Abstract
Factor XIII and fibrinogen are unusual among clotting factors in that neither is a serine protease. Fibrin is the main protein constituent of the blood clot, which is stabilized by factor XIIIa through an amide or isopeptide bond that ligates adjacent fibrin monomers. Many of the structural and functional features of factor XIII and fibrin(ogen) have been elucidated by protein and gene analysis, site-directed mutagenesis, and x-ray crystallography. However, some of the molecular aspects involved in the complex processes of insoluble fibrin formation in vivo and in vitro remain unresolved. The findings of a relationship between fibrinogen, factor XIII, and cardiovascular or other thrombotic disorders have focused much attention on these 2 proteins. Of particular interest are associations between common variations in the genes of factor XIII and altered risk profiles for thrombosis. Although there is much debate regarding these observations, the implications for our understanding of clot formation and therapeutic intervention may be of major importance. In this review, we have summarized recent findings on the structure and function of factor XIII. This is followed by a review of the effects of genetic polymorphisms on protein structure/function and their relationship to disease.
Factor XIII structure and function
Overall structure of factor XIII
Plasma factor XIII is a tetrameric molecule composed of 2 A-subunits of 83.2 kd and 2 B-subunits of 79.7 kd that are held together noncovalently in a heterologous tetramer of 325.8 kd.1-3 In addition, 50% of the total fibrin-stabilizing activity in blood is found in the platelet where factor XIII exists as a dimeric molecule composed of only A-subunits.4 The A-subunit contains the active site of the enzyme and is synthesized by hepatocytes, monocytes, and megakaryocytes.5-8 Analysis of the protein phenotype after liver and bone marrow transplantation showed that the A-subunit circulating in plasma is derived from both liver and bone marrow.9 The B-subunit serves as a carrier for the catalytic A-subunit in plasma, is synthesized by the liver, and is secreted as a monomer that binds free A in plasma.8
The A-subunit is divided into 4 domains, designated the β-sandwich, the catalytic core, barrel 1, and barrel 2 (Figure1).10,11 It contains an activation peptide of 37 amino acids that limits the access of the substrate to the active-site cysteine. The activation peptide from one subunit of the molecule crosses the opening of the active site on the other. This structure is stabilized by several hydrogen bonds and salt bridges between the activation peptide, the β-sandwich, and the catalytic core of one subunit and the catalytic core and β-barrel of the second subunit.11
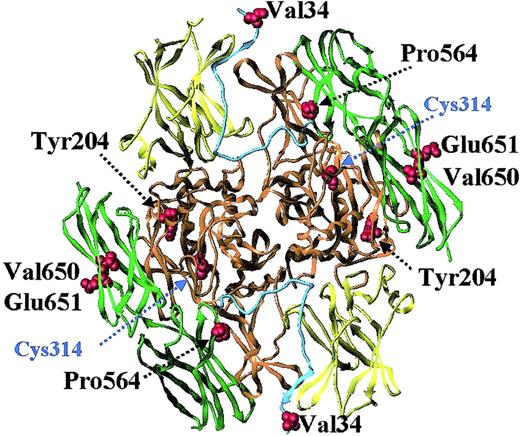
Factor XIII A-subunit structure and location of common coding polymorphisms.
The structure shown is that of recombinant factor A-subunit dimer, modeled with the use of x-ray crystallography coordinates from Weiss et al.11 The catalytic core region is colored orange; the beta-sandwich, yellow; the 2 beta-barrels, green; and the activation peptide, cyan. Highlighted are the active-site cysteine residue and 5 residues (Val34, Tyr204, Pro564, Val650, and Glu651) that show common variation in the general population (Table2).
Factor XIII A-subunit structure and location of common coding polymorphisms.
The structure shown is that of recombinant factor A-subunit dimer, modeled with the use of x-ray crystallography coordinates from Weiss et al.11 The catalytic core region is colored orange; the beta-sandwich, yellow; the 2 beta-barrels, green; and the activation peptide, cyan. Highlighted are the active-site cysteine residue and 5 residues (Val34, Tyr204, Pro564, Val650, and Glu651) that show common variation in the general population (Table2).
Recent x-ray crystallography studies of thrombin-treated factor XIII A-subunit have suggested that the activation peptides do not dissociate upon cleavage.12 Earlier studies described the kinetics of the release of the activation peptide by thrombin13 and proposed that this reaction was enhanced by fibrin. However, in these studies, the samples were acidified prior to the high-pressure liquid chromatography analysis, and the activation peptide release may have been a consequence of this acidification. Further studies are required to determine whether the activation peptide is actually released during activation and whether substrates can modulate this reaction.
Whereas the A-subunit contains 6 potential asparagine-linked glycosylation sites, none of these have carbohydrate attachments, as judged by staining with periodic acid Schiff base reagent14 or by mass spectrometry.3 In contrast, carbohydrate contributes to approximately 8.5% of the total molecular weight of the factor XIII B-subunit.15 The B-subunit is a modular protein composed of 10 repeated Sushi or glycoprotein-1 domains.16 17 Each Sushi domain contains 2 disulfide bridges that sustain its tertiary structure, amounting to a total of 40 cysteine residues and 20 disulfide bridges in the mature B-subunit protein. The main function of the B-subunit is the stabilization and transport of the hydrophobic A-subunit in the aqueous environment of human plasma.
Factor XIII activation
Thrombin cleavage of the A-subunits is necessary to activate the plasma tetramer (Figure 2) and dimeric platelet factor XIII.18 In addition to thrombin, activation by cleavage of the Arg37-Gly38 peptide bond may occur by other serine proteases; both endogenous platelet acid protease19 and calpain20 have been reported to activate factor XIII. Fibrin polymers are an important cofactor to generate factor XIIIa.13 21-25 A complex between thrombin, fibrin polymers, and plasma factor XIII accelerates cleavage of the A-subunit, which has important implications for hemostasis. Factor XIII is not activated until a critical mass of fibrin polymerizes, a delay that ensures the hemostatic plug has a supply of factor XIII as it forms. Plasma concentrations of factor XIII and fibrinogen are approximately 0.07 μM and 9 μM, respectively. Thus, the molar ratio of factor XIII to fibrinogen in plasma is in the order of 1:100. The generation of factor XIIIa in plasma can be triggered when as little as 1% to 2% of fibrinogen is converted to fibrin polymers. This indicates that factor XIIIa begins stabilizing fibrin polymers before a visible thrombus appears, as the latter requires the conversion of at least 20% of fibrinogen into fibrin.
Factor XIII tetrameric structure and activation.
Plasma factor XIII is a heterologous tetramer consisting of 2 A- and 2 B-subunits. The A-subunits contain the enzyme's active site, and the B-subunits serve a carrier function of the hydrophobic A-subunit in the aqueous environment of human plasma. Activation of factor XIII involves cleavage of the activation peptides from the A-subunit, which then may or may not dissociate from the complex. In a second step, calcium and fibrin induce the dissociation of the B-subunits from A to expose the active site's thiol group.
Factor XIII tetrameric structure and activation.
Plasma factor XIII is a heterologous tetramer consisting of 2 A- and 2 B-subunits. The A-subunits contain the enzyme's active site, and the B-subunits serve a carrier function of the hydrophobic A-subunit in the aqueous environment of human plasma. Activation of factor XIII involves cleavage of the activation peptides from the A-subunit, which then may or may not dissociate from the complex. In a second step, calcium and fibrin induce the dissociation of the B-subunits from A to expose the active site's thiol group.
Activation of platelet factor XIII by thrombin is very rapid.26 In contrast; there is a significant lag phase between thrombin cleavage and expression of the active site of plasma factor XIII. This lag phase in activation represents the time it takes for the B-subunits to dissociate from plasma factor XIIIa.25,27,28 Dissociation of the B- from the A-subunits is necessary to expose the active-site cysteine in plasma factor XIII A-subunit (Figure 2). There are no B-subunits bound to the fibrin clot, suggesting that B dissociates as fibrin gels.29 By contrast, more than 90% of the A-subunit remains bound to fibrin.
Localization of factor XIII to fibrin
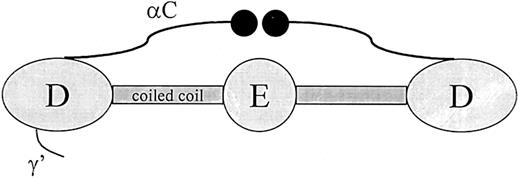
Thrombin-activated factor XIII (FXIIIa) binds fibrin through an interaction with the αC-domains. The αC-domain consists of residues Aα220 through Aα610, which protrude from the distal region (D-region) of fibrin and end in a globular domain (Figure3).30,31 In total, both αC domains contribute to around 25% of the mass of the protein. In fibrinogen, the αC-domain connecting polypeptides contribute a fourth strand to the coiled coil while the globular end structures are positioned near the central region (E-region). During fibrin formation, this conformation of the αC-domains changes drastically: the globular portions dissociate from the central region so that they are available for intermolecular interactions.31 The binding site for FXIIIa in the αC-domain has been localized to residues 241 through 476 with the use of a fragment produced by cyanogen bromide cleavage.32 Binding of FXIIIa to this region has been confirmed by inhibition of the interaction with an antibody directed against residues 389 through 402.32 In an earlier study, a similar fragment of the α-chain produced by plasmin digestion (residues 242-424) was found to regulate dissociation of the B-subunit from thrombin-cleaved plasma factor XIII.33 Combined, these studies suggest that FXIIIa binds to fibrin in the αC-domain–connecting polypeptide and that this interaction enhances dissociation of the factor XIII subunits.
Schematic representation of the fibrinogen molecule.
Fibrinogen consists of 6 polypeptide chains held together by disulfide bonds in a molecule with bilateral symmetry. Illustrated in the Figure are the central region (E), which contains the fibrinopeptides; the distal region (D); α-helical coiled-coil segments parts of which are included in both the D and E regions; αC-domain; and the γ′ segment, which contains a thrombin and factor XIII binding site.
Schematic representation of the fibrinogen molecule.
Fibrinogen consists of 6 polypeptide chains held together by disulfide bonds in a molecule with bilateral symmetry. Illustrated in the Figure are the central region (E), which contains the fibrinopeptides; the distal region (D); α-helical coiled-coil segments parts of which are included in both the D and E regions; αC-domain; and the γ′ segment, which contains a thrombin and factor XIII binding site.
Zymogen factor XIII binds fibrinogen in a different site than the active enzyme. This binding occurs through an interaction between the B-subunit and fibrinogen γ′ (Figure 3).34 Fibinogen γ′ is a product of alternative processing of the γ-chain transcript leading to a readthrough of the exon IX/intron I splice junction.35 The result of this alternative processing is a replacement of the last 4 amino acids at the carboxy terminus of the γ polypeptide with 20 different residues.35,36The γ′ extension is negatively charged and is found in about 10% of the fibrinogen in plasma. In addition to a binding site for factor XIII B-subunit, fibrinogen γ′ also contains a binding site for thrombin.37 Binding of factor XIII to γ′ may have important physiological consequences since an increase in γ′-chain concentration will increase the amount of factor XIII that is brought into the clot, and it has been postulated that alterations in the concentrations of the γ′-chain could be a potential risk factor for thrombosis.
Thrombin binding site
The reciprocal binding domains on factor XIII and thrombin are not well established. Serine proteases often use an exosite to orient the substrate and facilitate proteolysis. Recent studies provide evidence to suggest that exosites exist on both thrombin and factor XIII A-subunit that play a role in the binding and activation of factor XIII by thrombin.38 The B-subunits may sterically interfere with the cleavage of the A-subunit by thrombin since they can inhibit proteolysis of the activation peptide.25 28 It is possible that dissociation of the B-subunits by either soluble fibrinogen or fibrin complexes is involved in the exposure of the thrombin cleavage site on the A-subunits. One could hypothesize that as fibrin polymerizes, factor XIII attached to the γ′-chain extension on the D-region aligns with thrombin on the E-region to promote thrombin cleavage of factor XIII. Further studies are needed to define the structural domain(s) responsible for this well-controlled reaction.
Catalytic mechanism
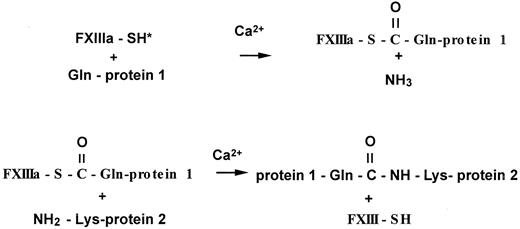
The enzymatic reaction catalyzed by factor XIIIa is classified as a transglutaminase reaction, and the enzyme is designated an R-glutaminyl-peptide: amine γ-glutamyltransferase (EC 2.3.2.13). A catalytic triad is formed by residues Cys314, His373, and Asp396 and participates in the formation of the isopeptide bond.10The first step in catalysis is the recognition of a select group of protein-bound glutamine residues,39 which is followed by formation of a thioester bond that releases ammonia from the glutamine (Figure 4).40 The second step of the reaction involves binding of the enzyme-glutamine substrate complex to a primary amine, which is either a protein-bound lysine, a polyamine, or another primary amine. The thioester intermediate is highly reactive, and there is rapid formation of the isopeptide bond (Figure 4).41 42 If there are no primary amines available in the active-site pocket, the enzyme-substrate complex will react with water, releasing the enzyme and converting glutamine to glutamic acid.
Cross-linking reaction catalyzed by activated factor XIII.
Activated factor XIII first forms a thioester bond with a select protein-bound glutamine residue, releasing ammonia. The thioester intermediate then reacts with a primary amine from a protein-bound lysine residue, a polyamine, or other primary amines, resulting in an amide or isopeptide bond.
Cross-linking reaction catalyzed by activated factor XIII.
Activated factor XIII first forms a thioester bond with a select protein-bound glutamine residue, releasing ammonia. The thioester intermediate then reacts with a primary amine from a protein-bound lysine residue, a polyamine, or other primary amines, resulting in an amide or isopeptide bond.
It has been shown that there are nonproline cis-bonds present in the molecule at residues Arg310-Tyr311 and Gln425-Phe426.11 On the basis of these findings, Weiss et al11 proposed that factor XIII can exist in 2 conformational states and that the conversion of these nonprolinecis-peptide bonds into the trans-configuration might provide the necessary conformational change to expose the active site for catalysis. Site-directed mutagenesis of either Arg310 or Tyr311 to Ala results in a mutant factor XIII molecule that is inactive,43 suggesting that productive catalysis cannot occur when this nonproline cis-peptide bond is disrupted. The importance of these bonds in the activation of factor XIII is a subject for future investigation.
The orientation of specific residues in the active site is beginning to be appreciated as more mutant forms of factor XIII are expressed and analyzed. Mutagenesis of residues on either side of the active-site cysteine results in substantial loss of the enzyme activity without altering the ability of factor XIII to recognize and bind fibrin.43 This suggests that the sites used for substrate binding are different or are less susceptible to disruption by the point mutations around the active-site cysteine (amino acid residues 310-317) than those required for catalytic activity.
Calcium and catalysis
Calcium ions are required for the activation of plasma factor XIII after thrombin cleavage.44,45 There is a calcium-dependent conformational change that causes the B-subunits to dissociate from thrombin-cleaved factor XIII. Calcium ions are also required for the first step in catalysis. The close proximity of the catalytic triad to the calcium-binding site suggests that calcium ions regulate conformational changes that accelerate catalysis.46 The catalytic mechanism uses calcium ions as a cofactor to hold the active site in proper conformation to trigger formation of the thioester intermediate with glutamine.
Platelet factor XIII A-subunit is rapidly activated at plasma calcium concentrations (2.5 mM). In contrast, calcium concentrations exceeding 10 mM are required for full expression of plasma factor XIII activity.44 45 This concentration is significantly higher than the one that exists in plasma and suggests that another cofactor regulates activation of plasma factor XIII. Indeed, fibrinogen reduces the calcium concentration required for dissociation of the B-subunits and facilitates factor XIII activation.
Nitric oxide regulation of factor XIII activity
A recent report by Catani et al47 demonstrates that nitric oxide donors can inhibit factor XIII activity by S-nitrosylation of the active-site cysteine. Regulation of factor XIII activity by nitric oxide at sites of vascular injury could influence clot formation and may provide a regulatory mechanism for inhibiting fibrin stabilization and enhancing fibrin degradation.
Factors regulating substrate specificity
The mechanism by which transglutaminases recognize their protein substrates remains unknown. Using recombinant chimeras of factor XIII and tissue transglutaminase, Hettasch et al48 reported that some of the properties required for the transglutaminases to recognize macromolecular substrates reside in the residues within the exon defining the active site of the molecule. This conclusion was based on the finding that exchanging the exon coding for the active site of factor XIII with the exon coding for the active site of tissue transglutaminase produced a recombinant transglutaminase that cross-linked fibrin in a pattern more characteristic of the tissue transglutaminase than of factor XIII. However, the efficiency of the cross-linking reaction was lower than that of either wild- type enzyme, indicating that regions outside the residues defined by exon 7 must also be important for macromolecular substrate recognition.
It is generally accepted that the second half of the cross-linking reaction with primary amines is not very specific. However, if one examines the enzyme's overall structure, there is some steric hindrance and constraints are placed on protein-bound lysine residues. The residue that precedes the donor lysine modulates the recognition of this lysine as a cross-linking site.49Even though studies with small peptide-bound glutamines, small peptide-bound lysines, and point mutations in recombinant factor XIII have provided information about the influence of single residues in modulating the cross-linking reaction, the macromolecular interactions that allow 3 large proteins to associate to produce intermolecular ε–(γ-glutamyl)lysine bonds remain to be defined.
The process of fibrin polymerization enhances cross-linking by aligning the molecules. In the presence of the tetrapeptide Gly-Pro-Arg-Pro, which inhibits fibrin polymerization, the glutamine sites in the carboxy terminal tail of the γ-chain are no longer aligned for rapid cross-linking by factor XIIIa.50 Further evidence that end-to-end alignment of the D-domains of fibrin plays an important role in the cross-linking reaction was provided by experiments performed by Lorand et al.51 The authors found that a double-headed ligand, produced from 2 Gly-Pro-Arg-Pro peptides linked to each end of a long polyethylene glycol molecule, could mimic the E-region in joining 2 D-regions and allow for γ-chain cross-linking to occur.51 In the absence of the double-headed ligand (or the E-region), no cross-linking takes place between 2 D-regions.
Substrates of factor XIIIa and biological consequences of cross-linking
Fibrin γ-chain
Activated FXIII introduces a number of cross-links in the fibrin clot, of which the first are formed between γ-chains of 2 neighboring fibrin molecules in the longitudinal orientation of the (proto)fibril.52,53 Cross-linking occurs within 5 to 10 minutes between Gln398 or 399 on the γ-chain of one fibrin molecule and Lys406 on the γ-chain of another,52 54 resulting in the formation of 2 antiparallel isopeptide bonds that connect the D-regions of 2 fibrinogen molecules longitudinally.
There has been some controversy regarding the spatial orientation of γ-chain cross-links. Some studies suggested that the isopeptide bond was formed between 2 fibrin molecules aligned end to end in the same strand of the protofibril,53,55 while others suggested that the γ-γ cross-link was oriented between 2 fibrin molecules aligned across the fibrin protofibril in atrans-orientation.56,57 Recent crystallography data of the cross-linked D-fragment indicate that the predominant orientation is the cis-configuration.52
Fibrin α-chain
Cross-linking of the fibrin α-chains occurs more slowly than cross-linking of the γ-chains. A number of residues have been reported to be involved in fibrin α-chain cross-linking. Studies of the incorporation of primary amines such as fluorescent dansylcadaverine have shown that glutamine residues involved in the cross-linking reaction of the α-chain include Gln221, Gln237, Gln328, and Gln366.58-60 Many lysine residues that potentially function as acceptor sites for the transglutaminase reaction have been reported, including Lys208, Lys219, Lys224, Lys418, Lys427, Lys429, Lys446, Lys448, Lys508, Lys539, Lys556, Lys580, Lys583, Lys601, and Lys606.61 62 The identification of lysine acceptor sites has been based mainly on incorporation studies of different glutamine-containing peptides such as dansyl-ε-aminocaproyl-Ala-Gln-Gln-Ile-Val, and the sites that are involved in α-chain cross-linking in vivo are not clear. The multiplicity of potential cross-linking sites provides the possibility for a highly complex and intricate cross-linking network to be formed between neighboring αC-domains in the fibrin clot.
The extent of α-chain cross-linking plays an important role in the regulation of fibrinolysis63,64 and viscoelastic properties of fibrin. Most investigators agree that α-chain cross-links appear to stabilize and promote the association of fibrin protofibrils into thick bundles of opaque fibers with higher tensile strength. The exact orientation of the α-chain cross-links in relationship to the fibrin network is, however, not thoroughly established. The α-chain cross-linked network appears to form a protective barrier that impedes the ability of plasmin to degrade the coiled-coil regions of the fibrin molecule maintaining clot structure. Plasmin must degrade fibrin in the coiled coil between the D- and E-regions to allow fibrin to release soluble fragments and lose its structure, and the extensive α-chain cross-linking interferes with this reaction.65 66
α2-Antiplasmin
A plasma glycoprotein that is the major physiological inhibitor of plasmin in vivo is α2-antiplasmin. Factor XIIIa rapidly cross-links α2-antiplasmin to the α-chain of fibrin.67,68 This cross-linking reaction occurs between Gln2 in the amino terminus of α2-antiplasmin69 and Lys303 in the fibrin α-chain.68 The α2-antiplasmin remains an efficient plasmin inhibitor when covalently cross-linked to fibrin, and incorporation of the inhibitor into the clot by factor XIII plays a major role in the regulation of the breakdown of fibrin.70 71
Fibronectin
Both the cellular and plasma forms of fibronectin are factor XIII substrates. Fibronectin can be cross-linked to both itself and collagen through Gln3 at the amino-terminal end of the molecule.72However, when fibrin is present, fibronectin-fibrin complexes are the preferred cross-linking products.73 Fibronectin is cross-linked to the α-chain of the fibrin molecule. Fibronectin can inhibit fibrin cross-linking and lead to the formation of soluble fibrin.74-76 Cross-linking of fibronectin to fibrin can alter the mechanical properties76-78 of the clot and promote cellular adherence as well as migration of cells into the clot.79 This may be important to facilitate the wound healing process.
Collagen
During vascular injury, fibrin clots may be covalently attached to collagen in the vessel wall by factor XIIIa, a reaction that may prevent the clot from being dislodged from the vessel wall. Collagen types I, II, III, and V can be cross-linked to fibronectin by factor XIIIa.80 Collagen provides the lysine residues necessary to form an isopeptide bond with Gln3 in fibronectin. The cross-linking of both fibronectin and collagen to fibrin suggests that these reactions could stabilize the extracellular matrix that forms at sites of tissue injury.
Other factor XIIIa substrates
Many other proteins are cross-linked by factor XIIIa. These substrates, which include inhibitors of fibrinolysis, von Willebrand factor, factor V, and platelet (glyco)proteins, are summarized in Table1, together with the potential physiological significance of their cross-linking.
Factor XIII substrates
| Substrate . | Cross-linking site . | Substances with which it is cross-linked . | Known or potential function . |
|---|---|---|---|
| Fibrin(ogen) γ-chain52-54 | Gln398, Gln399, and Lys406 | Itself and α-chain | Clot stabilization |
| Fibrin(ogen) α-chain58-62 | Gln221, Gln237, Gln328, Gln366, and 15 potential lysines from Lys208 to Lys606 | Itself and γ-chain | Clot stabilization |
| α2-Antiplasmin67-69 | Gln2 | Lys303 fibrin α-chain | Resistance to fibrinolysis |
| TAFI150 | Gln2, Gln5, Gln292 | Fibrin, itself | Resistance to fibrinolysis |
| PAI-2151 152 | — | Lys148, Lys230, Lys413 fibrin α-chain | Resistance to fibrinolysis |
| Fibronectin72 73 | Gln3 | Itself, fibrin, collagen | Migration of cells into the clot; wound healing |
| Collagen72 80 | — | Fibronectin, fibrin | Stabilization of extracellular matrix |
| Von Willebrand factor153 154 | — | Fibrin, collagen | Platelet adhesion to the clot |
| Vitronectin155 156 | Gln93 | — | — |
| Thrombospondin157 | — | Fibrin | — |
| Factor V158 159 | — | Fibrin, platelets | Increased thrombin generation at the clot surface |
| Actin160 161 | — | Fibrin | Clot retraction, stabilization of the platelet cytoskeleton |
| Myosin162 | — | Itself | Clot retraction, stabilization of the platelet cytoskeleton |
| Vinculin163 | — | Fibrin | Clot retraction, stabilization of the platelet cytoskeleton |
| αIIbβ3164 | — | Fibrin | Stabilization of the platelet—fibrin clot |
| Substrate . | Cross-linking site . | Substances with which it is cross-linked . | Known or potential function . |
|---|---|---|---|
| Fibrin(ogen) γ-chain52-54 | Gln398, Gln399, and Lys406 | Itself and α-chain | Clot stabilization |
| Fibrin(ogen) α-chain58-62 | Gln221, Gln237, Gln328, Gln366, and 15 potential lysines from Lys208 to Lys606 | Itself and γ-chain | Clot stabilization |
| α2-Antiplasmin67-69 | Gln2 | Lys303 fibrin α-chain | Resistance to fibrinolysis |
| TAFI150 | Gln2, Gln5, Gln292 | Fibrin, itself | Resistance to fibrinolysis |
| PAI-2151 152 | — | Lys148, Lys230, Lys413 fibrin α-chain | Resistance to fibrinolysis |
| Fibronectin72 73 | Gln3 | Itself, fibrin, collagen | Migration of cells into the clot; wound healing |
| Collagen72 80 | — | Fibronectin, fibrin | Stabilization of extracellular matrix |
| Von Willebrand factor153 154 | — | Fibrin, collagen | Platelet adhesion to the clot |
| Vitronectin155 156 | Gln93 | — | — |
| Thrombospondin157 | — | Fibrin | — |
| Factor V158 159 | — | Fibrin, platelets | Increased thrombin generation at the clot surface |
| Actin160 161 | — | Fibrin | Clot retraction, stabilization of the platelet cytoskeleton |
| Myosin162 | — | Itself | Clot retraction, stabilization of the platelet cytoskeleton |
| Vinculin163 | — | Fibrin | Clot retraction, stabilization of the platelet cytoskeleton |
| αIIbβ3164 | — | Fibrin | Stabilization of the platelet—fibrin clot |
TAFI indicates thrombin-activatable fibrinolysis inhibitor; PAI-2, plasminogen activator inhibitor 2.
Platelet factor XIII
Platelet factor XIII is localized in the cytoplasm and is composed of 2 A-subunits.1 The function of cytosolic platelet factor XIII is not well established. Plasma factor XIII can bind to glycoprotein IIb/IIIa on platelets, and this binding site can be cleaved by plasmin.81 Studies have indicated that normal platelets resuspended in factor XIII–free plasma catalyze the cross-linking of fibrin itself as well as the cross-linking of α2-antiplasmin to fibrin.82 Thus, factor XIII may further increase clot stabilization when released from platelets entrapped in fibrin clots.83 In addition, activated platelets may provide a surface for accelerating the cross-linking of fibrin polymers. Immunological studies have demonstrated that platelet-associated factor XIII is a marker of activation.84 Recently, Dale et al85 have shown that platelet factor XIII increases the procoagulant potential of activated platelets by cross-linking the primary amine serotonin to von Willebrand factor, factor V, and fibrinogen, which then localize to the platelet membrane via a serotonin receptor. The importance of platelet factor XIII in clot stabilization is illustrated by the fact that patients with A-subunit deficiency usually suffer a more severe bleeding diathesis than patients with B-subunit deficiency. In the latter case, factor XIII A-subunit is absent from plasma but normally present in platelets.
Factor XIII genes and polymorphisms
Structure of the factor XIII genes
The factor XIII A-subunit gene belongs to the transglutaminase family. The best-characterized members of this family are factor XIII, keratinocyte, tissue, and epidermal transglutaminases.86Erythrocyte protein band 4.2 has significant sequence homology to the transglutaminases and is also a member of the family.87The factor XIII A-subunit gene has been localized to chromosome 6p24-p25 and shows linkage with the major histocompatibilty complex.88,89 The gene codes for a mature protein of 731 amino acids, has 15 exons, and is more than 160 kilobases (kb) in size.90
The factor XIII B-subunit gene codes for a mature protein of 641 amino acids, is 28 kb in size, and is composed of 12 exons.17 It is located on chromosome 1q32-q32.1.91 The B-subunit contains a structural feature of 10 short consensus repeat units also known as Sushi domains.16 Sushi domains are a common structural feature of proteins associated with the regulation of the complement system.92,93 This family of proteins includes 5 complement control proteins: factor H, C4 binding protein, CR1, decay accelerating factor, and the membrane cofactor proteins. The genes for other Sushi domain proteins are located in the same chromosome 1q32 locus.94-98
Polymorphisms of the factor XIII A-subunit gene
Five common coding polymorphisms have been identified in the A-subunit (Figure 1; Table 2). A common G>T transition in codon 34 of the factor XIII A-subunit leading to a replacement of valine with leucine was found by investigators studying the molecular basis of factor XIII deficiency.99,100 This transition is not associated with factor XIII deficiency, but has been shown to change the function of factor XIII (see below). A tyrosine-to-phenylalanine polymorphism has been identified at residue 204 in the central domain of the factor XIII A-subunit.101Tyr204Phe has a frequency of 0.01 to 0.03, the lowest frequency in the general population of the 5 coding polymorphisms, and has been associated with an increased risk for recurrent miscarriage in women.102 A replacement of Pro564 with leucine in barrel 1 of the factor XIII A-subunit is responsible for the phenotypic discrimination on isoelectric focusing of factor XIII A*1A and 1B.103 Two further base changes, leading to a replacement of Val650 with isoleucine and Glu651 with glutamine in barrel 2, are responsible for the differences between the A*1A and 2A and A*1B and 2B phenotypes, respectively.103 Two polymorphisms have been identified in the noncoding regions of the gene, both occurring in the promoter region. One is a −246G>A transition,104,105 which is located close to SP-1 and MZF-1 protein-binding sites.106 Potential effects of this polymorphism on protein expression and factor XIII levels have not been investigated. The second polymorphism in the promoter is a short tandem repeat (AAAG)n approximately 800 to 900 base pairs upstream of the transcription start site.107 This tetranucleotide repeat occurs close to a GATA-1 binding site,106 but again, its potential functional effects are unknown.
Factor XIII polymorphisms
| Polymorphims . | Approximate allele frequency . | Location . | Association with disease . | Biochemical/molecular phenotype . |
|---|---|---|---|---|
| A-subunit | ||||
| Val34Leu | 0.25 | Activation peptide | CAD, stroke, DVT (see Table 3) | Increases activation rate—affects fibrin structure |
| Tyr204Phe | 0.01-0.03 | Catalytic core | Miscarriage,102stroke?146 | Specific activity? |
| Pro564Leu | 0.21 | Catalytic core | Stroke?146 | Specific activity? |
| Val650Ile | 0.06 | Beta-barrel | None | None |
| Glu651Gln | 0.27 | Beta-barrel | None | None |
| − 246G > A | — | Promoter | Unknown | Unknown |
| (AAAG)n | — | Promoter, around 800 to 900 bp upstream of transcription site | Unknown | Unknown |
| B-subunit | ||||
| His95Arg | 0.1 | 2ndSushi | VTE? | Increases subunit dissociation?124 |
| B∗1 | Varies in populations | Unknown | Unknown/unlikely? | Unknown |
| B∗2 | Varies in populations | Unknown | Unknown/unlikely? | Unknown |
| B∗3 | Varies in populations | Unknown | Unknown/unlikely? | Unknown |
| HS-1 Alu insertion | — | — | Unknown/unlikely? | Unknown |
| Polymorphims . | Approximate allele frequency . | Location . | Association with disease . | Biochemical/molecular phenotype . |
|---|---|---|---|---|
| A-subunit | ||||
| Val34Leu | 0.25 | Activation peptide | CAD, stroke, DVT (see Table 3) | Increases activation rate—affects fibrin structure |
| Tyr204Phe | 0.01-0.03 | Catalytic core | Miscarriage,102stroke?146 | Specific activity? |
| Pro564Leu | 0.21 | Catalytic core | Stroke?146 | Specific activity? |
| Val650Ile | 0.06 | Beta-barrel | None | None |
| Glu651Gln | 0.27 | Beta-barrel | None | None |
| − 246G > A | — | Promoter | Unknown | Unknown |
| (AAAG)n | — | Promoter, around 800 to 900 bp upstream of transcription site | Unknown | Unknown |
| B-subunit | ||||
| His95Arg | 0.1 | 2ndSushi | VTE? | Increases subunit dissociation?124 |
| B∗1 | Varies in populations | Unknown | Unknown/unlikely? | Unknown |
| B∗2 | Varies in populations | Unknown | Unknown/unlikely? | Unknown |
| B∗3 | Varies in populations | Unknown | Unknown/unlikely? | Unknown |
| HS-1 Alu insertion | — | — | Unknown/unlikely? | Unknown |
HS-1 indicates human specific-1; CAD, coronary artery disease; DVT, deep vein thrombosis; and VTE, venous thromboembolism.
Factor XIII Val34Leu
Factor XIII Val34Leu occurs in the activation peptide, 3 amino acids from the thrombin-cleavage site between Arg37 and Gly38 (Figure1). Val34Leu is relatively common, with an allele frequency of around 0.25 to 0.30 in the white population.108-110 The Leu allele frequency varies among different populations, being highest in whites (0.25-0.30) and American Indians (0.29), with a maximum of 0.40 among Pima Indians.109,110 In South Asians and in African populations, however, frequency of the Leu34 allele is lower, at around 0.13 and 0.17, respectively,109,110 and it reaches its lowest point in the Japanese at 0.01.110
Despite the fact that the transition of valine to leucine is a relatively conservative change—the only difference between the 2 amino acids being an additional CH2 group present in the leucine side-chain—the Val34Leu polymorphism has a significant effect on factor XIII function.111-115 Activation of Leu34 factor XIII by thrombin proceeds more rapidly than that of the Val34 variant.112-115 This effect is independent of the interaction between the A- and B-subunits, as both plasma and platelet Leu34 factor XIII are activated more rapidly than their Val34 counterparts.113 The catalytic efficiency of cleavage by thrombin is increased approximately 2.5-fold, from 0.2 μM−1sec−1 for the Val34 activation peptide to 0.5 μM−1sec−1 for Leu34.112Trumbo and Maurer115 found that the catalytic efficiency of thrombin cleavage of a synthetic peptide spanning factor XIII residues 28 through 41 is increased 5-fold when leucine rather than valine is at position 34 (0.068 versus 0.013 μM−1sec−1). Overall, the catalytic efficiency of this segment is approximately 10 times less than that of the entire factor XIII polypeptide, and it appears that the difference between the Leu34 and Val34 variants is accentuated by the absence of the remainder of the A-subunit.
The mechanism by which replacement of Val34 with leucine accelerates thrombin cleavage is not clear, but 2 studies have suggested that sterical/structural effects may play a role. Balogh et al114 reported that the Leu34 variant of a factor XIII segment from residues 32 to 42 showed greater interaction energy than the Val34 variant in a computer model of the molecular interaction with thrombin. Sadasivan and Yee116 analyzed the interaction between thrombin and a factor XIII peptide from residues 28 to 37 by x-ray crystallography. The study confirmed the critical role that residue 34 plays in the interaction between factor XIII and thrombin, and it was found that residues Val34 and Val29 are closer in the 3-dimensional structure than expected from their position in the secondary structure.116 The authors surmised that a bulkier side-chain at either residue 34 or residue 29 would alter the substrate peptide conformation.116 A recent study by Trumbo and Maurer117 on peptide (FXIII A-subunit 28-41) hydrolysis and conformation confirmed this; in this study, replacement of Val34 with Leu increased both kcat andKm of thrombin cleavage, and replacement of Val29 with Phe increased the Km. However, analysis of a double 34/29 mutant peptide showed that residue 34 of the factor XIII activation peptide plays a more influential role in thrombin interaction than residue 29, indicating that the main interaction between thrombin and factor XIII resides in the P4-P1 (Val/Leu34-Arg37) segment of the activation peptide.117
In the presence of polymerizing fibrin, the catalytic efficiencies of thrombin cleavage of the activation peptide are increased approximately 10-fold, but activation of factor XIII Leu34 remains faster than that of Val34, with a catalytic efficiency of 4.8 compared with 2.2 μM−1sec−1.112 An interesting observation is that release of the Leu34 activation peptide proceeds at a similar rate to that of fibrinopeptide A, whereas the rate of Val34 peptide cleavage is in tandem with that of fibrinopeptide B.112 These data suggest that factor XIII Leu34 is activated at the time of des-A fibrin formation, whereas the Val34 variant is activated when des-AB fibrin is formed.
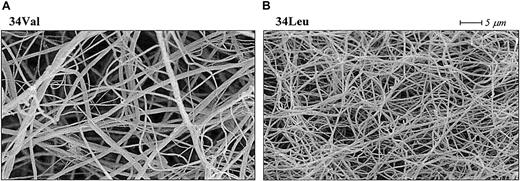
The molecular structure of fibrin is dynamic in the sense that it changes from thin protofibrils immediately after fibrinopeptide A cleavage to thicker fibers induced by lateral aggregation upon slower release of fibrinopeptide B.118-120 The alteration in factor XIII activation kinetics induced by the Val34Leu polymorphism could change this fibrin formation process, whereby cross-linking by factor XIII Leu34 acts as a fixative of the early, thinner des-A fibrin structure, inhibiting molecular rearrangement and lateral aggregation of the fibers. In agreement with this hypothesis, fibrin clots formed in the presence of Leu34 factor XIII have thinner fibers, smaller pores, and altered permeation characteristics when compared with fibrin clots formed in the presence of the Val34 variant (Figure5).112 In addition, Schroeder et al121 recently reported that clot formation time as measured by thromboelastography was significantly shortened in factor XIII Leu34 samples.
Effect of the factor XIII Val34Leu polymorphism on cross-linked fibrin structure.
Scanning electron micrographs of fibrin clots cross-linked by factor XIII Val34 (A) and factor XIII Leu34 (B). The clots were made from plasma samples homozygous for each Val34Leu allele. Fibrin cross-linked by the Leu34 variant of factor XIII, which is associated with a protective effect on thrombotic disorders, consists of thinner fibers and has altered permeation characteristics.112
Effect of the factor XIII Val34Leu polymorphism on cross-linked fibrin structure.
Scanning electron micrographs of fibrin clots cross-linked by factor XIII Val34 (A) and factor XIII Leu34 (B). The clots were made from plasma samples homozygous for each Val34Leu allele. Fibrin cross-linked by the Leu34 variant of factor XIII, which is associated with a protective effect on thrombotic disorders, consists of thinner fibers and has altered permeation characteristics.112
Polymorphisms of the factor XIII B-subunit gene
Factor XIII B-subunit shows 3 distinct alleles, B*1, B*2, and B*3 (Table 2).122 The noncoding region of the B-subunit gene contains a polymorphic human specific–1 Alu insertion.123In contrast to the factor XIII A-subunit polymorphisms, the molecular basis of the 3 most common B-subunit alleles has not been elucidated. Komanasin et al124 identified an A>G transition in codon 95 of the factor XIII B-subunit gene, which causes a replacement of histidine with arginine in the second Sushi domain. This polymorphism is relatively common, with an allele frequency of 0.10 in white subjects. It is unlikely that His95Arg would explain the phenotypic differentiation on isoelectric focusing, as both histidine and arginine have basic side-chains; however, this may need further investigation.
Factor XIII genetic polymorphisms and thrombotic disorders
Pathophysiology of thrombotic disorders
Clinical conditions associated with the development of thrombosis are a major cause of morbidity and mortality in the developed world. Among these are the atherothrombotic disorders (myocardial infarction, ischemic cerebrovascular disease, and peripheral vascular disease) and the venous thrombotic disorders, (deep vein thrombosis and pulmonary embolus). The development of atherothrombotic vascular disorders occurs over many decades and involves the interaction of classic atherogenic risk factors (diabetes, dyslipidemia, hypertension, etc) with abnormalities of the hemostatic system. Arterial disease develops in what is a high-pressure, high-flow system, and lipid deposition and smooth muscle hyperplasia occur in the arteries of subjects at risk, which ultimately leads to coronary atheroma formation. Later in life, plaques become unstable and rupture, exposing the highly prothrombotic lipid core, which activates the extrinsic coagulation cascade (factor VII, tissue factor), thereby initiating a series of proteolytic events culminating in thrombotic occlusion of coronary arteries. Ultimately myocardial infarction (MI) arises from the development of a cross-linked, fibrinolysis-resistant, platelet-rich fibrin clot. By contrast, venous thrombosis occurs in the context of a low-pressure, low-flow system in which damage to the vessel wall and atheroma formation are not etiological factors. Classically, venous thrombosis occurs in individuals who are genetically predisposed to thrombosis (protein C, protein S, antithrombin III deficiency); in the general population, it usually occurs secondary to environmental risk (surgery, pregnancy, malignancy) in which genetic influences play a role (factor V Leiden, prothrombin variants). A thrombotic occlusion in the venous system is low in platelets as compared with arterial disease and is composed mainly of cross-linked fibrin.
Factor XIII Val34Leu and coronary artery disease
Several studies have investigated the relationship between factor XIII Val34Leu and the risk of MI (Table3). The first was carried out in a cohort of consecutive individuals undergoing coronary angiography and reported a highly significant underrepresentation of the Leu34 allele in subjects with a history of MI as compared with angiographic subjects who had no history of MI and as compared with controls.108 These results suggested that possession of the Leu allele was protective against MI, an impression reinforced by the observation that, in carriers of the Leu allele, cardioprotection was lost in the presence of increasing degrees of insulin resistance as estimated by the homeostasis model assessment method125 and in the presence of high levels of the fibrinolytic inhibitor plasminogen activator inhibitor–1 (PAI-1).126 These findings indicated a major gene (factor XIII Leu34)–environment (insulin resistance) interaction that modulated vascular risk. A study from Finland confirmed the protective association of factor XIII Leu34 in a combined postmortem and coronary angiography study but failed to demonstrate an interaction with the PAI-1 4G/5G promoter genotype.127 Interestingly, the prevalence of the Leu allele was reported as being lowest in the Eastern Kainuu area, in which the highest risk of MI is found in Finland. These findings are similar to those described for Asians living in Britain, who both are at high vascular risk and have a low prevalence of the Leu34 allele.109 Two further studies have reported a protective effect of Leu34 against MI, with similar odds ratios of around 0.6 (Table 3).128,129 A third study, which investigated acute MI risk in young women, showed a relative risk of 0.8 for the Leu34 allele in this group of patients.130
Relationship between the factor XIIIA Val34Leu polymorphism and thrombotic disease
| Disease . | Association with disease . | No. subjects (controls) . | Origin of subjects . | Odds ratio . | Author . |
|---|---|---|---|---|---|
| MI | + | 398 (196) | Northern UK | 0.67 (0.54-0.85) | Kohler et al108 |
| MI | + | 470 | Finland | 0.59 (0.38-0.93) | Wartiovaara et al127 |
| MI | + | 150 (150) | Brazil | 0.6 (0.4-0.9) | Franco et al128 |
| MI | − | 201 (244) | Southern France | NA | Canavy et al131 |
| MI | − | 423 (479) | US | NA | Aleksic et al132 |
| MI | − | 191 | UK Asians | NA | Warner et al134 |
| MI | + | 120 (120) | Northern Italy | 0.59 | Gemmati et al129 |
| ICH | + | 130 (130) | Northern Italy | 1.74 | Gemmati et al129 |
| BI | + | 120 | Northern Italy | 0.60 | Gemmati et al129 |
| MI3-150 | + | 68 | US | 0.8 | Reiner et al130 |
| BI3-150 | − | 41 (345) | US | NA | Reiner et al130 |
| MI | − | 101 | Southern Spain | NA | Corral et al133 |
| DVT | − | 97 | Southern Spain | NA | Corral et al133 |
| CVD | − | 104 | Southern Spain | NA | Corral et al133 |
| DVT | + | 226 (254) | Northern UK | 0.63 (0.38-0.82) | Catto et al136 |
| DVT | + | 189 (189) | Brazil | 0.16 (0.05-0.5) for homozygous Leu | Franco et al137 |
| DVT | + | 154 (308) | Austria | 0.7 (0.5-1.0) | Renner et al138 |
| DVT | − | 273 (288) | Hungary | NA | Balogh et al114 |
| DVT | − | 427 (1045) | Southern Italy | NA | Margaglione et al139 |
| ICH | + | 612 (436) | Northern UK | — | Catto et al143 |
| BI | + | 456 (456) | France | 0.58 (0.44-0.75) | Elbaz et al145 |
| ICH | − | 201 (201) | Southern Spain | NA | Corral et al144 |
| RAO | + | 108 (313) | Austria | 0.22 for homozygous Leu | Weger et al147 |
| Disease . | Association with disease . | No. subjects (controls) . | Origin of subjects . | Odds ratio . | Author . |
|---|---|---|---|---|---|
| MI | + | 398 (196) | Northern UK | 0.67 (0.54-0.85) | Kohler et al108 |
| MI | + | 470 | Finland | 0.59 (0.38-0.93) | Wartiovaara et al127 |
| MI | + | 150 (150) | Brazil | 0.6 (0.4-0.9) | Franco et al128 |
| MI | − | 201 (244) | Southern France | NA | Canavy et al131 |
| MI | − | 423 (479) | US | NA | Aleksic et al132 |
| MI | − | 191 | UK Asians | NA | Warner et al134 |
| MI | + | 120 (120) | Northern Italy | 0.59 | Gemmati et al129 |
| ICH | + | 130 (130) | Northern Italy | 1.74 | Gemmati et al129 |
| BI | + | 120 | Northern Italy | 0.60 | Gemmati et al129 |
| MI3-150 | + | 68 | US | 0.8 | Reiner et al130 |
| BI3-150 | − | 41 (345) | US | NA | Reiner et al130 |
| MI | − | 101 | Southern Spain | NA | Corral et al133 |
| DVT | − | 97 | Southern Spain | NA | Corral et al133 |
| CVD | − | 104 | Southern Spain | NA | Corral et al133 |
| DVT | + | 226 (254) | Northern UK | 0.63 (0.38-0.82) | Catto et al136 |
| DVT | + | 189 (189) | Brazil | 0.16 (0.05-0.5) for homozygous Leu | Franco et al137 |
| DVT | + | 154 (308) | Austria | 0.7 (0.5-1.0) | Renner et al138 |
| DVT | − | 273 (288) | Hungary | NA | Balogh et al114 |
| DVT | − | 427 (1045) | Southern Italy | NA | Margaglione et al139 |
| ICH | + | 612 (436) | Northern UK | — | Catto et al143 |
| BI | + | 456 (456) | France | 0.58 (0.44-0.75) | Elbaz et al145 |
| ICH | − | 201 (201) | Southern Spain | NA | Corral et al144 |
| RAO | + | 108 (313) | Austria | 0.22 for homozygous Leu | Weger et al147 |
MI indicates myocardial infarction; ICH, intracranial hemorrhage; BI, brain infarction; TIA, transient ischemic attack, DVT, deep vein thrombosis; CVD, cerebrovascular disease; RAO, retinal artery occlusion; and NA, not applicable.
Subjects were all young women.
Four publications have reported no association between possession of Leu34 and risk of MI. These include patients recruited from southern France,131 the United States,132 and southern Spain,133 and Asian Indian patients recruited from the United Kingdom (Table 3).134 One explanation for the discrepancies in the studies could have been linkage disequilibrium between Val34Leu and other polymorphisms in the factor XIII A-subunit gene. However, a study from Kohler et al135failed to show any association between 3 other coding polymorphisms (Pro564Leu, Val650Ile, and Glu651Gln; Table 2) and MI.
Factor XIII Val34Leu and venous thrombotic disorders
Six studies have investigated the relationship between factor XIII Val34Leu and venous thrombosis (Table 3). Three have shown significant protective associations similar to that described for MI,136-138 while 3 demonstrated no association.114,133,139 Two studies have addressed the question as to whether interactions occur between factor XIII Val34Leu and factor V Leiden.137,140 Neither study was positive although the study by Franco et al137 hinted at a weak interaction with increasing age. A study by Carter et al141 reported an interaction between a common polymorphism in the fibrinogen Aα gene (Thr312Ala) and factor XIII Val34Leu that negates the protective effect afforded by Val34Leu. Additionally, studies of embolic stroke indicate that the Ala312 allele is associated with a poor prognosis in high-risk subjects with atrial fibrillation to indicate that it may influence embolic disorders.142 The plausibility of these observations lies in the fact that position 312 in the Aα fibrinogen chain is very close to the factor XIII cross-linking and α2-antiplasmin incorporation sites. The biochemical consequences of these interactions are the subject of current investigation.
Factor XIII Val34Leu, cerebrovascular disease, and other associations
In comparison with MI and venous thromboembolism, the relationship between factor XIII Val34Leu and cerebrovascular disease has been investigated to a lesser extent (Table 3). The original paper on this subject by Catto et al143 demonstrated a higher prevalence of the Leu allele in subjects with primary intracranial hemorrhage (ICH) and no association with ischemic stroke. While the findings in ICH support the concept of a gene that can be both protective against thrombosis and involved in the pathogenesis of bleeding, the study involved only 62 patients with ICH. The findings in relation to ICH were not supported by a much larger study from Spain,144and this group reported no association between possession of Leu34 and ischemic stroke.133 However, a large, well-matched case-control study of cerebral infarction reported a major protective effect of Leu34, with interactions with smoking that modified risk of stroke.145 These findings were supported by a smaller study from Italy.129 A more recent study from the United States did not find a protective effect of Leu34 in a limited number of young women with cerebral infarction.130 Instead, the authors found that the Phe204 allele of factor XIII, which previously has been linked with an increased risk for miscarriage,102was associated with a mild increased risk of ischemic stroke. In an earlier report from the same authors, however, the Phe204 and Leu564 alleles was associated with an increased risk of hemorrhagic stroke.146 These apparently contradictory findings indicate that the relationship between factor XIII polymorphisms and cerebrovascular disease may be more complex, and further studies are warranted that take into account other genetic and environmental factors involved in the pathogenesis of the disease. With regard to other thrombotic disorders, a recent report from Austria has found a protective effect of homozygous factor XIII Leu34 on retinal artery thrombosis,147 with an odds ratio similar to that reported previously for the homozygous genotype in relation to deep vein thrombosis (Table 3).
Conclusion
The structure of the fibrin clot and the effects of cross-linking by factor XIII on fibrin structure are immensely complex. Fibrin clot structure in plasma samples from different individuals shows considerable variation, suggesting that many factors, both genetic and environmental, play a role in determining the stability and resistance of the clot to fibrinolysis. FactorXIII Val34Leu is a genetic determinant of fibrin structure/function, and fibrinogen Aα Thr312Ala and Bβ Arg448Lys are potential candidates, but the effects of additional determinants may be major. The concentrations of fibrinogen, metal ions, and many other factors may affect fibrin clot structure and interact with the genetic polymorphisms. Incorporation of constitutive proteins such as fibronectin into the clot may play an additional role in determining molecular structure. The true complexity of clot formation is only now becoming apparent, and future developments in this area may have profound effects on our understanding of the basic science and its translation into clinical practice.
Activation of factor XIII by thrombin and the roles that fibrin formation and the factor XIII B-subunit play in this reaction are intriguing processes. To date, apart from x-ray crystallography and kinetic data of the interaction between thrombin and factor XIII–derived peptides, little is known about the molecular and structural details of factor XIII activation. It is known that activation of factor XIII is regulated mainly by fibrin polymerization and the clot formation process, but the exact molecular mechanisms have so far remained unresolved.
Two animal studies have indicated the potential therapeutic relevance of factor XIII modulation. In ferrets, factor XIII–mediated fibrin-fibrin and α2-antiplasmin–fibrin cross-linking caused pulmonary emboli that were resistant to endogenous and exogenous tissue plasminogen activator–induced fibrinolysis.148 In a canine model of coronary thrombosis, inhibition of cross-linking by factor XIII promoted tissue plasminogen activator–induced thrombolysis of the clots.149
The laboratory and clinical epidemiological studies presented in this review indicate that genetic and environmental modulation of the processes involved in fibrin structure/function are of importance in clot formation. Recent studies on factor XIII Val34Leu suggest that therapeutic targets exist within the final processes of the coagulation cascade that may be of clinical value in the management of thrombotic disorders and, potentially, in enhancing therapeutic and physiological fibrinolysis. The challenge is to acquire a fuller understanding of the biochemical processes and to support the development of research programs that may lead to therapeutic advances in this field.
Supported by British Heart Foundation grants PG/98104 (P.J.G., R.A.S.A.), PG/99082 (P.J.G., R.A.S.A.), and FS2000023 (P.J.G., R.A.S.A.); Medical Research Council grants G9900904 (P.J.G., R.A.S.A.) and G0000624 (P.J.G., R.A.S.A.); National Institutes of Health grant HL30954 (J.W.W.); National Cancer Institute grant CA 71753 (C.S.G.); Duke University SPORE in Breast Cancer grant CA68438 (C.S.G.); National Heart, Lung and Blood Institute grant HL28391 (C.S.G.); and an American Heart Association (North Carolina Affiliate) Grant-in-Aid (T.S.L.).
References
Author notes
Robert A. S. Ariëns, Academic Unit of Molecular Vascular Medicine, G-Floor, Martin Wing, The General Infirmary, Leeds LS1 3EX, United Kingdom; e-mail:r.a.s.ariens@leeds.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal