, and fluorescence by eGFP is plotted on the horizontal axis. Analyses are gated to include only those cells that label positive for the CD45 antigen, as depicted in Figure 7. Boxed areas in this figure are the events indicating cells that are eGFP positive. There are no such events in panel A but 42% of the human cells are positive in panel B.
Flow cytometric analyses of the expression of eGFP by high side scatter human CD45+ cells in the bone marrow of NOD/SCID mice that underwent transplantation with normal human CD34+PBSCs transduced with lentivector–eGFP.
Shown are representative dot plots of analyses of eGFP expression in human (CD45+) cells engrafted in chimeric bone marrow from NOD/SCID mice that underwent transplantation with (A) 4-day cultured but nontransduced normal CD34+PBSCs and (B) lentivector–eGFP-transduced normal CD34+PBSCs. Side scatter is plotted on the vertical axis, and fluorescence by eGFP is plotted on the horizontal axis. Analyses are gated to include only those cells that label positive for the CD45 antigen, as depicted in Figure 7. Boxed areas in this figure are the events indicating cells that are eGFP positive. There are no such events in panel A but 42% of the human cells are positive in panel B.
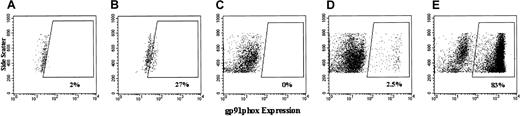
Flow cytometric analyses of the expression of human gp91phox by high side scatter human CD45+ cells in the bone marrow of NOD/SCID mice that received transplants of human CD34+PBSCs.
Shown are representative dot plots of analyses of gp91phoxexpression in human (CD45+) cells engrafted in chimeric bone marrow from NOD/SCID mice that underwent transplantation with 4-day cultured but nontransduced X-CGD CD34+PBSCs (A,C), lentivector–gp91phox-transduced X-CGD CD34+PBSCs (B); MFGS–gp91phox-transduced X-CGD CD34+PBSCs (D), and 4-day cultured but nontransduced normal CD34+PBSCs (E). Side scatter is plotted on the vertical axis, and labeling with antihuman gp91phox is plotted on the horizontal axis. Analyses are gated to include only those cells that label positive for the CD45 antigen, as depicted in Figure 7. Boxed areas represent the events that fall into the gp91phox-positive region. Labeling and analyses in the different panels were performed on different days and resulted in a variability of dot plots. Of note is that the PCR-measured copy number of vector insert per engrafted human cell in each mouse correlates proportionately with the percentage of human cells expressing transgene by flow cytometry in each mouse, though silencing would alter the apparent ratio.
Flow cytometric analyses of the expression of human gp91phox by high side scatter human CD45+ cells in the bone marrow of NOD/SCID mice that received transplants of human CD34+PBSCs.
Shown are representative dot plots of analyses of gp91phoxexpression in human (CD45+) cells engrafted in chimeric bone marrow from NOD/SCID mice that underwent transplantation with 4-day cultured but nontransduced X-CGD CD34+PBSCs (A,C), lentivector–gp91phox-transduced X-CGD CD34+PBSCs (B); MFGS–gp91phox-transduced X-CGD CD34+PBSCs (D), and 4-day cultured but nontransduced normal CD34+PBSCs (E). Side scatter is plotted on the vertical axis, and labeling with antihuman gp91phox is plotted on the horizontal axis. Analyses are gated to include only those cells that label positive for the CD45 antigen, as depicted in Figure 7. Boxed areas represent the events that fall into the gp91phox-positive region. Labeling and analyses in the different panels were performed on different days and resulted in a variability of dot plots. Of note is that the PCR-measured copy number of vector insert per engrafted human cell in each mouse correlates proportionately with the percentage of human cells expressing transgene by flow cytometry in each mouse, though silencing would alter the apparent ratio.
Figure 9 shows representative examples of dot plot analyses of gp91phox transgene expression in the CD45+human X-CGD-cell–gated population from NOD/SCID chimeric marrow without (Figure 9A-C) or with transduction with lentivector–gp91phox (Figure 9B) or MFGS–gp91phox (Figure 9D). As a control, native expression of gp91phox in normal human cells in the NOD/SCID chimeric marrow is also shown (Figure 9E). It is important to note that the intensity of labeling with anti-gp91phox antibody in the gp91phox-expressing population (boxed areas), as determined by the mean fluorescence intensity per cell, was greater in the normal (Figure 9E) and the MFGS–gp91phox-transduced (Figure 9D) groups than in the lentivector–gp91phox group (Figure 9B). Although Figure 9 demonstrates this in vivo in the transduced cells from one mouse in each of the groups shown, this pattern of expression level has been seen in every experiment we conducted comparing the lentivector–CMV–gp91phox-transduced cells versus MFGS–gp91phox-transduced cells, whether ex vivo or in the NOD/SCID model in vivo. This indicates that the expression of gp91phox from the MFGS LTR promoter was significantly greater than the expression from the CMV internal promoter in the SIN lentivector. Of note is that this was not the case with eGFP, in which the cellular expression of eGFP was the same using SIN lentivector or MFGS vector.
Effect of vector type and internal promoter on expression of gp91phox
Because the level of transduction of NOD/SCID-repopulating human CD34+PBSCs with our lentivectors was encouraging but the expression of gp91phox per cell from the internal CMV promoter was low, it was important to determine the basis of this observation and to determine possible corrective measures. We recently constructed our SIN lentivector–gp91phox with alternative internal promoters (human PGK or human EF1α promoters) to improve gp91phox expression. We used these vectors in a series of experiments with our K562–X-CGD model, described in “Materials and methods,” to assess these issues. Of importance to the current analysis, VSV-G–pseudotyped lentivector–CMV-gp91phox and amphotropic MFGS–gp91phox transduced this K562–X-CGD model efficiently, and relative amounts and differences in the expression of gp91phox transgene in these X-CGD–type K562 cells measured by mean fluorescence intensity (anti-gp91phox monoclonal antibody) by flow cytometry were similar to the amounts and differences seen in the transduced X-CGD CD34+ cells. As shown in Table1, we compared genomic vector insert copy number (TaqMan PCR of genomic DNA), mRNA transcript numbers (TaqMan PCR on reverse-transcribed mRNA), mean fluorescence intensity (by flow cytometry) of gp91phox transgene expression in the transgene-positive population, and PMA-stimulated oxidase activity (chemiluminescence light units).
Analyses performed on the unsorted bulk-transduced population
| Vector used to transduce K562-X-CGD cells . | Insert copy no. per gp91phox- positive K562-X-CGD cell . | Vector mRNA transcripts per genomic insert copy . | Mean fluorescence intensity of the gp91phox-positive cell population by flow cytometry . | PMA stimulated oxidase activity (LU/40 min/105gp91phox-positive cells) . |
|---|---|---|---|---|
| MFGS-gp91phox | 3.7 | 1427 | 383 | 1988 |
| Lenti-CMV-gp91phox | 3.2 | 176 | 58 | 508 |
| Lenti-EF1α-gp91phox | 4.4 | 1328 | 214 | 1815 |
| Vector used to transduce K562-X-CGD cells . | Insert copy no. per gp91phox- positive K562-X-CGD cell . | Vector mRNA transcripts per genomic insert copy . | Mean fluorescence intensity of the gp91phox-positive cell population by flow cytometry . | PMA stimulated oxidase activity (LU/40 min/105gp91phox-positive cells) . |
|---|---|---|---|---|
| MFGS-gp91phox | 3.7 | 1427 | 383 | 1988 |
| Lenti-CMV-gp91phox | 3.2 | 176 | 58 | 508 |
| Lenti-EF1α-gp91phox | 4.4 | 1328 | 214 | 1815 |
More than 70% of cells in all 3 transduction groups expressed gp91phox transgene. Identical results were found when gp91phox-expressing cells were purified by flow cytometry sorting first, and then insert copy number analyses and other determinations were performed. Not shown is that there was no improvement of expression levels when PGK replaced CMV promoter in the lentivector–gp91phox.
We conclude that the primary source of the differences in gp91phox expression per cell that we saw between lentivector–CMV-gp91phox and MFGS–gp91phoxwas related to lower production of mRNA per integrated vector copy insert. Furthermore, replacement of the CMV promoter with EF1α promoter in the same lentivector significantly enhanced the production of mRNA encoding the gp91phox transgene, leading to higher levels of gp91phox protein and significantly greater oxidase activity.
Results of lentivector–eCFP and MFGS–eGFP cotransduction of G0 sorted CD34+PBSCs
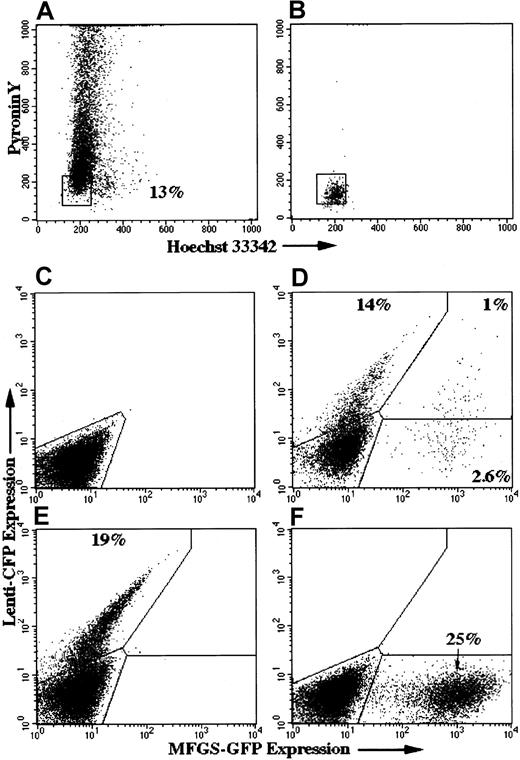
Based on the work of others, the basis for low correction or marking of human cells with amphotropic MFGS vector in vivo in NOD/SCID mice despite high ex vivo transduction is likely attributed to the failure of this vector relative to lentivector to transduce the most primitive cells. It would be helpful to verify this hypothesis in ex vivo culture. Human hematopoietic stem cells that contribute to long-term human cell engraftment are predominantly in G0, and in ex vivo culture do not cycle until after the 3rd day.33 40-42 We compared VSV-G pseudotyped lentivector and amphotropic MFGS transduction of those normal CD34+PBSCs remaining in G0 at 18 hours of culture in proliferation-permissive growth medium. G0 cells sorted to high purity, as described in “Materials and methods” (Figure 10A-B), were transduced at 24 hours and 48 hours after sorting. MFGS vector achieved higher ex vivo transduction rates in the bulk unsorted CD34+PBSCs in this experiment than lentivector (Figure 10E-F), as expected based on our other experiments described above. However, lentivector greatly outperformed MFGS vector in the transduction of sorted G0CD34+PBSCs ex vivo (Figure 10D), a result that correlates more closely with our NOD/SCID data than the ex vivo transduction of the bulk unsorted CD34+PBSCs and that provides a biologic basis for our results.
Lentivector but not oncoretrovirus MFGS-vector efficiently transduced G0-sorted CD34+PBSCs.
CD34+PBSCs were sorted for the G0 cell-cycle phase using Hoechst 33342 (DNA dye) and Pyronin Y (RNA dye) 18 hours after the initiation of culture in proliferation-permissive growth medium. The gate for sorting G0 cells (low DNA, low RNA content) were selected as depicted in panel A. Postsort analysis (B) confirms the accuracy of the cell-sorting procedure. Sorted G0 CD34+PBSC cells were cotransduced with lentivector–CFP and MFGS–GFP at 24 hours and 48 hours after sorting and were analyzed on day 10 by flow cytometry. Gates were drawn according to simultaneously performed single lentivector–CFP and MFGS-GFP transductions on unsorted (E,F) and naive CD34+PBSCs (C). Although the MFGS vector achieved higher transduction levels in the unsorted population (25% vs 19%), the lentivector outperformed the MFGS vector in transduction of the G0 sorted cells (D).
Lentivector but not oncoretrovirus MFGS-vector efficiently transduced G0-sorted CD34+PBSCs.
CD34+PBSCs were sorted for the G0 cell-cycle phase using Hoechst 33342 (DNA dye) and Pyronin Y (RNA dye) 18 hours after the initiation of culture in proliferation-permissive growth medium. The gate for sorting G0 cells (low DNA, low RNA content) were selected as depicted in panel A. Postsort analysis (B) confirms the accuracy of the cell-sorting procedure. Sorted G0 CD34+PBSC cells were cotransduced with lentivector–CFP and MFGS–GFP at 24 hours and 48 hours after sorting and were analyzed on day 10 by flow cytometry. Gates were drawn according to simultaneously performed single lentivector–CFP and MFGS-GFP transductions on unsorted (E,F) and naive CD34+PBSCs (C). Although the MFGS vector achieved higher transduction levels in the unsorted population (25% vs 19%), the lentivector outperformed the MFGS vector in transduction of the G0 sorted cells (D).
Discussion
This study demonstrates for the first time efficient transduction of NOD/SCID-repopulating human X-CGD CD34+PBSCs by a VSV-G–pseudotyped 3rd-generation SIN lentivector–gp91phoxassociated with significant correction of the X-CGD defect in human neutrophils. Another important unique feature of our studies is the demonstration that the NOD/SCID model can be used to study mature human neutrophils differentiating in vivo and expressing the X-CGD phenotype derived from human X-CGD patient stem cells. This validates the more general principle of using the NOD/SCID model to study other human hematopoietic cell diseases, particularly those in which there is no large-animal model.
Our study provides important new information for considering the transition of lentivector from the laboratory to the clinic. First, we used lentivector that incorporates 3rd-generation safety features.18 19 Second, our studies were performed using G-CSF–mobilized CD34+PBSCs, which are the target of choice for most hematopoietic clinical applications of gene therapy but have rarely been the reported target of lentivector in NOD/SCID model studies. Third, we look at outcomes in which the vector transgene is a therapeutic protein, gp91phox, which corrects the genetic defect of X-linked CGD.
The same 3rd-generation SIN lentivector–eGFP used in our study also was used by Guenechea et al22 in their studies to demonstrate efficient transduction of NOD/SCID-repopulating CD34+Lin− stem cells selected from human cord blood or bone marrow. In their studies transduction efficiencies of 15% were achieved ex vivo when human bone marrow or cord blood stem cells were incubated with vector over a 24-hour period, even without prestimulation with cytokines. When their transduction was limited to only a single 5-hour period, the transduction rate ex vivo was 5%. We found a similarly low transduction efficiency of 3rd-generation SIN lentivector when CD34+PBSCs were transduced for only a single 7-hour period under conditions that limited cell division. When we incubated CD34+PBSCs overnight with VSV-G–pseudotyped lentivector in our proliferation-limiting conditions, we found a substantial decrease in cell viability that we did not see in the absence of VSV-G vector. It is well known that CD34+ from PBSCs, relative to those from bone marrow or cord blood, already are at a substantial disadvantage in terms of their engraftment potential in the NOD/SCID-repopulating model.43-46 This cell loss, together with the need for relatively large numbers of CD34+PBSCs per mouse to achieve evaluable levels of human cell engraftment, limited our ability to explore the NOD/SCID engraftment potential of CD34+PBSCs transduced for 24 hours with lentivector under cell division–limiting conditions. However, our studies of the functional correction of the X-CGD oxidase defect by the lentivector–gp91phox versus the MFGS–gp91phoxex vivo under conditions limiting cell division by CD34+PBSCs demonstrated that lentivector–gp91phox achieves significant correction of the oxidase defect, whereas MFGS–gp91phox does not. This is concordant with a number of studies indicating that cell division is an absolute requirement for transduction with oncoretrovirus vectors but not lentivectors.8-11,13,47 48 We found that the addition of multiple growth factors promotes the survival of CD34+PBSCs and enhances transduction with the lentivector. Although extended ex vivo culture may reduce NOD/SCID-repopulating potential, our choice of 4 days of culture in growth factors did not eliminate NOD/SCID-repopulating cells because we were able to achieve 15% to 60% human cell chimerism with cultured CD34+PBSCs. Given that lentivector transduction of CD34+PBSCs under conditions limiting cell division was relatively low and that the MFGS vector did not transduce at all under those conditions, we designed studies to assess the performance of lentivector and MFGS oncoretrovirus vectors in transducing NOD/SCID-repopulating CD34+PBSCs using ex vivo conditions permissive for lentivector and MFGS transduction.
As shown in Figures 4 and 5, after 3 days of transduction under proliferation-permissive conditions, the high-titer, amphotropic oncoretrovirus vector MFGS transduced more CD34+ cells than the VSV-G–pseudotyped 3rd-generation SIN lentivector, as assessed during prolonged culture ex vivo, regardless of the transgene in the construct (eGFP or gp91phox). When these same cells were injected into irradiated NOD/SCID mice, the percentage of human cells containing vector insert by real-time PCR and expressing the transgene 2 to 3.5 months after engraftment was from almost 10- to 100-fold greater with the lentivector than with the oncoretrovirus vector (20% vs 2.4% for expression of the gp91phox therapeutic transgene; 29% vs 0.3% for the eGFP marker gene). This is in accordance with study results indicating that amphotropic oncoretrovirus vectors are less efficient at transduction of the primitive stem cells that encompass the NOD/SCID-engrafting group, even under proliferation-permissive conditions.49,50 One factor in the poor performance of the MFGS oncoretrovirus vector in the transduction of NOD/SCID-repopulating cells in the current study might have been amphotropic vector pseudotyping. Of note is that 2 published studies have reported significant transduction of NOD/SCID-repopulating human CD34+PBSCs with GALV-pseudotyped oncoretrovirus vector.51,52 Other investigators have explored the potential of VSV-G pseudotyping of a variety of oncoretrovirus vectors and have not found this maneuver to greatly enhance transduction of the most primitive hematopoietic stem cells.53
Important information can be derived from our observation that amphotropic MFGS vector achieved some long-term transgene expression in the human CD34+PBSCs engrafted in the NOD/SCID mouse–human chimera model. The absolute requirement of cell division for integration of oncoretrovirus cDNA preintegration complex allows us to conclude that a small number of NOD/SCID-repopulating CD34+PBSCs were stimulated to go through at least one cell division before engraftment during transduction under proliferation-permissive conditions. Furthermore, the high level of overall engraftment of human cells in the NOD/SCID mice in our studies demonstrated that proliferation-permissive conditions of ex vivo culture for 4 days can preserve NOD/SCID-repopulating potential. Our studies of the transduction of CD34+PBSCs remaining in G0 at 18 hours of culture in proliferation-permissive conditions suggest a close numerical correlation between the MFGS or lentivector transduction rates of this subpopulation ex vivo and the results we saw in vivo in the NOD/SCID mice.
The 3rd-generation SIN lentivector that we used in this study was reliant on the activity of an internal promoter (CMV) to achieve transgene expression in the target cell population. By contrast, the MFGS oncoretrovirus vector relied on the activity of the virus LTR to achieve transgene expression. Expression of the gp91phoxtherapeutic gene product was significantly higher per MFGS–gp91phox gene-corrected cell (approaching that of native gp91phox expression in normal neutrophils) than the 3rd-generation SIN lentivector–CMV-gp91phox gene-corrected cell, as indicated by the mean fluorescence intensity by flow cytometry (Figure 9). This is true ex vivo and in vivo in the NOD/SCID chimeras. Similarly low expression per cell of gp91phox transgene mediated by a SIN lentivector with the CMV internal promoter was reported to occur after transduction of the X-CGD PLB985 cell line,54 and we found the same result in our K562–X-CGD model. By contrast, we have found the expression of eGFP to be uniformly high per cell with both types of vectors. This emphasizes that it may be necessary to individually assess the expression of each therapeutic transgene in a particular vector system.
Studies from Dr Dinauer's laboratory have demonstrated that the expression of only 15% to 20% of normal amounts of gp91phox per cell in gp91phox-deficient myeloid cells is associated with higher relative levels of restoration of oxidant production per cell.55 Thus, it may not be necessary to achieve full restoration of gp91phoxprotein to levels seen in normal neutrophils to obtain a clinically beneficial level of correction of oxidase function. Nonetheless, it would be desirable to be able to achieve the highest level of gp91phox transgene expression possible from the 3rd-generation SIN lentivector system. In the current study we found that the expression of gp91phox protein per cell from our lentivector CMV promoter construct is probably inadequate for clinical application. Our studies with the K562–X-CGD cell line suggest that the basis for this observation is low mRNA production mediated by the CMV promoter. EF1α promoter in SIN lentivectors has been reported to achieve significant improvement of transgene expression when hematopoietic progenitors are the target.23,25 56 We also demonstrate that the substitution of human EF1α promoter for CMV significantly enhanced correction of the deficiency in gp91phox expression from lentivector. In preliminary studies, we found that lentivector–EF1α-gp91phox also performs significantly better with regard to gp91phoxprotein production in transduced CD34+PBSCs.
In summary, we show for the first time that 3rd-generation SIN lentivector–gp91phox provides significant correction of the X-CGD functional defect in oxidase activity and efficiently transduces NOD/SCID-repopulating X-CGD CD34+PBSCs, resulting in long-term persistence of gp91phox expression in human X-CGD neutrophils in vivo.
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2001-12-0165.
Supported by Deutsche Forschungsgemeinschaft grant BR 2057/1 (S.B.) and National Institutes of Health grant PO1 CA70970 (C.I.C.).
J.R. and S.B. contributed equally to this manuscript.
A.A.B., T.D., and M.K. are or have been employees of Cell Genesys, Inc, which has a commercial interest in the lentivector system described in these studies. Johns Hopkins University holds patents on CD34 monoclonal antibodies and related inventions. C.I.C. is entitled to a share of the sales royalty received by the University under licensing agreements between the University, Becton Dickinson Corp, and Baxter HealthCare Corp. The terms of these arrangements have been reviewed and approved by the University in accordance with its conflict-of-interest policies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Harry L. Malech, Laboratory of Host Defenses, National Institute of Allergy and Infectious Diseases, Bldg 10, Rm 11N113, 10 Center Dr, MSC 1886, Bethesda, MD 20892-1886; e-mail:hmalech@nih.gov.



![Fig. 6. Expression of the gp91phox therapeutic gene or eGFP marker gene in human cells in vivo in NOD/SCID mice that underwent transplantation with lentivector or oncoretrovirus transduced CD34+PBSC. / Shown is the percentage of the high side scatter CD45+human cells (granulocytes) detected in engrafted NOD/SCID mouse–human chimeric marrow that express transgene (human gp91phox for transplantation with transduced X-CGD CD34+PBSCs or eGFP for transplantation with transduced normal CD34+PBSCs [3 experiments; N = total number of mice]). Approximately 107 CD34+PBSCs were transplanted by tail-vein injection into irradiated (325 cGy) NOD/SCID mice. Marrow was harvested at 2 to 3.5 months after transplantation. All mice analyzed for this study demonstrated 15% to 60% human cell chimerism. Experiments in which CD34+PBSCs were transduced with lentivector, ■; those transduced with MFGS, ▪. NOD/SCID mice also underwent transplantation with similarly cultured, but nontransduced X-CGD CD34+PBSCs or normal CD34+PBSCs (not shown here, but see Figures 7-9). There was significant human cell chimerism in the marrow of these latter groups of animals (more than 15%), but no human cells expressing gp91phox were detected in mice that received transplants of nontransduced X-CGD CD34+PBSCs and no human cells expressing eGFP detected in mice that received transplants of nontransduced normal CD34+PBSCs. In mice engrafted with normal CD34+PBSCs, 70% to 85% of the high side scatter CD45+ human cells expressed native gp91phox.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/13/10.1182_blood-2001-12-0165/4/m_h82423568006.jpeg?Expires=1767839933&Signature=KoRMAd5Wncuo4PrvRhwrWwwk-BmnhuAeO6IUtv9gfFTmowqTQTk4llJcWT8aC8jfzGsXmRpyot4CZ4DWtJe1QGiH8-1KiFwn7iE~ca3xyBa3j8i7WyYMSg5c7Fe2cUwZiDx715jjYl5rF8S5S7F7koreE0muLo72qDH7bgNFhqdAI4KnG0B6kUJ0tuzzw5InuhMkdA5vLsq3uspL7uozrxzw3KsGMPGVxLj5mj3O~qpD6OID0GSzUlTk3~DWytxb3pQuhFVlqgO2MN-FGkUTOpWEMzjEQJ4U3R-MUYZ3oDpwq56TnbJFU0oeEmYDSKptxjBKwG05OShZ6UpeufhGnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal