The human CD34 gene is expressed on early progenitor and stem cells in the bone marrow. Here we report the isolation of the human CD34 locus from a human P1 artificial chromosome (PAC) library and the characterization and evaluation of this genomic fragment for expression of reporter genes in stable cell lines and transgenic mice. We show that a 160-kb fragment spanning 110 kb of the 5′ flanking region and 26 kb of the 3′ flanking region of theCD34 gene directs expression of the human CD34gene in the bone marrow of transgenic mice. The expression of human CD34 transgenic RNA in tissues was found to be similar to that of the endogenous murine CD34 gene. Colony-forming cell assays showed that bone marrow cells staining positive for human CD34 consist of early progenitor cells in which expression of CD34 decreased with cell maturation. In order to test the construct for its ability to express heterologous genes in vivo, we used homologous recombination in bacteria to insert the tetracycline-responsive transactivator protein tTA. Analysis of transgenic human CD34-tTA mice by cross breeding with a strain carrying Cre recombinase under control of a tetracycline-responsive element demonstrated induction of Cre expression in mice in a pattern consistent with the expression of the human CD34 transgene.
Introduction
The human CD34 gene encodes a type I transmembrane glycoprotein expressed on early hematopoietic stem and progenitor cells1,2 and endothelial cells forming small vessels.3,4 Expression of the CD34 antigen decreases as hematopoietic stem cells differentiate and it is absent on all mature blood cells.1,2,5 CD34 expressed on endothelial cells may play a role in leukocyte adhesion and it has been hypothesized that it also plays a role in stem/progenitor cell localization/adhesion in the bone marrow. A recent study suggests that expression of CD34 on progenitor cells might be important for proliferation by contact-mediated inhibition.6 Despite being the object of extensive research, little is known about the regulation of expression and the biologic function of CD34.
The expression of murine CD34 exhibits a pattern similar to that of the human gene,7,8 which suggests that the regulation of both genes might involve similar molecular mechanisms. However, differences became apparent when a CD34− fraction within the murine bone marrow was shown to be capable of long-term reconstitution.9 In addition, bone marrow side population (SP) cells, which are defined by a 2-color fluorescence-activated cell sorting (FACS) analysis using Hoechst 33342 dye, are highly enriched for stem cells and are CD34−/lo.10,11 Further studies have shown that the expression of murine CD34 on stem cells varies with activation status and age.12-14 Finally, studies from our own laboratory demonstrate that in resting bone marrow, humanCD34 transgenes are expressed in repopulating stem cells, whereas endogenous murine CD34 is not.15 These studies suggest that human and murine CD34 might be differentially expressed within the stem cell population.
Experiments on the regulation of human globin,16CD2,17 the murine T-cell receptor α/δ,18 immunoglobin heavy chain,19 or lysozyme20 identified distally located DNA elements that are important for high-level, cell type–specific expression. In light of our own data, in which we had failed to obtain high-level, position-independent human CD34 expression in stable cell lines and transgenic mice using either single elements, such as the promoter and the 3′ enhancer, or a mini gene containing both elements,21 we considered the possibility that expression of the human CD34 gene might require the combinatorial action of multiple proximal and long-range elements for cell type–specific, high-level CD34 expression. To localize critical distal control elements of CD34, we isolated large P1 artificial chromosome (PAC) clones encompassing the human CD34 gene and tested their ability to confer high-level expression of humanCD34 in vivo. Here we show that a 160-kb PAC clone is capable of driving cell type–specific expression in transgenic mice and that this expression cassette can be used to express heterologous genes in transgenic mouse models. This result is of particular interest as controlled expression in CD34+ cells is important for experiments addressing the expression of heterologous genes in stem cells, which may ultimately find applications in gene therapy.
Materials and methods
PAC library screening
There were 2 sets of polymerase chain reaction (PCR) primers specific to the human CD34 gene designed to amplify the extreme 5′ and 3′ ends of the human CD34gene.22 The 5′ primer set consisted of (sense) 5′-ATCACCACTCCTTCCTATCCTT-3′ and (antisense) 5′-CTTTTATTGAGCTACCTTATTAACT-3′, and amplified a 197-bp fragment spanning nucleotides 53 406 to 53 603 of Genbank entry AL035091, located 5 kb upstream from the human CD34 transcription start site.22 A second set of primers (sense: 5′-AGAAGAGATGAGGTGTGAGGAT-3′; antisense: 5′-GGATCCACAAGAATGAGCATGTA-3′) amplified a 248-bp product corresponding to nucleotides 16 649 and 16 919 (Genbank accession number AL035091), located 32 kb downstream of the transcription start site and 5.7 kb downstream from the core 3′ enhancer21 (GenBank accession number AF047373). These 2 sets of primers were used to screen a human genomic PAC library (Genome Systems, St Louis, MO). There were 5 clones identified, 3 of which (PAC-88L2 [GS#9213], PAC-7H11 [GS#9211], and PAC-54A19 [GS#9212]) were analyzed.
PAC DNA preparation and manipulation
Single colonies of bacteria harboring PAC clones were grown in 5 mL of LB (Luria-Bertani) medium containing 50 μg/mL kanamycin and grown overnight. A quantity of 2 mL of this culture was used to start a 200-mL culture. After 2 hours, 200 μL isopropylthio-β-D-galactoside (IPTG; 0.5 M) was added and the culture allowed to grow for another 5 hours. Cells were divided into 50-mL tubes and the bacterial cells were harvested. DNA was prepared using the Midi Plasmid kit (Qiagen, Valencia, CA) according to the manufacturer's protocol with some modifications. In short, elution of PAC DNA from the columns was performed with preheated (65°C) elution buffer. The DNA was subjected to extractions with phenol and chloroform:isoamyl alcohol 24:1 prior to ethanol precipitation. DNA was recovered by spooling around a glass rod. Mapping of the PAC clones was performed by restriction enzyme analysis. A quantity of 1 μg DNA was digested with the appropriate restriction enzymes in a total volume of 30 μL. The digested DNA fragments were separated by field inversion gel electrophoresis (FIGE) for 3 hours at room temperature at 200 V (MJ Research program 2) followed by 17 hours at 10°C using program 3.
Generation of stable transfected lines
The murine myeloid progenitor line 416B23 was cultured in Dulbecco modified Eagle medium (DMEM) with 15% fetal bovine serum (FBS). Stable clones were generated by electroporation of 6 × 106 cells at 270 V and 960 microfarads (μF) with 20 μg supercoiled PAC-88L2 DNA together with 0.2 μg NaeI-linearized PGKpuro.24 Cells were plated on 96-well plates and selected for stable clones with 2 μg/mL puromycin 48 hours later.
Southern blot analysis
Genomic DNA was restricted with BamHI and separated on agarose gels. The DNA was transferred to a positively charged nylon membrane (Biotrans Plus; ICN, Irvine, CA), hybridized, and washed as described.25 Stable chromosomal incorporation of the humanCD34 gene was detected in BamHI-digested genomic DNA using a 0.5-kb HindIII/BamHI fragment from the 3′ untranslated region of human CD34 as a probe.21
PAC modification by homologous recombination
To generate the recombination cassette, the human CD34 promoter and 5′ untranslated region (bp -391 to +261) were amplified by PCR and cloned upstream of the tTA gene,26 which was followed by an internal ribosome entry site (IRES) element. The 3′ homology arm consisted of a 3.2-kb fragment containing human CD34 exon 1 and 2.3 kb of intron 1, with an internal insertion (1.8 kb downstream from the end of exon 1) of Sh ble (the phleomycin/zeocin resistance gene) driven by a prokaryotic EM7 promoter and flanked by Flp recombination target (FRT) sequences.27 This cassette was inserted into the human CD34 locus by homologous recombination as described27,28 using NS3516 bacteria harboring PAC-88L2. Following selection on medium containing 50 μg/mL kanamycin and 50 μg/mL zeocin, bacteria harboring a homologous recombined PAC including the zeocin gene were electroporated with plasmid 705-FLP27 and the excision of the zeocin gene between FRT sequences was achieved by transient expression of Flp recombinase. Details of the subcloning steps are available upon request.
Linearization and preparation of the PAC clone for generation of transgenic mice
PAC DNA was linearized by digestion with SfiI in a total volume of 300 μL. Complete digestion of plasmid DNA was confirmed by FIGE through a 0.8% agarose gel for 24 hours as described above. The digested DNA was extracted with phenol and subsequently extracted with chloroform:isoamyl alcohol 24:1. Centrifugation was performed at low speed (2500 rpm) in order to avoid shearing of the DNA. The DNA was precipitated with ethanol at 4°C and recovered by spooling with a glass rod. The air-dried pellet was resuspended in 0.1 × Tris-EDTA (TE). Transgenic mice were generated by injection of DNA into the fertilized oocytes of FVB/N mice followed by implantation in the uteri of pseudo-pregnant foster mice according to standard procedures.
Generation of Tet-O-cre mice and CD34tTA-Tet-O-cre double-transgenic mice
Plasmid pBS 185 containing the cre gene downstream of the human cytomegalovirus (CMV) promoter/enhancer29 was digested with XhoI and HindIII releasing the cre gene and the MT-I polyadenylation signal. The fragment was subcloned into the SalI-HindIII sites of plasmid pUHD 13-330 to create pUHD 13-3cre. Subsequent digestion with restriction enzymes XhoI andHindIII and subcloning of this fragment into respective sites of pBluescript SK generated the transgenic plasmid consisting of the cre gene with the MT-I polyadenylation signal under control of the tetracycline-responsive element (TRE). The plasmid was digested with XhoI and HindIII to release the transgenic construct from the vector backbone and injected into fertilized oocytes. Double-transgenic mice (CD34tTA-Tet-O-cre) were generated by cross breeding of CD34tTA mice with Tet-O-cre mice.
Genotyping
Transgenic mice were identified by either Southern blot analysis (for the identification of founder animals) or PCR with the 3′ primer set used to isolate the PACs described above. SuperMix polymerase (GIBCO BRL, Grand Island, NY) was used with the amplification conditions consisting of 35 cycles at 94°C for 40 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. For Southern blot analysis, the probe used was a 0.5-kb fragment from the 3′ enhancer region of the human CD34 gene generated by digestion with BamHI and HindIII that detects a 2-kb genomic fragment. Genotyping was performed as described previously.31 Integrity of the human CD34 reporter gene sequence was determined by subsequent hybridization to probes spanning the entire genomic sequence of the gene. The probes used for this purpose were a 0.48-kb XbaI fragment generated by digestion of a 6.0-kb EcoRI fragment containing exons 2, 3, and 4 of human CD34; a 0.75-kb CD34 exon 1/intron 1 junction fragment generated by digestion withEcoRI/SstI;21 and a 1.2-kb fragment by cleavage with BamHI/SstI.22Tet-O-cre transgenic mice as well as CD34tTA-Tet-O-cre transgenic mice were identified by PCR with primers specific for thecre gene. The upstream primer sequence was 5′-ATGTTCAATTTACTGACCG-3′, and the downstream primer sequence was 5′-CGCCGCATAACCAGTGAAAC-3′. Amplification conditions consisted of 35 cycles of 30 seconds denaturation at 94°C, 30 seconds annealing at 51°C, and 30 seconds extension at 72°C yielding an amplification product of 355 bp.
RNA isolation and Northern blot analysis
Total RNA from cell lines was isolated by guanidine isothiocyanate extraction32 and blotted to a positively charged nylon membrane (Biotrans Plus; ICN, East Hills, NY) or MagnaGraph (Micron Separations, Westborough, MA) as described.21 The blots were washed at a final stringency of 65°C in 0.1 × standard saline citrate (SSC)/0.1% sodium dodecyl sulfate (SDS). RNA from murine tissue was prepared by guanidine isothiocyanate extraction followed by cesium chloride gradient centrifugation,33,34 and expression analysis was performed as described.31 A 0.5-kbHindIII/BamHI fragment from the 3′ enhancer of human CD34 was used as a probe for expression of human CD34 cDNA,21 and a 1.26-kb PstI/XhoI fragment from the full-length cDNA7 was used for endogenous murine CD34. To ensure uniform loading levels and integrity of RNA samples, blots were stripped and rehybridized to probes specific for 18S RNA or glyceraldehyde phosphate dehydrogenase (GAPDH).35-37 The intensity of specific signals was quantitated with ImageQuant software (Molecular Dynamics, Sunnyvale, CA) and the values for human versus the endogenous murine CD34 were determined. The expression pattern and the amount of human CD34 RNA expression in various human tissues was determined using a polyA+ selected mRNA blot (Clontech, Palo Alto, CA) using the same probes as described above.
Analysis of protein expression by flow cytometry
Bone marrow cells were flushed from tibias and femurs with phosphate- buffered saline (PBS), filtered through a nylon mesh, washed once with PBS, and incubated with the appropriate antibodies in PBS containing 5% calf serum for 30 minutes on ice in a total volume of 100 μL. The suspension was underlayed with 0.5 mL calf serum and centrifuged for 5 minutes at 1000 rpm. The cell pellet was washed once with PBS prior to staining with the secondary antibody. All analyses were performed on a dual-laser FACS (Becton Dickinson, Franklin Lakes, NJ). The data were analyzed with the CellQuest program. Primary antibodies included HPCA-2 anti–human CD34 (Becton Dickinson), anti–murine CD34 (Pharmingen, San Diego, CA), and anti–murine B220 (Caltag, San Francisco, CA). All other antibodies (α–Ter 119, α–Gr-1, α–Mac-1, α-Thy1, α–c-kit, α–Sca-1) were purchased from Pharmingen.
Tissue processing and immunohistochemistry
Mice were killed following CO2 inhalation, and organs were removed and fixed in neutral buffered 10% formalin at room temperature for 16 hours prior to embedding in paraffin and sectioning. Tissue from wild-type littermates was carried along as control. Sections were deparaffinized and then subjected to antigen retrieval in a microwave oven. Endogenous peroxidase activity was blocked by incubation in 1% hydrogen peroxide. Staining for human CD34 was performed with QBend 10 (Vector Laboratories, Burlingame, CA), a monoclonal antibody against human CD34 raised in mouse against murine epitopes, diluted 1:50, along with the M.O.M. kit (Vector Laboratories) to suppress nonspecific binding. Stable diaminobenzidine (ResGen, Huntsville, AL) was used as chromogen substrate, and the sections were counterstained with hematoxylin solution (Sigma).
Colony-forming unit (CFU) assay
Bone marrow cells from femurs and tibias of 4-month-old human CD34 transgenic mice were suspended in PBS with 2% FBS and stained with phycoerythrin-conjugated anti–human CD34 antibody (Becton Dickinson) and fluorescein isothiocyanate (FITC)–conjugated anti–mouse CD34 antibody (Pharmingen). Afterward, cells were washed twice with PBS and resuspended in PBS containing 2 μg/mL propidium iodide (PI). PI-negative cells from the hCD34+/mCD34−, hCD34+/mCD34+, hCD34−/mCD34+ and hCD34−/mCD34− cell populations were sorted using a high-speed cell sorter (Moflo-MLS; Cytomation, Fort Collins, CO). Cells were inoculated in Methocult H4100 (Stem Cell Technologies) supplemented with 30% fetal calf serum, 1% bovine serum albumin, 20 ng/mL stem cell factor (SCF), 20 ng/mL interleukin 3 (IL-3), 10 ng/mL IL-11, 10 ng/mL granulocyte macrophage–colony-stimulating factor (GM-CSF), 1 U/mL erythropoietin, 10 ng/mL thrombopoietin (R&D Systems, Minneapolis, MN), 2 mM L-glutamine (Stem Cell Technologies), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO BRL). Colony numbers were counted after 5 to 6 days culture.
Reverse transcription (RT) and expression analysis by RT-PCR
Reverse transcription was performed using SuperScript reverse transcriptase (GIBCO BRL) with 100 ng of random primer hexamers according to the manufacturer's suggestions. In order to exclude DNA contamination, RNA was treated with DNase I, amplification grade (GIBCO BRL) following the manufacturer's guidelines prior to reverse transcription. The efficiency of DNase I treatment was determined by PCR in the absence of reverse transcription, using 1 μL per reaction. First strand synthesis was subsequently performed on negative samples. A quantity of 2 μL of each cDNA reaction was used for the detection of cre gene expression. The same primers and amplification conditions as described under genotyping for Tet-O-cre mice were used. Efficiency of reverse transcription was determined by amplification of murine GAPDH with upstream primer 5′-GGTGCTGAGTATGTCGTGGAGTCTA-3′ and downstream primer 5′-CCTGGTTCACCACCTTCTTCTTGATGTC-3′, which amplify a 523-bp sequence of the murine cDNA.38 Reaction conditions for this PCR consisted of 25 cycles, with 30 seconds denaturation at 94°C, 30 seconds annealing at 68°C, and 1 minute extension at 72°C.
Quantitation of expression by real-time PCR
Multiplex PCR with amplification of 18S RNA in the same tube for quantitation of cre expression, TaqMan analysis, and subsequent calculations were performed with an ABI Prism 7700 sequence detection system (Perkin Elmer, Foster City, CA), which detects the signal from a fluorogenic internal probe. A quantity of 100 ng cDNA for each sample was subjected to PCR with primers 5′-TAACCAGTGAAACAGCATTGCTGTC-3′ and 5′-GGCAGTAAAAACTATCCAGCAGCAACAT-3′ for cre expression according to protocols provided by the manufacturer of the TaqMan system (ABI, Foster City, CA). The sequence of the double-labeled oligonucleotide used as probe was FAM-AGCCCGGACCGAACGATGAAGC-TAMRA. Amplification of 18S RNA was performed in the same reaction tubes as an internal standard with an alternatively labeled probe to distinguish its product from that derived from cre RNA (multiplex PCR). All experiments were performed in duplicate for each standard and tissue. Expression in the liver was used for calibration to determine relative quantitation of expression in other tissues.
Results
Isolation and characterization of large human genomic CD34 clones
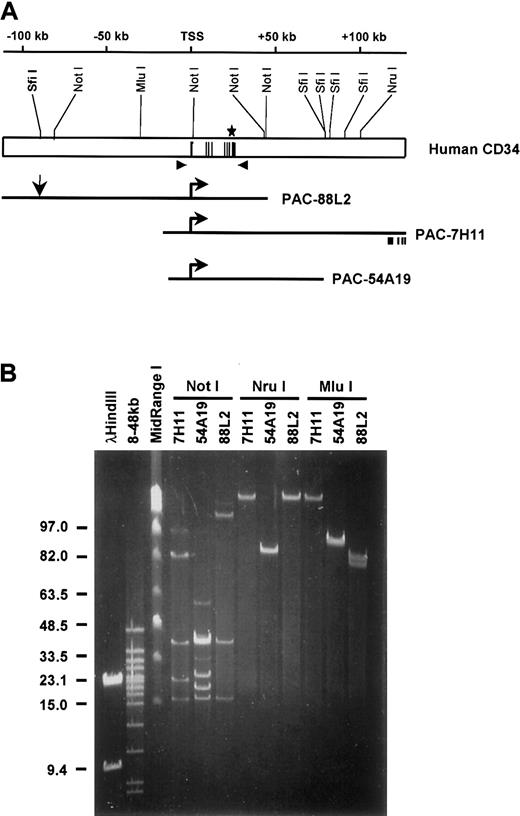
Several cis regulatory elements of the human CD34 gene have been previously identified.21,22,39-42 However, neither these elements alone or in combination were sufficient to direct expression of the stably integrated reporter genes.21 This led us to hypothesize that additional control elements are responsible for proper regulation of CD34, and that they are located at a relatively large distance from the gene. To obtain larger genomic clones of human CD34, we designed 2 sets of CD34 specific primers to screen a human genomic PAC library (a commercial service of Genome Systems) (Figure 1A). One set of primers amplified a DNA region located 5.0 kb upstream from the transcription start site, whereas the second set amplified a sequence 5.7 kb downstream from the core 3′ enhancer.21 40We obtained 3 PAC clones (PAC-7H11, PAC-54A19, and PAC-88L2; Figure 1A) and mapped them by digestion with rare cutting restriction enzymes (Figure 1B). The DNA was resolved on agarose gels using FIGE (Figure1B) followed by Southern blot analysis with hybridization to T7-, SP6-, or CD34-specific oligonucleotide probes. All clones contained the entire CD34 gene, but differed in the length of the 5′ and 3′ flanking sequences (Figure 1A). PAC-88L2 contained the largest insert (160 kb) and the longest 5′ flanking region (110 kb). PAC-7H11 extended most to the 3′ end encompassing 106 kb of the 3′ flanking region. Both clones, PAC-88L2 and PAC-7H11, overlapped over a region of 72 kb. PAC-54A19 had the shortest insert with a total of 86 kb containing only 18 kb of the 5′ flanking region and 26 kb of the 3′ flanking sequence. The entire inserts of PAC-88L2 and PAC-7H11 were used to construct bacterial clone contigs by the Sanger Centre Chromosome 1 Mapping Group (Sanger Centre, Hinxton, Cambridgeshire, United Kingdom) and were sequenced as part of the Human Genome Project. Information about these clones can be viewed at the web site of the Sanger Centre, http://www.sanger.ac.uk/HGP/Chr1/. The sequences of the 88L2 and the 7H11 clones were also deposited to GenBank under accession numbers AL035091 and AL035209, respectively. A BLAST homology search showed that the 3′ end of PAC-7H11 contains exons 11-14 of theCD46 gene, which is in opposite transcriptional orientation to the CD34 gene.
Restriction map of the human CD34 locus and PAC clones.
(A) The top diagram depicts the human CD34 genomic locus. Numbering is relative to the transcription start site (TSS).22 Black lines represent the 8 exons. Shown above are the restriction sites mapped in panel B. Arrowheads indicate the location of the 5′ and 3′ primer sets used to screen the PAC library (the 3′ primer set was used for genotyping). Shown below are diagrams of the inserts of the 3 human CD34 PAC clones; the horizontal arrows refer to the transcription start site. Vertical arrow represents the location of theSfiI restriction site used to linearize PAC-88L2 prior to generation of transgenic mice. Also shown is the relative location of exons 11-14 of the CD46 gene, which is included in the 3′ end of PAC-7H11 in opposite transcriptional orientation to theCD34 gene. (B) A representative example of field inversion gel electrophoresis (FIGE) which was employed to map 1 μg DNA from each PAC clone. Migration of Lambda HindIII and MidRange I markers (NEB, Beverly, MA) are shown on the left. The lane marked “8-48 kb” shows markers ranging from 8 kb to 48 kb (BioRad, Hercules, CA).
Restriction map of the human CD34 locus and PAC clones.
(A) The top diagram depicts the human CD34 genomic locus. Numbering is relative to the transcription start site (TSS).22 Black lines represent the 8 exons. Shown above are the restriction sites mapped in panel B. Arrowheads indicate the location of the 5′ and 3′ primer sets used to screen the PAC library (the 3′ primer set was used for genotyping). Shown below are diagrams of the inserts of the 3 human CD34 PAC clones; the horizontal arrows refer to the transcription start site. Vertical arrow represents the location of theSfiI restriction site used to linearize PAC-88L2 prior to generation of transgenic mice. Also shown is the relative location of exons 11-14 of the CD46 gene, which is included in the 3′ end of PAC-7H11 in opposite transcriptional orientation to theCD34 gene. (B) A representative example of field inversion gel electrophoresis (FIGE) which was employed to map 1 μg DNA from each PAC clone. Migration of Lambda HindIII and MidRange I markers (NEB, Beverly, MA) are shown on the left. The lane marked “8-48 kb” shows markers ranging from 8 kb to 48 kb (BioRad, Hercules, CA).
A 160-kb human CD34 PAC clone contains regulatory element(s) sufficient to express human CD34 in stable cell lines and transgenic mice
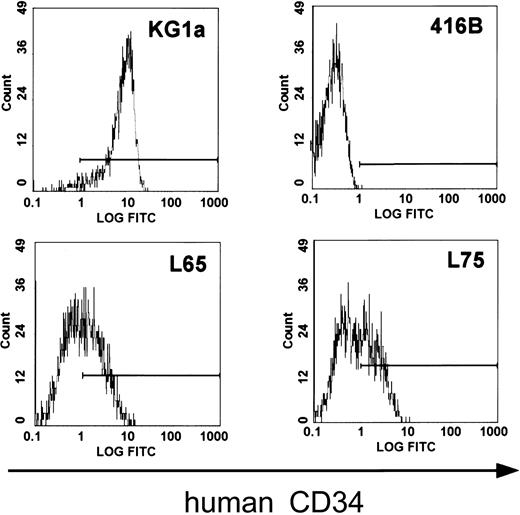
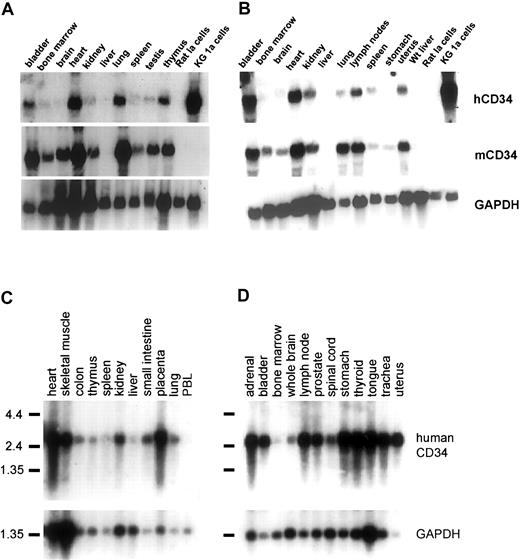
In order to determine whether all critical regulatory elements of the human CD34 gene are contained in clone 88L2, we generated stable lines in murine CD34+ progenitor 416B cells,23 and demonstrated that this large fragment contains necessary regulatory elements for high-level CD34 mRNA expression.21 In addition, as shown in Figure2, multiple stable lines expressed human CD34 protein on the cell surface. These results suggested that the critical control elements necessary for mRNA and protein expression are indeed located a large distance away from the CD34 gene. To test whether these elements are responsible for tissue- and cell type–specific CD34 gene expression in vivo, we injected purified PAC DNA and obtained 4 transgenic founder animals, from which 3 lines were established. The remaining fourth animal did not have germ line transmission. The copy number was either single (lines 8 and 14) or 2 copies (line 29). Northern blot analysis of mRNA from F1 animals containing a single copy of the transgene demonstrated detectable levels of human CD34 mRNA in almost all of the tissues examined (Figure3). Highest levels of expression were found in the bladder, heart, lymph nodes, and lung, whereas the lowest level among the tissues examined was seen in the liver, consistent with a previously published report.43 The pattern of expression of human CD34 transgenic RNA resembled that of the endogenous murine gene, with less than 2-fold differences as determined by quantitation of the specific RNAs, indicating that both genes were expressed in a similar fashion.
Murine progenitor 416B cells transfected with PAC 88L2 express the human CD34 protein on the cell surface.
Fluorescence intensity is displayed on the x-axis. The horizontal line indicates fluorescence above the intensity of isotype control. Human KG1a cells express the CD34 protein and served as a positive control (top left), and untransfected 416B cells served as a negative control (top right). The staining pattern for human CD34 of 2 representative clones of 416B cells (L65, bottom left; L75, bottom right) transfected with 88L2 is shown below.
Murine progenitor 416B cells transfected with PAC 88L2 express the human CD34 protein on the cell surface.
Fluorescence intensity is displayed on the x-axis. The horizontal line indicates fluorescence above the intensity of isotype control. Human KG1a cells express the CD34 protein and served as a positive control (top left), and untransfected 416B cells served as a negative control (top right). The staining pattern for human CD34 of 2 representative clones of 416B cells (L65, bottom left; L75, bottom right) transfected with 88L2 is shown below.
Coordinate expression of human and murine CD34 RNA in human CD34 transgenic mice and human tissues.
(A-B) Northern blot analysis of human transgenic and endogenous murine CD34 in tissues of mice from 2 different founder lines. (A) Line 14. (B) Line 29. A quantity of 20 μg total RNA was loaded for each lane and hybridized with a 0.5-kb probe from the 3′ enhancer of the humanCD34 gene,21,40 stripped and rehybridized to a 1.26-kb fragment generated by digestion of murine CD34 cDNA7 with PstI and XhoI, and then finally to a GAPDH probe to control for relative loading. Rat 1a cells served as a negative control, and human KG1a cells served as a positive control for human CD34.39 The human and murine CD34 probes do not cross-hybridize with RNA from the other species under stringent conditions.21 Bladder, heart, lymph node, and lung show the highest levels of expression for both genes. (C) The human CD34 and GAPDH probes were successively hybridized to a human tissue mRNA blot (Clontech).
Coordinate expression of human and murine CD34 RNA in human CD34 transgenic mice and human tissues.
(A-B) Northern blot analysis of human transgenic and endogenous murine CD34 in tissues of mice from 2 different founder lines. (A) Line 14. (B) Line 29. A quantity of 20 μg total RNA was loaded for each lane and hybridized with a 0.5-kb probe from the 3′ enhancer of the humanCD34 gene,21,40 stripped and rehybridized to a 1.26-kb fragment generated by digestion of murine CD34 cDNA7 with PstI and XhoI, and then finally to a GAPDH probe to control for relative loading. Rat 1a cells served as a negative control, and human KG1a cells served as a positive control for human CD34.39 The human and murine CD34 probes do not cross-hybridize with RNA from the other species under stringent conditions.21 Bladder, heart, lymph node, and lung show the highest levels of expression for both genes. (C) The human CD34 and GAPDH probes were successively hybridized to a human tissue mRNA blot (Clontech).
In order to compare the pattern of human CD34transgene expression with that of the human CD34 gene in human tissues, we determined the mRNA expression in multiple human organs (Figure 3B). In each instance, human CD34 RNA was expressed at high levels in the bladder, heart, lymph nodes, and lung, whereas the lowest expression was seen in the liver. Therefore, on a whole-tissue level, the expression of the transgene is not only similar to that of the endogenous murine gene, but moreover, resembles that of human tissues.
Human CD34 is expressed in vascular endothelial cells
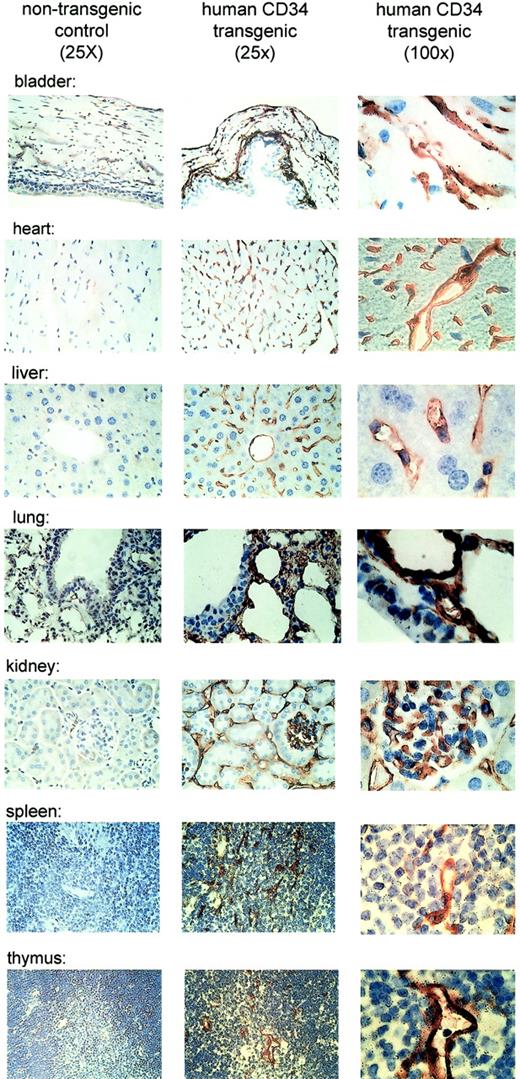
Several studies have investigated the expression of murine as well as human CD34 in a variety of tissues by immunohistochemistry.3,4,7 44 In those studies, the expression of the protein was found to be predominantly in vascular endothelial cells. In order to determine if the expression of the humanCD34 transgene follows a similar pattern, we performed immunohistochemistry with a monoclonal antibody specific for humanCD34. Again, expression of the transgene was consistent with previous studies for the murine and the human gene. We observed expression of human CD34 (Figure4) in vascular endothelial cells of the tissues.
Human CD34 is expressed in endothelial cells of tissues of transgenic mice.
Expression of human CD34 was detected by immunohistochemistry using a monoclonal antibody against human CD34. The left panels demonstrate lack of staining in tissues from a nontransgenic littermate as a control (original magnification, × 25). The middle panels demonstrate expression of human CD34 protein in transgenic mice at low magnification (original magnification, × 25). The right panels demonstrate expression of the protein in vascular endothelial cells at high magnification (original magnification, × 100).
Human CD34 is expressed in endothelial cells of tissues of transgenic mice.
Expression of human CD34 was detected by immunohistochemistry using a monoclonal antibody against human CD34. The left panels demonstrate lack of staining in tissues from a nontransgenic littermate as a control (original magnification, × 25). The middle panels demonstrate expression of human CD34 protein in transgenic mice at low magnification (original magnification, × 25). The right panels demonstrate expression of the protein in vascular endothelial cells at high magnification (original magnification, × 100).
Expression of the transgene in the bone marrow decreases with cell maturation
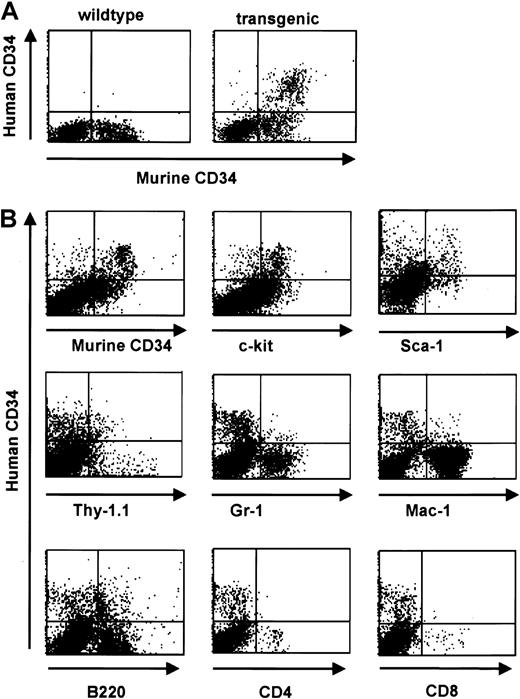
Consistent with a relatively low level of RNA expression in total bone marrow (Figure 3), FACS analysis revealed protein expression of the human CD34 transgene in 4% to 5% of total bone marrow cells of transgenic animals. This pattern was consistently seen in all 3 founder lines. The percentage of cells staining positive for the endogenous gene was slightly higher (8%-9% of total bone marrow cells) (Figure 5). We identified 3 populations of CD34+ cells: hCD34+/mCD34−, hCD34+/mCD34+, and hCD34−/mCD34+. In order to examine the expression of human CD34 in committed and mature cells we performed 2-color staining for surface markers defining mature hematopoietic cells. We found that expression of the exogenous human CD34gene decreased with cell maturation in concordance with published studies of the expression of the endogenous murine CD34 gene during ongoing cell maturation,1,2,8,45-47 whereas double-positive populations were found when staining for Sca-1 and c-kit, markers found on cells corresponding to a more immature phenotype.48,49 A notable exception was the coexpression with the B-cell–specific marker B220 on a subset of cells. This finding likely represents the normal expression pattern of CD34 in pro-B cells as determined by coexpression with CD43, CD10, and CD19,15 in concordance with the expression of human CD34 in an early B-cell progenitor subset.50 A small number of cells were found to coexpress the transgene and Mac-1, consistent with results obtained by transplantation assays demonstrating that CD34+/Mac-1hi as well as CD34+/Mac-1lo cells were capable of long-term reconstitution.51 In parallel studies, we were able to demonstrate that the human CD34 transgene is expressed on cells capable of competitive reconstitution.15 Therefore, we conclude that the human CD34 transgene is expressed on stem cells and multilineage progenitor cells.
Human CD34 is coexpressed with early hematopoietic surface markers in bone marrow and down-regulated with differentiation.
(A) Flow cytometric analysis of human and murine CD34 staining of bone marrow cells from a transgenic mouse (right panel) and a wild-type littermate (left panel). (B) Flow cytometric analysis of bone marrow cells from human CD34 transgenic animals demonstrates a decrease in transgene expression with differentiation. Coexpression of lineage-specific cell surface markers was performed to define the expression pattern with cell maturation. Double-positive populations were detected when costaining for markers found on cells corresponding to a more immature phenotype (Sca-1, c-Kit, and Thy1) whereas a decrease in double-staining cells was detected when costaining for markers defining more mature phenotypes (myeloid markers Gr-1 and Mac-1, and T-cell markers CD4 and CD8). A significant percentage of human CD34+ cells are Thy1 positive; murine stem cells have a Thy1lo phenotype.48 A small percentage of the human CD34+ cells express Mac-1, which has also been detected on murine stem and progenitor cells,59-61 and the pan-B cell marker B220, which is expressed on pro-B cells62 and has also been detected on murine CD34+ cells.47
Human CD34 is coexpressed with early hematopoietic surface markers in bone marrow and down-regulated with differentiation.
(A) Flow cytometric analysis of human and murine CD34 staining of bone marrow cells from a transgenic mouse (right panel) and a wild-type littermate (left panel). (B) Flow cytometric analysis of bone marrow cells from human CD34 transgenic animals demonstrates a decrease in transgene expression with differentiation. Coexpression of lineage-specific cell surface markers was performed to define the expression pattern with cell maturation. Double-positive populations were detected when costaining for markers found on cells corresponding to a more immature phenotype (Sca-1, c-Kit, and Thy1) whereas a decrease in double-staining cells was detected when costaining for markers defining more mature phenotypes (myeloid markers Gr-1 and Mac-1, and T-cell markers CD4 and CD8). A significant percentage of human CD34+ cells are Thy1 positive; murine stem cells have a Thy1lo phenotype.48 A small percentage of the human CD34+ cells express Mac-1, which has also been detected on murine stem and progenitor cells,59-61 and the pan-B cell marker B220, which is expressed on pro-B cells62 and has also been detected on murine CD34+ cells.47
The human CD34 transgene is expressed on multipotential progenitor cells in the bone marrow
We had identified 3 CD34+ cell populations within the transgenic bone marrow by FACS analysis: hCD34+/mCD34−, hCD34+/mCD34+, and hCD34−/mCD34+. We evaluated the in vitro differentiation potential within these populations by methylcellulose asssay (Table 1). The highest total number of total colonies (colony-forming units [CFU-Cs]) arose from the hCD34+/mCD34+ population, among which 14% were formed by multipotent myeloid progenitor cells (CFU–granulocytes, erythrocytes, monocyte/macrophages, and megakaryocytes [GEMMs]). The hCD34+/mCD34−cell population gave rise to a small number of colonies, among which a proportionately higher number of CFU-GEMMs were found as compared with the hCD34+/mCD34+ population, suggesting that these colonies may be derived from mCD34− stem cells.9 hCD34−/mCD34+ cells gave rise to colonies at a very low frequency and hCD34−/mCD34− cells gave rise to no colonies. We conclude that the human CD34 transgene is expressed on early multipotent progenitor or stem cells within the murine bone marrow.
Colony assay for each cell population
| Cell population . | CFU-GEMM . | CFU-C . |
|---|---|---|
| hCD34+mCD34− | 16.0 ± 8.0 | 53.3 ± 8.3 |
| hCD34+ mCD34+ | 306.7 ± 23.1 | 2120.0 ± 144.2 |
| hCD34−mCD34+ | 0.0 ± 0.0 | 256.7 ± 15.3 |
| hCD34− mCD34− | 0.0 ± 0.0 | 0.3 ± 0.5 |
| Cell population . | CFU-GEMM . | CFU-C . |
|---|---|---|
| hCD34+mCD34− | 16.0 ± 8.0 | 53.3 ± 8.3 |
| hCD34+ mCD34+ | 306.7 ± 23.1 | 2120.0 ± 144.2 |
| hCD34−mCD34+ | 0.0 ± 0.0 | 256.7 ± 15.3 |
| hCD34− mCD34− | 0.0 ± 0.0 | 0.3 ± 0.5 |
Colony numbers are from 10 000 cells of each cell population. Experiments were done in triplicate and figures are given as the mean ± SEM.
CD34 long-range control elements can direct the expression of heterologous genes in transgenic mice
To test if the elements enclosed within the 160-kb sequence are sufficient for driving the expression of heterologous genes in stem cells and early progenitor cells, we modified clone 88L2 by inserting the sequence of the transactivator gene tTA (part of the tetracycline-responsive expression system)26 downstream from the promoter of hCD34. An IRES was added at the 3′ end of the tTA gene followed by hCD34 sequences beginning with exon 1, so that this construct could potentially express both tTA and human CD34 proteins (Figure 6).
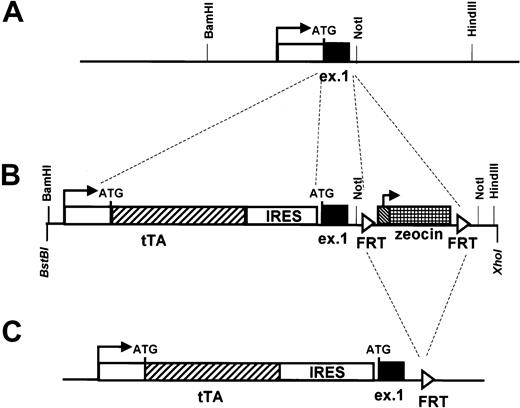
Modification of the human CD34 transgene by homologous recombination in bacteria to allow expression of the tetracycline transactivator (tTa).
The original PAC clone 88L2 (A) was modified to allow for the expression of the transactivator protein in tissues targeted by regulatory elements of the human CD34 locus. Shown is the transcription start site (horizontal arrow) and exon 1 (“ex 1”) with the location of the ATG translational start. The white box indicates the 5′ untranslated region, and the black box indicates the 27 amino acids of the human CD34 protein encoded by exon 1. Also shown are flankingBamHI, NotI, and HindIII sites located 1075 bp upstream and 360 bp and 2.7 kb downstream, respectively, from the transcription start site. The 1026-bp tTAgene26 was inserted directly at the promoter of the humanCD34 gene, such that the first ATG codon of theCD34 gene became the first codon of tTA, using the recombination cassette shown in panel B. An IRES element inserted downstream of tTA was followed by the sequence for humanCD34. A fragment containing FRT recombinase sites27 flanking the zeocin antibiotic resistance gene was inserted into human CD34 intron 1 and served as a selectable marker for homologous recombination. FRT recombinase was then used to delete the zeocin gene in the final transgene, shown in panel C.
Modification of the human CD34 transgene by homologous recombination in bacteria to allow expression of the tetracycline transactivator (tTa).
The original PAC clone 88L2 (A) was modified to allow for the expression of the transactivator protein in tissues targeted by regulatory elements of the human CD34 locus. Shown is the transcription start site (horizontal arrow) and exon 1 (“ex 1”) with the location of the ATG translational start. The white box indicates the 5′ untranslated region, and the black box indicates the 27 amino acids of the human CD34 protein encoded by exon 1. Also shown are flankingBamHI, NotI, and HindIII sites located 1075 bp upstream and 360 bp and 2.7 kb downstream, respectively, from the transcription start site. The 1026-bp tTAgene26 was inserted directly at the promoter of the humanCD34 gene, such that the first ATG codon of theCD34 gene became the first codon of tTA, using the recombination cassette shown in panel B. An IRES element inserted downstream of tTA was followed by the sequence for humanCD34. A fragment containing FRT recombinase sites27 flanking the zeocin antibiotic resistance gene was inserted into human CD34 intron 1 and served as a selectable marker for homologous recombination. FRT recombinase was then used to delete the zeocin gene in the final transgene, shown in panel C.
To determine whether the homologous recombination protocol had introduced undesirable rearrangements, bidirectional Southern blot analysis of DNA isolated from PAC CD34tTA digested with 6 different restriction enzymes was performed. The 2 blots were hybridized to either the original PAC-88L2 or the modified PAC-CD34tTA DNA as probes. Both hybridization patterns were identical and demonstrated no aberrant fragments (data not shown). The predicted structure of the transgene after homologous recombination is shown in Figure 6C.
The 160-kb expression cassette drives the expression of a heterologous transgene in a manner similar to that of human CD34 in vivo
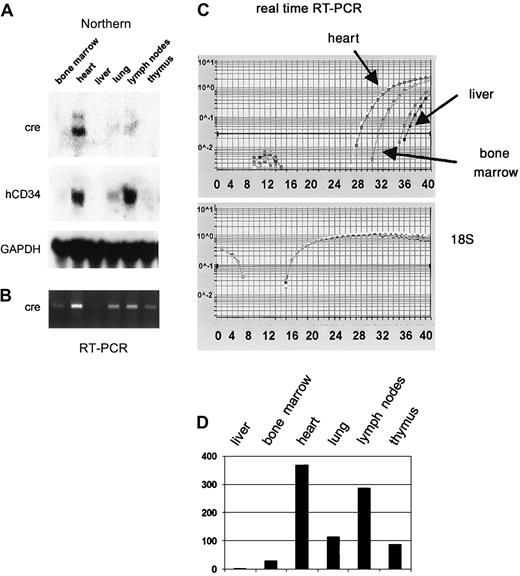
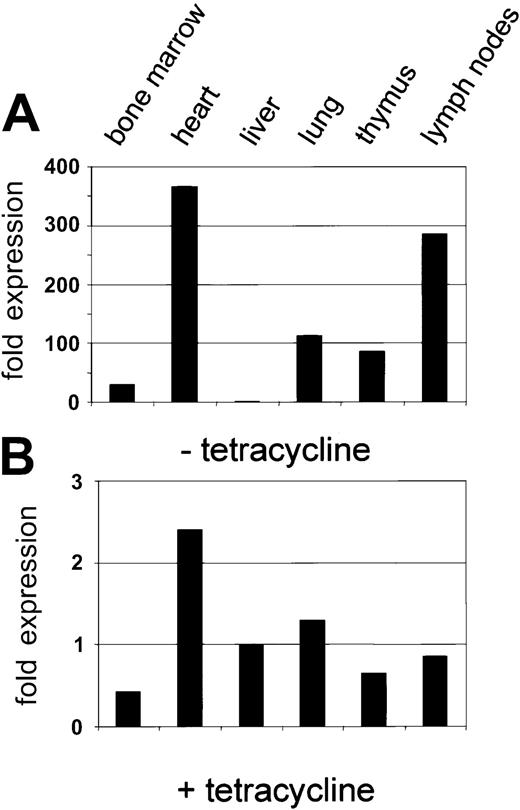
Injection of the purified PAC-CD34tTA DNA resulted in one transgenic founder animal, from which a line was established. In order to determine if a functional transactivator protein is expressed and to analyze the expression pattern, CD34tTA mice were cross-bred with a second line of mice carrying a transgene consisting of thecre gene under the control of the tetracycline-responsive element Tet-O-cre. In this line of mice, the cre gene was used as an indicator to check for expression of a functional transactivator protein. In addition, the double-transgenic animals (CD34tTA-Tet-O-cre) will be useful for further studies aimed at CD34-specific gene disruption. Within this system, expression of a functional transactivator protein in double-transgenic animals would be predicted to activate expression of the cre reporter gene in tissues targeted by the transactivator construct within 5 days of induction, which is accomplished by withdrawal of tetracycline administration. In addition, expression of the transresponder gene (cre) would be predicted to be suppressed in the presence of the antibiotic, thus demonstrating inducible expression ofcre in double-transgenic animals. We have previously observed inducible expression of cre RNA in mice double transgenic for Tet-O-Cre and tTA under the control of the mouse mammary tumor virus promoter/enhancer (C.S.H. and D.G.T., unpublished data, August 2000). RNA was isolated from various tissues and Northern blot analysis was performed. As shown in Figure7A, expression of cre was detected in the heart, lung, and lymph nodes, in concordance with results obtained with hCD34 transgenic animals, in which these organs were shown to express high levels of the human CD34 reporter gene (Figure 3). In order to determine if cre is also expressed in bone marrow, liver, and thymus at levels undetectable by Northern blot analysis, we performed RT-PCR from various tissues of double-transgenic mice with primers specific for the cre gene. As shown in Figure 7B,cre expression was detected in all tissues analyzed, although the expression in the liver was extremely low. In order to compare levels of expression, we used quantitative real-time PCR, which was consistent with the data obtained with the conventional PCR approach. Highest levels were found in the heart, in which RNA levels were more than 300-fold higher than in liver. Bone marrow expression was 30-fold higher than liver (Figure 7C-D). In order to test if expression of the transgene can be suppressed by the administration of tetracycline, double-transgenic mice were given the antibiotic in the drinking water (0.5 g/L) for 5 days. The animals were euthanized and the RNA expression was determined and compared with the levels found in induced mice. There was a more than 100-fold suppression of expression in all tissues analyzed (Figure 8). We conclude from these results that the 160-kb PAC fragment does indeed contain all elements necessary for the expression of heterologous genes in the bone marrow and other tissues similar to the pattern of the human CD34 gene.
The cre gene is expressed in CD34-tTA/Tet-O-cre double-transgenic mice in a manner similar to that of human CD34.
(A) Northern blot analysis demonstrates that a functional transactivator protein is expressed in the heart, lung, and lymph nodes of double-transgenic animals as indicated by the expression of cre RNA. The blot was stripped and rehybridized to human CD34, which is part of the transgenic construct (Figure 6) and demonstrates a similar expression pattern, and then to murine GAPDH. (B) Expression analysis with RT-PCR confirms expression of cre in bone marrow, liver, and thymus. (C) Multiplex real-time PCR with amplification of 18S RNA in the same tube as cre was performed for relative quantitation of expression levels among different tissues. The top graph shows the curves for amplification of the cre gene, whereas the bottom graph demonstrates equal amounts of cDNA in all samples as determined by amplification of 18S RNA. (D) Expression levels of cre RNA relative to 18S within different tissues calculated based on results shown in panel C. Expression in liver, which showed the lowest levels, was arbitrarily assigned to a value of 1.
The cre gene is expressed in CD34-tTA/Tet-O-cre double-transgenic mice in a manner similar to that of human CD34.
(A) Northern blot analysis demonstrates that a functional transactivator protein is expressed in the heart, lung, and lymph nodes of double-transgenic animals as indicated by the expression of cre RNA. The blot was stripped and rehybridized to human CD34, which is part of the transgenic construct (Figure 6) and demonstrates a similar expression pattern, and then to murine GAPDH. (B) Expression analysis with RT-PCR confirms expression of cre in bone marrow, liver, and thymus. (C) Multiplex real-time PCR with amplification of 18S RNA in the same tube as cre was performed for relative quantitation of expression levels among different tissues. The top graph shows the curves for amplification of the cre gene, whereas the bottom graph demonstrates equal amounts of cDNA in all samples as determined by amplification of 18S RNA. (D) Expression levels of cre RNA relative to 18S within different tissues calculated based on results shown in panel C. Expression in liver, which showed the lowest levels, was arbitrarily assigned to a value of 1.
Inducible expression of cre by human CD34 elements: suppression by tetracycline.
(A) Expression of the cre transgene in the tissue of a double-transgenic mouse (CD34tTA-Tet-O-cre) in the absence of tetracycline. The expression level for liver was used to quantitate relative expression in other tissues as described in the legend to Figure 7. (B) Suppression of cre expression in the presence of tetracycline. A double-transgenic mouse was kept on tetracycline for 5 days before it was killed. The level of Cre RNA in liver in the absence of tetracycline (Figure 7A) was used to quantitate relative cre RNA. Note that the relative scale is approximately 100-fold lower than that in Figure 7A.
Inducible expression of cre by human CD34 elements: suppression by tetracycline.
(A) Expression of the cre transgene in the tissue of a double-transgenic mouse (CD34tTA-Tet-O-cre) in the absence of tetracycline. The expression level for liver was used to quantitate relative expression in other tissues as described in the legend to Figure 7. (B) Suppression of cre expression in the presence of tetracycline. A double-transgenic mouse was kept on tetracycline for 5 days before it was killed. The level of Cre RNA in liver in the absence of tetracycline (Figure 7A) was used to quantitate relative cre RNA. Note that the relative scale is approximately 100-fold lower than that in Figure 7A.
Discussion
The goal of this study was to generate a construct that allows for the expression of heterologous genes in stem cells and early progenitor cells in transgenic mice. Such a construct will be an invaluable tool for a broad range of studies, including the establishment of models of leukemia in mice, which requires targeting of the transgene to stem and/or early progenitor cells. Furthermore, modification of the construct by insertion of the cre gene instead of thetTA gene will allow for cell type–specific recombination of genes in hematopoietic stem cells and early progenitor cells. We show here that regulatory elements contained within the largest of the PAC clones, 88L2, are indeed sufficient to serve this purpose. The RNA expression pattern of the exogenous human CD34 reporter gene, as well as of the tTA gene in the modified transgenic construct, are similar with respect to their tissue distribution and in this respect resemble the pattern of the endogenous murine CD34. The protein was found to be expressed in vascular endothelial cells by immunohistochemistry. FACS analysis demonstrated correct expression of the transgene on the cell surface of early progenitor cells, which decreases with maturation of the cells. It is of interest that 3 subpopulations of cells expressing CD34 were identified in the murine bone marrow. The majority of these cells were found to be positive for human as well as for murine CD34, whereas a smaller population of cells stained solely for either the endogenous murine CD34 or the exogenous human molecule. Some studies have supported the hypothesis that hematopoietic stem cells in the mouse express the murine CD34 molecule.8 However, more recent studies have suggested that hematopoietic stem cells of adult mice are CD34−.9,11-14,52,53 We have attempted to define these populations with respect to their ability to form multipotential progenitor colonies in vitro. The vast majority of total colonies grew out of the hCD34+/mCD34+population, whereas hCD34−/mCD34+ cells gave rise to a significantly lower number. However, multipotent myeloid progenitor colonies (CFU-GEMMs) grew out only from the hCD34+ populations, and were proportionately higher in the hCD34+/mCD34− population, whereas none were observed arising from hCD34−/mCD34+ cells. This result indicates that cells with the capacity for self-renewal and differentiation into various lineages are contained within the 2 populations that express human CD34, suggestive of expression of the transgene on hematopoietic stem cells. We have tested and verified this hypothesis in experiments by transplanting cells from these 3 subpopulations in vivo.15 Thus, the 160-kb transgenic construct appeared to be a likely candidate for directing expression of heterologous genes in stem cells.
Furthermore, the human CD34 PAC appears to be a likely candidate for directing expression of heterologous genes to stem cells in a way that should ensure expression of the transgene similar to that of the humanCD34 gene, which is characterized by down-regulation of expression with ongoing cell differentiation. This is somewhat different from that of other constructs aimed at targeting stem cells, including the H-2K promoter,54,55 Sca-1 regulatory elements,56 and the vav promoter.57 The expression of transgenes under control of these constructs was not limited to the stem and progenitor cell compartment, but was found either on more mature hematopoietic cells and/or a broad variety of other cell types.56-58
To test the ability of the human CD34 elements to direct expression of a heterologous gene, we modified this 160-kb construct by inserting the sequence of the transactivator gene of the tetracycline-responsive expression system in a manner that allows for expression of tTA. The data presented here demonstrate that transgenic mice express the hybrid transgene RNA in the same tissues as the unmodified human CD34 construct. Furthermore, analysis of double-transgenic animals (CD34tTA-Tet-O-cre), which express the cre gene upon transactivation by tTA, clearly demonstrated that a functional tTA protein is expressed. We are currently conducting experiments aimed at the further delineation of important regulatory elements by performing analysis of deletion mutants of the 160-kb clone described herein (see the accompanying article by Okuno et al63 that describes analysis of human CD34 deletion mutants in transgenic mice, beginning on page 4420).
We thank Maris Fenyus for expert animal husbandry and genotyping; other labmates from the Tenen lab for their support; Joel Lawitts for production of transgenic mice; Bertie Göttgens and Tony Green for assistance with sequence comparisons; Keith Reimann for assistance with flow cytometry; Victoria Petkova for assistance with real-time PCR; and Mary Singleton and Alison Lugay for expert assistance with preparation of the manuscript.
Prepublished online as Blood First Edition Paper, August 15, 2002; DOI 10.1182/blood-2002-02-0355.
Supported by The Golden Family Foundation (H.S.R.), an international (JC 2000) fellowship from the Jose Carreras Leukemia Foundation (C.S.H.), and by National Institutes of Health grant DK48660 (D.G.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel G. Tenen, Harvard Institutes of Medicine, Room 954, 77 Ave Louis Pasteur, Boston, MA 02115; e-mail:dtenen@caregroup.harvard.edu, or Hanna Radomska, e-mail: hradomsk@caregroup.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal