This study evaluates the prognostic significance of genetic abnormalities (detected at or shortly after presentation), clinical stage, lymphocyte morphology, CD38 expression, and IGVHgene status in 205 patients with chronic lymphocytic leukemia (B-CLL). Deletion of chromosome 11q23, absence of a deletion of chromosome 13q14, atypical lymphocyte morphology, and more than 30% CD38 expression are significantly associated with the presence of unmutatedIGVH genes. Advanced stage, male sex, atypical morphology, more than 30% CD38 expression, trisomy 12, deletion of chromosome 11q23, loss or mutation of the p53 gene, and unmutatedIGVH genes are all poor prognostic factors in a univariate analysis. However, only 98% or more homology of IGVH genes to the germline sequence, loss or mutation of the p53 gene, and clinical stage retain prognostic significance in a multivariate analysis. The median survival of patients with mutated IGVHgenes, unmutated IGVH genes, and loss or mutation of thep53 gene regardless of IGVH gene status is 310, 119, and 47 months, respectively. These data should facilitate the design of new trials for the management of patients presenting with advanced disease or poor prognosis early stage disease.
Introduction
Currently, 70% to 80% of patients with chronic lymphocytic leukemia (CLL) present with a low tumor burden and are frequently diagnosed from a blood count performed for an incidental reason.1 Treatment of these patients at presentation with chlorambucil does not prolong survival.2,3 Therapeutic options for patients with progressive disease and those who present with advanced disease have recently increased.4 5 The introduction of new and more reliable prognostic factors may be valuable in 2 clinical situations. First, they may facilitate the design of randomized clinical trials to determine whether early intensive treatment of patients with a low tumor burden and poor risk factors can prolong survival. Second, they may influence the choice of initial treatment and subsequently the need for and benefit of additional treatments for patients with advanced or progressive disease.
Both the French and Spanish CLL study groups identified good risk factors such as a normal hemoglobin, low lymphocyte count, nondiffuse bone marrow infiltration, and lymphocyte doubling time of more than 1 year, which identified a subgroup of patients with smoldering CLL from within Binet stage A or Rai stage 0 or 1 disease.6,7 However, disease in 10% of these patients still progresses and requires treatment. Serum markers including β2-microglobulin,8 soluble CD23,9 and serum thymidine kinase10 can all identify patients who present with low tumor burden who are at high risk of disease progression.
Genetic studies using chromosome analysis or interphase fluorescence in situ hybridization (FISH) have identified recurring abnormalities with prognostic significance.11,12 Patients with a normal karyotype or deletion of 13q14 as the sole genetic abnormality have a better prognosis than those with a complex karyotype or deletion of 11q23 or 17p13. In addition del11q23 is associated with bulky lymphadenopathy13 and a high incidence of residual disease following autologous transplantation,14 whereas structural abnormalities of 17p13 resulting in loss or mutation of thep53 gene correlate with resistance to purine analogues.15 16
In 1999, our group17 and Damle et al18demonstrated that patients with mutated IGVH genes had significantly superior survival compared with patients with unmutatedIGVH genes. This finding also applies to patients presenting with Binet stage A disease and has subsequently been corroborated by other studies.19,20 Expression of CD38 has recently been shown to have prognostic value in multivariate analyses, which have included clinical stage, cytogenetic abnormalities, and β2-microglobulin levels.21-23 CD38 expression is associated with unmutated IGVH genes, but results are discordant in approximately 30% of cases and it is still controversial as to whether CD38 expression has prognostic significance in patients with known IGVH gene status.24-26 In addition, CD38 expression may vary during the course of the disease,23,24 and karyotype evolution occurs in 15% to 40% of cases.23 27
The aim of this study is to evaluate the prognostic significance of genetic abnormalities and CD38 expression, assessed at or close to presentation, on the survival of patients whose IGVH genes have been sequenced.
Patients and methods
Patients
This study is based on samples obtained with consent from 205 patients with CLL and known IGVH gene status; 188 were from a consecutive series of 382 patients seen at the Royal Bournemouth Hospital between October 1983 and May 2001. A total of 194 cases were excluded because no IGVH gene data were available, due either to the lack of a suitable sample or because contamination with normal B cells did not allow direct sequencing of the sample tested. The Bournemouth cases have been supplemented by 17 cases from among 120 cases referred to this center from other hospitals for cytogenetic analysis. The additional cases, for which full clinical and laboratory data were available, were included because of their cytogenetic abnormalities; 58% had a structural abnormality of chromosome 17p13 and 33% had deletion of 11q23. The stage at presentation of the 188 Bournemouth cases, 17 referral cases, and the 194 excluded cases was similar, with 83%, 76%, and 89%, respectively, being diagnosed as Binet stage A.
In all cases, clinical diagnosis of CLL was based on standard morphologic and immunophenotypic criteria. Atypical lymphocyte morphology was defined as more than 10% prolymphocytes or more than 15% lymphoplasmacytoid or cleaved cells. Clinical follow-up for all patients ranged from 4 to 370 months, with a median follow-up of 74 months, and for live patients from 8 to 370 months with a median follow-up of 88 months. Ten patients have been followed for more than 20 years.
Patients were considered to have progressive disease if they had at least one of the following parameters: a lymphocyte doubling time of less than 1 year, progression to a more advanced Binet stage, development of systemic symptoms or Richter syndrome, or a downward trend of hemoglobin or platelet count to below the normal range. Patients with progressive or advanced disease were treated predominantly with chlorambucil as first-line therapy and since 1991 with fludarabine for chronically relapsing or resistant disease. Patients who died during the period of follow-up have been categorized either as dying from a CLL-related cause or a non–CLL-related cause. Categorization was made independently by 2 assessors (D.G.O. and T.J.H.), who then discussed and resolved any disagreements.
Plan of investigation
IGVH gene data were available for all patients. Cytogenetic data from karyotype analysis were available on all except 1 of the 205 study patients. Karyotype analysis was performed at presentation for 189 patients; however, 15 presented prior to October 1983 and had a median follow-up of 89 months prior to analysis. None of these patients had been treated prior to the initial karyotype analysis, and all had stable stage A disease. Serial karyotype analysis was performed on 145 of the 204 cases, allowing evaluation of karyotype evolution. The cytogenetic abnormalities detected on sequential testing were not included in the evaluation of prognostic factors at presentation. Although karyotype analysis is valuable as a global technique, in view of its relatively low sensitivity in detecting genetic abnormalities in CLL, additional techniques for assessment of the most common abnormalities, del11q, del13q, trisomy 12, and p53 abnormalities, were used. The choice of technique was largely dictated by the availability of suitable samples taken at the time of presentation.
Interphase FISH was used to detect trisomy 12 and deletions at 11q23 in all cases where these abnormalities had not been detected by conventional cytogenetics; 138 and 178 cases, respectively, were thus examined.
Interphase FISH was also used to detect loss of the p53 gene in 76 cases in which conventional cytogenetics had not detected abnormality at 17p, and in 13 cases that had a translocation or deletion of chromosome 17 (12 cases) or known p53 mutation (1 case).
Expression of p53 protein was assessed by flow cytometry in 145 cases, including all those that were not examined by FISH.
The nonisotopic RNase cleavage assay (NIRCA, AMS Biotechnology, Oxford, United Kingdom)28 was used to screen for p53 mutation in 3 of the referred cases, 1 of which had a del17p, 1 a normal karyotype, and 1 in which no metaphases were obtained for karyotype analysis.
Direct sequence analysis of the p53 gene was performed in all but one of the cases that showed abnormality by FISH or flow cytometry and one case in which a positive NIRCA result was the only indication of p53 abnormality.
Southern blot analysis was used as an additional method to detect loss at 13q14 in 144 cases where sufficient tumor DNA was available. This included 24 of the 34 cases with a karyotypic abnormality of 13q14 and 120 additional cases.
Expression of CD38 was analyzed by flow cytometry in the 171 cases for which a suitable sample taken at or close to the time of presentation was available.
Cytogenetics
Cultures of peripheral blood were established using phorbol 12-myristate 13-acetate (TPA) as a mitogen, harvested after 3, 4, and 5 days of incubation and processed by standard cytogenetic techniques to obtain metaphases for analysis. Karyotype analysis of a minimum of 30 metaphases was carried out on Giemsa-trypsin-Leishman (GTL)–banded slides.
Interphase FISH
Separate hybridizations were carried out for loci on chromosomes 11, 12, and 17. For chromosomes 12 and 17, commercial probes were used (Vysis UK, London, United Kingdom). An alpha satellite DNA probe CEP12, directly labeled with SpectrumOrange, was used to detect aneuploidy of chromosome 12. LSIp53, together with CEP17 alpha satellite DNA probe labeled with SpectrumOrange and SpectrumGreen (Vysis), respectively, were used to evaluate chromosome deletion at 17p13.1. For chromosome 11, CEPH yacs 755b11 and 801b11 were labeled by nick translation with SpectrumOrange dUTP and SpectrumGreen dUTP (Vysis), respectively, according to the manufacturer's protocol. Hybridization was to peripheral blood lymphocytes, which were separated by density gradient centrifugation, treated with hypotonic solution (KCl) and fixed with methanol-acetic acid, or to cells from our archive of fixed TPA stimulated lymphocyte cultures, which were stored at −20°C after karyotype analysis. A minimum of 200 interphase nuclei was assessed from each hybridization.
Southern blot
Southern analysis for deletion at 13q14 was carried out as described previously.29 Ten micrograms of both tumor and control DNA was digested with the restriction enzymes EcoRI or HindIII and size fractionated on a 1% agarose gel before transfer to Hybond-N membrane (Amersham Pharmacia Biotech, Uppsala, Sweden). The resulting membrane was then hybridized with the probe p9E4.3, which recognizes a 4.3-kb EcoRI fragment within a region 10 kb centromeric of D13S319, a marker frequently deleted in CLL.
The membrane was also hybridized with a control probe, p105-15A, located at 5q11.2, which serves as an internal hybridization standard. Following hybridization the resulting autoradiographs were examined using a scanning densitometer (Amersham Pharmacia Biotech). Tumor samples were assessed as heterozygously deleted when the ratio of test to control probe was 40% to 75% that of the control sample on the membrane. Tumor samples were assessed as homozygously deleted at the p9E4.3 13q14 locus if the ratio of the test to control probe was less than 20% compared to the control samples.
p53 expression
Ficol-separated peripheral blood lymphocytes were fixed in 0.5% paraformaldehyde for 30 minutes at 4°C followed by 80% ethanol at −20°C. Cells were washed in phosphate-buffered saline (PBS) with 1% human serum albumin (Bio-Products Laboratory, Elstree, Herts, United Kingdom) and 0.05% Tween 20 (Sigma, Poole, Dorset, United Kingdom), before being labeled with 5 μL 1:20 dilution of unconjugated p53 antibody (clone D0-7, Dako, Glostrup, Denmark). A fluorescein isothiocyanate (FITC)–labeled sheep antimouse secondary antibody (Novacastra Laboratories, Newcastle, United Kingdom) diluted 1:200 was then applied. Isotype controls were run in parallel. Five thousand events were collected from each sample on a Becton Dickinson (San Jose, CA) FACSCalibur flow cytometer and analyzed using Cell Quest software. Eleven patients, who had no p53 mutations on sequencing, were used to establish the cutoff value for p53 by flow cytometry. The cutoff value (median plus 2 SDs) for this group was 30%, and values more than 30% were considered to be positive.30
p53 mutational analysis
Complementary DNA (cDNA) was synthesized by reverse transcription using an oligo (dT) primer (Promega, Madison, WI) from RNA extracted from Ficol-separated peripheral blood lymphocytes. A 1.1-kb fragment of the coding region was amplified by polymerase chain reaction (PCR) using primers p53S2 (5′GAGCCGCAGTCAGATCCTAG3′) and p53CT (5′GTCTGAGTCAGGCCCTTCTG3′). Direct sequencing using 6 internal primers 1: 5′CAGACCTATGGAAACTACTTCCTG3′; 2: 5′TCGGATAAGATGCTGAGGAG3′; 3: 5′CCTATGAGCCGCCTGAGGTT3′; 4: 5′CTTCCAGTGTGATGATGGTG3′; 5: 5′AACATCTCGAAGCGCTCACG3′; 6: 5′AACATCTTGTTGAGGGCAGG3′), which were designed to optimize the coverage of exons 3 to 10, was performed on an ABI 310 sequencer (Applied Biosystems, Foster City, CA). Sequence data were compared with wild-type sequence and any mutations identified were confirmed by duplicate PCR and sequence analysis.
IGVH gene analysis
Immunoglobulin variable region genes were sequenced as previously described.17 The preferred source material was RNA. cDNA was synthesized as above and amplified by PCR using a mixture of oligonucleotide 5′ primers specific for each leader sequence of the VH1 to VH6 families or a consensus 5′ FW1 region primer, together with either a consensus 3′ primer complementary to the germ line JH regions or a 3′ primer complementary to the constant region. Clonal sequences were determined by sequencing amplicons from at least 2 independent PCR reactions. The majority of samples were sequenced directly using the automated DNA sequencer. Nucleotide sequences were aligned to EMBL/GenBank and current databases (V-BASE sequence directory,31 using MacVector 4.0 sequence analysis software; International Biotechnologies, New Haven, CT, and Lasergene; DNASTAR, Madison, WI). Percentage homology was calculated by counting the number of mutations between the 5′ end of FR1 and the 3′ end of FR3.
Analysis of CD38 expression
Cells (106) from cryopreserved specimens or fresh whole blood were incubated for 15 minutes with 5 μL of the following antibodies: FITC-labeled anti-CD5 (clone DK23, Dako), phycoerythrin (PE)–labeled anti-CD38 (clone HB7, Becton Dickinson), and R-phycoerythrin-cyanine 5 (RPE-Cy5) labeled anti-CD19 (clone HD37, Dako). Red cells were lysed with FACSlyse (Becton Dickinson) and a minimum of 10 000 cells was acquired in the Cell Quest program on a FACSCalibur flow cytometer (Becton Dickinson). Each sample was run with an appropriate isotype control (FITC-labeled mouse IgG1 and PE-labeled mouse IgG1, both from Dako) and this was used to define the negatively stained cells. In each case the dot-plot was gated on the lymphoid gate on the side scatter-forward scatter (SCC-FSC) plot. Within this gate the markers were set on the isotype control to define the negative population. A single cell was regarded as CD38+ if its position lay outside this marker. The tumor population was defined by gating of the lymphoid population on the SCC-FSC plot, followed by gating of the CD5+/CD19+ population. The percentage of CD38+ cells in this gate was then determined. A cutoff point for CD38 expression of 30% was chosen because we have previously been shown this to give the highest Youden index.24
Statistical methods
Data were analyzed using SPSS for Windows Version 10. The association between prognostic factors and IGVH gene status was summarized using odds ratios with 95% CIs. For each level of each factor the odds ratio for having unmutated VH genes has been expressed relative to a reference level. The P values for the unadjusted associations were calculated using the χ2 test for association, incorporating Yates continuity correction when the prognostic factor had only 2 levels. Because these prognostic factors are statistically interrelated, logistic regression was used to determine those factors that were independently associated with IGVH gene status. A forward stepping modeling procedure (using a 5% significance level) was chosen, so that the ratio of number of patients with unmutated genes to the number of prognostic factors in the model at each step was as high as possible. Because patients are excluded from this modeling procedure if they have missing data for any of the factors, the odds ratios and P values for the final model were then recalculated using only those factors that were significant. The remaining factors were then added to this model singly to confirm that their P values remained high.
Survival functions have been estimated using the product-limit (Kaplan-Meier) method. Ten-year mortality (together with its SE) and median survival times (95% CI) have been used as summary statistics for the survival curves. The advantage of the former is that it can be calculated even when survival times are long relative to length of follow-up. The effect of prognostic factors on survival has been summarized using hazard ratios with 95% CIs, and for each level of each factor these have been expressed relative to a reference level. For univariable analysis, survival curves have been compared between groups using the log-rank test. Because these prognostic factors are interrelated the Cox proportional hazards model has been used to identify the independent prognostic factors. The method for reaching the final model is the same as described for logistic regression above. Interactions between prognostic factors were also tested.
The 10-year mortality rate from non–CLL-related causes has been calculated by subtracting the 10-year mortality for CLL related deaths from the 10-year mortality for all deaths.
Because there is controversy as to which percentage homology to the nearest germline sequence most reliably separates patients into good or poor risk groups, the Youden index32 for accurate identification of deaths was calculated for IGVH gene homologies from 94% to 99% in 1% increments. The sensitivities and specificities, which are given equal weight in calculating the index for each cut-point (x), were estimated from the 10-year mortality rates for patients above and below x, and the proportion of patients above x. The best cutoff point has been chosen on the basis of having the highest Youden index (sensitivity + specificity − 1).
Results
Patient data
We have studied 205 patients with CLL; 118 (58%) were men and 87 were women. At presentation 169 (82%) were Binet stage A, 21 (10%) stage B, and 15 (7%) stage C. A total of 137 (67%) patients had stable disease. There were 77 deaths during 1665 total years follow-up of which 46 deaths were considered to be disease related. The 10-year mortality from CLL-unrelated deaths was similar for patients with mutated IGVH genes and those with unmutated VHgenes (13% versus 9%).
The IGVH genes were mutated in 123 cases (60%) and unmutated in 82. Lymphocyte morphology was evaluable in 199 patients, of whom 68 (34%) had atypical morphology. CD38 expression was tested in 171 cases of whom 71 (42%) had more than 30% expression of CD38.
Cytogenetic and genetic data
An abnormal karyotype was detected at or near the time of presentation in 141 (69%) patients. The prevalence of trisomy 12 and structural abnormalities of chromosomes 11q23, 13q14, and 17p13 detected by karyotype analysis was 29%, 10%, 17%, and 6%, respectively. Surprisingly interphase FISH detected trisomy 12, del11q23, and loss of the p53 gene in only 1.4%, 2.2%, and 0%, respectively, of the cases that lacked these abnormalities karyotypically. For the 13q14 locus Southern analysis using a 13q14.3 probe showed either heterozygous or homozygous loss in all 24 patients with a karyotypic abnormality of 13q14 and detected deletion of this locus in 54 of 120 (45%) cases with no evidence of 13q14 abnormality by conventional chromosome analysis.
The overall incidence of trisomy 12 and structural abnormalities of 13q14, 11q23, and 17p13 was therefore 30%, 62%, 19%, and 6%, respectively. The true incidence of 13q14 loss may be underestimated because 60 cases without karyotypic abnormality of 13q14 were not tested by Southern blot analysis.
Fifteen patients had loss or mutation of p53 (Table1). All 12 cases with a translocation or deletion of 17p13 were shown to have lost one p53 allele by FISH. Increased expression of p53 protein was found in 6 of 10 cases with a structural abnormality of 17p and in 1 of 145 cases with no 17p abnormality detected by karyotype (Figure1). This case (patient 7) had a translocation of chromosome 17 with a q arm breakpoint and retained both p53 alleles by FISH. Mutational analysis was performed in 14 of 15 cases and a p53 mutation was found in 12 of 14 cases; 10 had a point mutation, 1 had a frameshift mutation, and 1 had a silent mutation.
p53 analysis and IGVH gene status
| Patient . | Chromosome abnormality . | FISH . | Flow % positive cells . | Sequence analysis . | V genes (% homology) . | Survival (mo) . | |||
|---|---|---|---|---|---|---|---|---|---|
| Exon . | Codon . | Nucleotide . | Amino acid . | ||||||
| 1 | i17q | +− | NT | NT | 100 | 20 | |||
| 2 | i17q | +− | 57 | 8 | 273 | CGT > TGT | Arg → Cys | 100 | 109 |
| 3 | t17p13 | +− | 25 | 7 | 248 | CGG > CAG | Arg → Gln | 100 | 53 |
| 4 | t17p13 | +− | 6 | 7 | 250 | 1-bp ins | Frameshift | 100 | 42 |
| 5 | t17p13 | +− | 93 | 5 | 158 | CGC > GGC | Arg → Gly | 100 | 31 |
| 6 | t17p13 | +− | 73 | 5 | 138 | GCC > CCC | Ala → Pro | 100 | 32 |
| 7 | t17q | ++ | 90 | 7 | 248 | CGG > CAG | Arg → Gln | 100 | 21 |
| 14* | del17p | +− | NT | 7 | 248 | CGG > GGG | Arg → Gly | 100 | 28 |
| 15* | Normal | NT | NT | 5 | 163 | TAC > TGC | Tyr → Cys | 100 | 96 |
| 8 | t17p13 | +− | 55 | 3 | 22 | CTA > CTG | Silent | 97 | 80 (alive) |
| 9 | t17p13 | +− | 15 | NAD | 97 | 66 | |||
| 10 | t17p13 | +− | 90 | 5 | 141 | TGC > CGC | Cys → Arg | 95 | 45 |
| 11* | NT | +− | NT | 6 | 281 | GAC > GAA | Asp → Glu | 91 | 21 (alive) |
| 12 | r17 | NT | 60 | 5 | 162 | ATC > AAC | Ile → Asn | 91 | 58 |
| 13 | t17p13 | +− | 16 | NAD | 91 | 79 (alive) | |||
| Patient . | Chromosome abnormality . | FISH . | Flow % positive cells . | Sequence analysis . | V genes (% homology) . | Survival (mo) . | |||
|---|---|---|---|---|---|---|---|---|---|
| Exon . | Codon . | Nucleotide . | Amino acid . | ||||||
| 1 | i17q | +− | NT | NT | 100 | 20 | |||
| 2 | i17q | +− | 57 | 8 | 273 | CGT > TGT | Arg → Cys | 100 | 109 |
| 3 | t17p13 | +− | 25 | 7 | 248 | CGG > CAG | Arg → Gln | 100 | 53 |
| 4 | t17p13 | +− | 6 | 7 | 250 | 1-bp ins | Frameshift | 100 | 42 |
| 5 | t17p13 | +− | 93 | 5 | 158 | CGC > GGC | Arg → Gly | 100 | 31 |
| 6 | t17p13 | +− | 73 | 5 | 138 | GCC > CCC | Ala → Pro | 100 | 32 |
| 7 | t17q | ++ | 90 | 7 | 248 | CGG > CAG | Arg → Gln | 100 | 21 |
| 14* | del17p | +− | NT | 7 | 248 | CGG > GGG | Arg → Gly | 100 | 28 |
| 15* | Normal | NT | NT | 5 | 163 | TAC > TGC | Tyr → Cys | 100 | 96 |
| 8 | t17p13 | +− | 55 | 3 | 22 | CTA > CTG | Silent | 97 | 80 (alive) |
| 9 | t17p13 | +− | 15 | NAD | 97 | 66 | |||
| 10 | t17p13 | +− | 90 | 5 | 141 | TGC > CGC | Cys → Arg | 95 | 45 |
| 11* | NT | +− | NT | 6 | 281 | GAC > GAA | Asp → Glu | 91 | 21 (alive) |
| 12 | r17 | NT | 60 | 5 | 162 | ATC > AAC | Ile → Asn | 91 | 58 |
| 13 | t17p13 | +− | 16 | NAD | 91 | 79 (alive) | |||
NT indicates not tested; NAD, no abnormality detected; ++, 2 signals; +−, 1 signal.
Indicates patients who were tested by NIRCA.
Flow cytometric analysis of p53 protein expression in case 7 of Table 1.
In an indirect staining protocol, a monoclonal antibody (clone DO-7), which recognizes an epitope comprising amino acids 20-25 of the p53 molecule, was used for detection of p53 expression. An isotypic control was used as the first layer followed by a second layer, FITC-conjugated sheep antimouse IgG F(ab′)2.
Flow cytometric analysis of p53 protein expression in case 7 of Table 1.
In an indirect staining protocol, a monoclonal antibody (clone DO-7), which recognizes an epitope comprising amino acids 20-25 of the p53 molecule, was used for detection of p53 expression. An isotypic control was used as the first layer followed by a second layer, FITC-conjugated sheep antimouse IgG F(ab′)2.
Karyotype evolution was detected in 37 of 145 cases; of these 3 acquired del11q23, 2 acquired a structural abnormality of 17p, and 1 acquired a structural abnormality of 13q.
Association between prognostic variables and IGVHgene status
To determine which percentage homology to the nearest germ lineIGVH sequence most reliably separates patients into good or poor risk groups, the Youden index was used. The highest value of the Youden index32 occurred for a cutoff of 98% whether CLL-related deaths (Table 2) or all deaths were analyzed.
Sensitivity and specificity for CLL-related deaths up to 120 months
| Cut-point for homology . | % with homology at or above cut-point . | Sensitivity, % . | Specificity, % . | Youden index, % . |
|---|---|---|---|---|
| ≥ 94 | 72 | 94 | 36 | 30 |
| ≥ 95 | 64 | 94 | 47 | 40 |
| ≥ 96 | 56 | 89 | 55 | 45 |
| ≥ 97 | 48 | 90 | 66 | 56 |
| ≥ 98 | 40 | 86 | 76 | 62 |
| ≥ 99 | 36 | 75 | 78 | 53 |
| Cut-point for homology . | % with homology at or above cut-point . | Sensitivity, % . | Specificity, % . | Youden index, % . |
|---|---|---|---|---|
| ≥ 94 | 72 | 94 | 36 | 30 |
| ≥ 95 | 64 | 94 | 47 | 40 |
| ≥ 96 | 56 | 89 | 55 | 45 |
| ≥ 97 | 48 | 90 | 66 | 56 |
| ≥ 98 | 40 | 86 | 76 | 62 |
| ≥ 99 | 36 | 75 | 78 | 53 |
Table 3 shows the relationship betweenIGVH gene status and other prognostic variables. All except the presence of karyotypic evolution and chromosome 17 abnormalities were significantly associated with IGVH gene status (P = < .05). However, for these 2 nonsignificant factors, the odds ratios were still high and the CIs were wide, indicating a possible type 2 error (incorrectly concluding that the factor is not important).
Association between IGVH gene status and other prognostic factors
| Factor . | Mutated N = 123 . | Unmutated N = 82 . | Odds ratio (95% CI) . | P . |
|---|---|---|---|---|
| Sex-male | 49% (60) | 71% (58) | 2.5 (1.4, 4.6) | .003 |
| Abnormal karyotype | 60% (74) | 83% (67) | 3.2 (1.6, 6.3) | .001 |
| Karyotype evolution present | 18% (19) | 29% (18) | 1.9 (0.9, 3.9) | .15 |
| Atypical morphology | 18% (21) | 59% (47) | 6.6 (3.5, 12.7) | < .001 |
| CD38 ≥ 30% | 25% (27) | 72% (44) | 8.0 (3.9, 16.2) | < .001 |
| Disease stage | < .001 | |||
| A | 93% (114) | 67% (55) | 1.0 | |
| B | 2% (3) | 22% (18) | 12.4 (3.5, 44.0) | |
| C | 5% (6) | 11% (9) | 3.1 (1.1, 9.2) | |
| Chromosome 13 abnormal | 53% (64) | 29% (24) | 0.4 (0.2, 0.7) | .002 |
| Chromosome 12 abnormal | 22% (26) | 45% (35) | 2.9 (1.6, 5.5) | .001 |
| Chromosome 11 abnormal | 8% (9) | 20% (16) | 3.1 (1.3, 7.4) | .02 |
| Chromosome 17 abnormal | 5% (6) | 14% (9) | 2.8 (1.0, 8.4) | .09 |
| Factor . | Mutated N = 123 . | Unmutated N = 82 . | Odds ratio (95% CI) . | P . |
|---|---|---|---|---|
| Sex-male | 49% (60) | 71% (58) | 2.5 (1.4, 4.6) | .003 |
| Abnormal karyotype | 60% (74) | 83% (67) | 3.2 (1.6, 6.3) | .001 |
| Karyotype evolution present | 18% (19) | 29% (18) | 1.9 (0.9, 3.9) | .15 |
| Atypical morphology | 18% (21) | 59% (47) | 6.6 (3.5, 12.7) | < .001 |
| CD38 ≥ 30% | 25% (27) | 72% (44) | 8.0 (3.9, 16.2) | < .001 |
| Disease stage | < .001 | |||
| A | 93% (114) | 67% (55) | 1.0 | |
| B | 2% (3) | 22% (18) | 12.4 (3.5, 44.0) | |
| C | 5% (6) | 11% (9) | 3.1 (1.1, 9.2) | |
| Chromosome 13 abnormal | 53% (64) | 29% (24) | 0.4 (0.2, 0.7) | .002 |
| Chromosome 12 abnormal | 22% (26) | 45% (35) | 2.9 (1.6, 5.5) | .001 |
| Chromosome 11 abnormal | 8% (9) | 20% (16) | 3.1 (1.3, 7.4) | .02 |
| Chromosome 17 abnormal | 5% (6) | 14% (9) | 2.8 (1.0, 8.4) | .09 |
Because many of these prognostic factors are statistically associated, logistic regression was used to determine which factors were independently associated with VH gene status (Tables4 and5). Deletion of chromosome 11q23, absence of chromosome 13q14 loss, atypical lymphocyte morphology, and more than 30% expression of CD38 were all independently associated with unmutated IGVH genes. The remaining factors were not statistically significant, but this needs to be interpreted with care because the adjusted odds ratios for sex and chromosome 17 abnormalities were still large (both > 2), and the CIs for all 6 factors were wide, particularly for chromosome 17 abnormalities, which had a low prevalence. This means that the prognostic value of these factors remains uncertain.
Logistic regression; independent factors associated withIGVH gene status
| Factors . | % with unmutated V gene status (n) . | Unadjusted odds ratio (95% CI) . | Odds ratio (95% CI) after adjusting for other 3 independent predictors . | P after adjusting for other 3 independent predictors . |
|---|---|---|---|---|
| CD38 | ||||
| < 30% | 17% (17) | — | — | |
| ≥ 30% | 62% (44) | 8.0 (3.9, 16.2) | 5.4 (2.3, 12.4) | < .001 |
| Chromosome 11 abnormality | ||||
| N | 37% (63) | — | — | |
| Y | 64% (16) | 3.1 (1.3, 7.4) | 5.2 (1.5, 18.3) | .010 |
| Morphology | ||||
| Typical | 25% (33) | — | — | |
| Atypical | 69% (47) | 6.6 (3.5, 12.7) | 4.6 (1.9, 11.0) | .001 |
| Chromosome 13 abnormality | ||||
| N | 50% (58) | — | — | |
| Y | 27% (24) | 0.4 (0.2, 0.7) | 0.4 (0.2, 0.8) | .02 |
| Factors . | % with unmutated V gene status (n) . | Unadjusted odds ratio (95% CI) . | Odds ratio (95% CI) after adjusting for other 3 independent predictors . | P after adjusting for other 3 independent predictors . |
|---|---|---|---|---|
| CD38 | ||||
| < 30% | 17% (17) | — | — | |
| ≥ 30% | 62% (44) | 8.0 (3.9, 16.2) | 5.4 (2.3, 12.4) | < .001 |
| Chromosome 11 abnormality | ||||
| N | 37% (63) | — | — | |
| Y | 64% (16) | 3.1 (1.3, 7.4) | 5.2 (1.5, 18.3) | .010 |
| Morphology | ||||
| Typical | 25% (33) | — | — | |
| Atypical | 69% (47) | 6.6 (3.5, 12.7) | 4.6 (1.9, 11.0) | .001 |
| Chromosome 13 abnormality | ||||
| N | 50% (58) | — | — | |
| Y | 27% (24) | 0.4 (0.2, 0.7) | 0.4 (0.2, 0.8) | .02 |
Logistic regression; other factors associated withIGVH gene status
| Other factors . | % With unmutated V gene status (n) . | Unadjusted odds ratio (95% CI) . | Odds ratio (95% CI) after adjusting for 4 independent factors . | P after adjusting for the 4 independent factors . |
|---|---|---|---|---|
| Chromosome 17 abnormality | ||||
| N | 35% (56) | — | — | |
| Y | 60% (9) | 2.8 (1.0, 8.4) | 3.7 (0.5, 25.2) | .19 |
| Stage | ||||
| A | 33% (55) | — | — | |
| B | 86% (18) | 12.4 (3.5, 44.0) | 2.6 (0.5, 12.3) | |
| C | 60% (9) | 3.1 (1.1, 9.2) | 3.1 (0.7, 14.6) | .21 |
| Sex | ||||
| Female | 28% (24) | — | — | |
| Male | 49% (58) | 2.5 (1.4, 4.6) | 2.1 (0.9, 5.0) | .09 |
| Karyotype abnormality | ||||
| N | 22% (14) | — | — | |
| Y | 48% (67) | 3.2 (1.6, 6.3) | 1.4 (0.5, 3.7) | .58 |
| Chromosome 12 abnormality | ||||
| N | 31% (43) | — | — | |
| Y | 57% (35) | 2.9 (1.6, 5.5) | 1.3 (0.5, 3.3) | .56 |
| Karyotype evolution | ||||
| N | 34% (45) | — | — | |
| Y | 49% (18) | 1.9 (0.9, 3.9) | 1.1 (0.4, 3.0) | .90 |
| Other factors . | % With unmutated V gene status (n) . | Unadjusted odds ratio (95% CI) . | Odds ratio (95% CI) after adjusting for 4 independent factors . | P after adjusting for the 4 independent factors . |
|---|---|---|---|---|
| Chromosome 17 abnormality | ||||
| N | 35% (56) | — | — | |
| Y | 60% (9) | 2.8 (1.0, 8.4) | 3.7 (0.5, 25.2) | .19 |
| Stage | ||||
| A | 33% (55) | — | — | |
| B | 86% (18) | 12.4 (3.5, 44.0) | 2.6 (0.5, 12.3) | |
| C | 60% (9) | 3.1 (1.1, 9.2) | 3.1 (0.7, 14.6) | .21 |
| Sex | ||||
| Female | 28% (24) | — | — | |
| Male | 49% (58) | 2.5 (1.4, 4.6) | 2.1 (0.9, 5.0) | .09 |
| Karyotype abnormality | ||||
| N | 22% (14) | — | — | |
| Y | 48% (67) | 3.2 (1.6, 6.3) | 1.4 (0.5, 3.7) | .58 |
| Chromosome 12 abnormality | ||||
| N | 31% (43) | — | — | |
| Y | 57% (35) | 2.9 (1.6, 5.5) | 1.3 (0.5, 3.3) | .56 |
| Karyotype evolution | ||||
| N | 34% (45) | — | — | |
| Y | 49% (18) | 1.9 (0.9, 3.9) | 1.1 (0.4, 3.0) | .90 |
Prognostic factors
Male sex, advanced stage, atypical morphology, unmutatedIGVH genes, CD38 expression, trisomy 12, and deletion at 11q23 or 17p13 but not at 13q14 are all poor prognostic factors in a univariate analysis of CLL-related deaths (Table6). The same parameters remain significant when all deaths are evaluated and also when the 17 cases referred from other centers are excluded.
Proportional hazards regression for deaths from CLL-related causes
| . | 10-y death rate (SE) . | Unadjusted hazard ratio (95% CI) . | Log-rank test (P) . | Hazard ratio (95% CI) after adjusting for other 2 independent predictors . | P after adjusting for other 2 independent predictors . |
|---|---|---|---|---|---|
| Independent predictors | |||||
| V gene status | |||||
| Mutated | 6% (3%) | — | — | ||
| Unmutated | 55% (7%) | 14.6 (6.3, 33.9) | < .001 | 6.9 (2.6, 18.3) | < .001 |
| Disease stage | |||||
| A | 15% (3%) | — | — | ||
| B or C | 77% (10%) | 10.2 (5.4, 19.1) | < .001 | 4.6 (2.1, 10.2) | < .001 |
| Chromosome 17 abnormality | |||||
| N | 17% (4%) | — | — | ||
| Y | 100% | 9.1 (4.0, 20.7) | < .001 | 3.3 (1.4, 7.8) | .006 |
| Other factors | |||||
| Karyotype abnormality | |||||
| N | 4% (3%) | — | — | ||
| Y | 35% (5%) | 5.5 (2.1, 14.0) | < .001 | 2.2 (0.8, 6.3) | .13 |
| Chromosome 11 abnormality | |||||
| N | 20% (4%) | — | — | ||
| Y | 45% (12%) | 3.6 (1.8, 7.1) | < .001 | 2.0 (0.8, 5.1) | .14 |
| Karyotype evolution | |||||
| N | 13% (4%) | — | — | ||
| Y | 50% (10%) | 4.1 (2.1, 8.1) | < .001 | 2.0 (0.8, 5.0) | .16 |
| Sex | |||||
| Female | 13% (4%) | — | — | ||
| Male | 36% (6%) | 2.2 (1.2, 4.0) | .01 | 1.7 (0.7, 4.1) | .21 |
| CD38 | |||||
| Less than 30% | 8% (3%) | — | |||
| More than 30% | 39% (8%) | 4.4 (1.9, 9.8) | < .001 | 1.7 (0.6, 4.4) | .29 |
| Morphology | |||||
| Typical | 11% (3%) | — | — | ||
| Atypical | 49% (8%) | 4.2 (2.2, 7.8) | < .001 | 1.1 (0.5, 2.5) | .85 |
| Chromosome 13 abnormality | |||||
| N | 31% (5%) | — | — | ||
| Y | 17% (5%) | 0.6 (0.3, 1.1) | .07 | 0.8 (0.4, 1.8) | .58 |
| Chromosome 12 abnormality | |||||
| N | 18% (4%) | — | — | ||
| Y | 38% (8%) | 2.0 (1.0, 3.7) | .03 | 0.7 (0.3, 1.5) | .34 |
| . | 10-y death rate (SE) . | Unadjusted hazard ratio (95% CI) . | Log-rank test (P) . | Hazard ratio (95% CI) after adjusting for other 2 independent predictors . | P after adjusting for other 2 independent predictors . |
|---|---|---|---|---|---|
| Independent predictors | |||||
| V gene status | |||||
| Mutated | 6% (3%) | — | — | ||
| Unmutated | 55% (7%) | 14.6 (6.3, 33.9) | < .001 | 6.9 (2.6, 18.3) | < .001 |
| Disease stage | |||||
| A | 15% (3%) | — | — | ||
| B or C | 77% (10%) | 10.2 (5.4, 19.1) | < .001 | 4.6 (2.1, 10.2) | < .001 |
| Chromosome 17 abnormality | |||||
| N | 17% (4%) | — | — | ||
| Y | 100% | 9.1 (4.0, 20.7) | < .001 | 3.3 (1.4, 7.8) | .006 |
| Other factors | |||||
| Karyotype abnormality | |||||
| N | 4% (3%) | — | — | ||
| Y | 35% (5%) | 5.5 (2.1, 14.0) | < .001 | 2.2 (0.8, 6.3) | .13 |
| Chromosome 11 abnormality | |||||
| N | 20% (4%) | — | — | ||
| Y | 45% (12%) | 3.6 (1.8, 7.1) | < .001 | 2.0 (0.8, 5.1) | .14 |
| Karyotype evolution | |||||
| N | 13% (4%) | — | — | ||
| Y | 50% (10%) | 4.1 (2.1, 8.1) | < .001 | 2.0 (0.8, 5.0) | .16 |
| Sex | |||||
| Female | 13% (4%) | — | — | ||
| Male | 36% (6%) | 2.2 (1.2, 4.0) | .01 | 1.7 (0.7, 4.1) | .21 |
| CD38 | |||||
| Less than 30% | 8% (3%) | — | |||
| More than 30% | 39% (8%) | 4.4 (1.9, 9.8) | < .001 | 1.7 (0.6, 4.4) | .29 |
| Morphology | |||||
| Typical | 11% (3%) | — | — | ||
| Atypical | 49% (8%) | 4.2 (2.2, 7.8) | < .001 | 1.1 (0.5, 2.5) | .85 |
| Chromosome 13 abnormality | |||||
| N | 31% (5%) | — | — | ||
| Y | 17% (5%) | 0.6 (0.3, 1.1) | .07 | 0.8 (0.4, 1.8) | .58 |
| Chromosome 12 abnormality | |||||
| N | 18% (4%) | — | — | ||
| Y | 38% (8%) | 2.0 (1.0, 3.7) | .03 | 0.7 (0.3, 1.5) | .34 |
Using the Cox proportional hazards regression to determine which of these prognostic factors were independent predictors of mortality, only clinical stage, unmutated IGVH genes, and chromosome 17 abnormalities remain significant when deaths from CLL- related causes (Table 6) and all deaths (data not shown) are assessed. The hazard ratio remains high for some factors including del11q23 and CD38 expression, which were not statistically significant, and because sample size is relatively small the possibility of a type 2 error exists. The adjusted hazard ratios for IGVH gene status and chromosome 17 status were significantly higher in stage A patients than in stage B and C patients combined (P = .01 for each of the interaction events).
When the analysis is confined to stage A patients, the hazard ratio forIGVH gene status (after adjusting for chromosome 17 status) is 17.9 (3.9-82.7; P < .001) and the hazard ratio for chromosome 17 status (after adjusting for IGVH gene status) was 11.2 (3.2-35.9; P < .001).
Median survival data
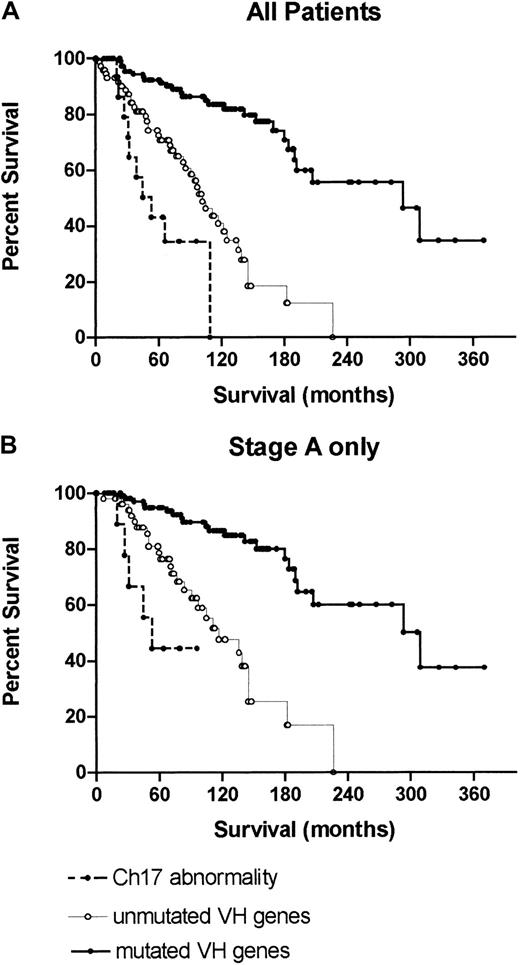
The median survival (95% CI) of patients included in the study was 180 months (range, 146-214 months) for all deaths and 293 months (range, 212-374 months) for disease-related deaths. The impact of cytogenetic and genetic abnormalities on survival is shown in Figure2. The median survival of patients with a normal karyotype and no evidence of trisomy 12, or deletions of 11q23, 13q14, or 17p13 using additional molecular techniques was 209 months. Patients with deletions of 13q14 as the sole abnormality had a median survival of 292 months, those with trisomy 12, 122 months, those with del11q23, 117 months, and those with loss or mutation of thep53 gene, 47 months. Figure 3A illustrates the superior survival of patients with mutatedIGVH genes (median survival, 310 months) compared to cases with unmutated IGVH genes (median survival, 119 months) and demonstrates the poor prognosis of patients with p53 loss or mutation, regardless of IGVH gene status.
Survival of patients.
A comparison of the survival of patients with del/t13q14 as the sole abnormality detected by karyotype or Southern blotting; +12 or del11q23 detected by karyotype analysis or interphase FISH; loss or mutation of p53 detected by interphase FISH or mutation analysis; and patients with no abnormality using any of the above techniques. The median survival of patients with no abnormality, del/t13q14 as the sole abnormality, +12, del11q23, or p53 loss/mutation was 292, 209, 122, 117, and 47 months, respectively.
Survival of patients.
A comparison of the survival of patients with del/t13q14 as the sole abnormality detected by karyotype or Southern blotting; +12 or del11q23 detected by karyotype analysis or interphase FISH; loss or mutation of p53 detected by interphase FISH or mutation analysis; and patients with no abnormality using any of the above techniques. The median survival of patients with no abnormality, del/t13q14 as the sole abnormality, +12, del11q23, or p53 loss/mutation was 292, 209, 122, 117, and 47 months, respectively.
Overall survival of patients with loss or mutation of p53 regardless of IGVH gene status compared to patients with mutated or unmutated IGVH genes with no p53 abnormality.
(A) All patients; (B) stage A patients only. The median survival of patients with mutated or unmutated IGVH genes or a p53 abnormality was 310, 119, and 47 months, respectively, for all patients, and 327, 115, and 54 months, respectively, for stage A patients.
Overall survival of patients with loss or mutation of p53 regardless of IGVH gene status compared to patients with mutated or unmutated IGVH genes with no p53 abnormality.
(A) All patients; (B) stage A patients only. The median survival of patients with mutated or unmutated IGVH genes or a p53 abnormality was 310, 119, and 47 months, respectively, for all patients, and 327, 115, and 54 months, respectively, for stage A patients.
Similar differences in survival were seen in patients presenting with stage A disease (Figure 3B).
Discussion
This study confirms that unmutated IGVH genes, high expression of CD38, atypical lymphocyte morphology, trisomy 12, del11q23, and loss or mutation of the p53 gene are all poor prognostic factors when evaluated in a univariate analysis of a large series of patients with CLL presenting predominantly with Binet stage A disease. Care was taken to ensure that only samples taken at or close to the time of presentation were tested. This is of particular importance when assessing the prognostic significance of p53 abnormalities, which are frequently acquired during the course of the disease. A potential cause of bias was the inclusion of cases from other centers because of their cytogenetic abnormalities. However, only presentation data were used, and the prognostic factors determined by univariate analyses remained unchanged when these cases were excluded.
Whether the degree of somatic hypermutation has prognostic significance in CLL is a controversial issue. Initial studies used a cutoff of 98% homology to the nearest germline sequence to allow for the presence of polymorphisms in unmutated IGVH genes. Stilgebauer et al33 have found that in their hands 96% homology provides a better separation of patients with differing prognoses. However, their use of a PCR primer within the framework 1 region rather than theIGVH gene leader sequence may have resulted in an apparent increase in the percentage of mutations, because the frequency of mutation in the 5′ framework 1 region is very low (personal observation of Z.A.D. in more than 250 IGVH gene sequences). Applying the Youden index to our own data shows that 98% homology is the optimum cutoff regardless of whether all deaths or only CLL-related deaths are analyzed. This result is consistent with the concept that the better prognosis of patients with somatic hypermutation reflects the origin of the disease from a postgerminal center B cell. We have previously noted a close correlation between trisomy 12 and unmutatedIGVH genes in patients with clinically aggressive disease and between structural abnormalities of chromosome 13q14 and mutatedIGVH genes in patients with benign disease.34These findings raised the question as to whether or not the difference in survival between the 2 IGVH gene subsets was a consequence of differing susceptibility of each subgroup to acquire subsequent genetic abnormalities with prognostic significance. Pettitt et al35 have recently shown that failure to up-regulate p21 expression in response to in vitro irradiation identifies cases with p53 dysfunction due either to mutations of the p53 orATM (ataxia telangiectasia mutated) genes. They have subsequently shown that p53 dysfunction is confined to patients with less than 5% divergence from the nearest germline IGVH gene and hypothesize that p53 and ATM mutations are at least partly responsible for the poor prognosis of patients with unmutatedIGVH genes.36
Our current study confirms the association between trisomy 12 and del11q23 with unmutated IGVH genes and between structural abnormalities of 13q14 and mutated IGVH genes. We were unable to demonstrate a significant association between IGVHgene status and loss or mutation of the p53 gene although the hazard ratio is high and a type 2 error, reflecting the small patient sample and low prevalence of the latter, is possible. However, the finding of p53 mutations in cases with heavily mutatedIGVH genes is clearly documented. We did not find CD38 expression to have independent prognostic significance despite showing that CD38 expression was an independent prognostic factor in patients of known IGVH gene status in a recent analysis of 145 patients, all of whom were included in this larger study.24 After adjusting for disease stage and V gene status the previous study found an odds ratio (95% CI) of 3.1 (1.1-8.8; P = .04) between CD38- and CLL-related deaths. The present analysis, after adjusting for these same 2 factors, produced a smaller odds ratio with a narrower CI, reflecting the increased sample size (odds ratio = 2.1 [range, 0.9-4.8];P = .09). Further adjustment for chromosome 17 status reduced the odds ratio further to 1.7 (0.6-4.4; P = .29; Table 6). Thus the apparent difference in results between the 2 sets of analyses is not substantial, is of borderline statistical significance, and can partly be explained by the further adjustment for chromosome 17 status. Multivariate analysis of all potential factors in a much larger group of patients is required to clarify the true prognostic significance of CD38.
The most important finding in this study was that p53 loss or mutation had independent prognostic significance in patients with knownIGVH gene status. In 13 of 15 patients, of whom 9 had stage A disease, the p53 abnormalities were detected at presentation confirming recent data that clinically significant genetic abnormalities may be found in Binet stage A disease.37 We used a combination of karyotype analysis, interphase FISH, and flow cytometry to screen for abnormalities of 17p13 and p53 dysfunction. Karyotypic analysis and interphase FISH were of comparable sensitivity and superior to flow cytometry, but none of these assays identified all the cases with a p53 mutation. A more sensitive screening test, direct genomic sequencing, or DNA microarray analysis should ideally be used.38
Despite an overall poor prognosis there was considerable variation in the survival of patients presenting with a p53 mutation. A much larger series of cases with p53 mutation will be required to determine whether these differences reflect the size of the p53 clone, the site of the mutation and function of the mutant protein, IGVH gene status, or other unknown variable.
The failure of del11q23 to emerge as an independent prognostic factor is surprising in view of previous studies demonstrating the poor prognosis of patients with this abnormality, although the CI for the hazard ratio is wide and so might be a type 2 error. Also, in our current study bulky lymphadenopathy was confined to those patients with del11q who had unmutated IGVH genes (data not shown). Schaffner et al39 found ATM mutations in only a subset of patients with del11q23, and Stankovic et al40 detected loss of heterozygosity in only 2 of 16 cases with an ATM mutation suggesting that deletions involving 11q23 may have differing molecular consequences with varying prognostic significance.
In summary, our results indicate that knowledge of IGVH gene status and p53 dysfunction should facilitate the design of clinical trials to assess the potential of new treatments. In particular younger patients with unmutated IGVH genes or p53 mutations should be considered for intensive or novel treatments regardless of clinical stage. Whereas IGVH gene status remains constant throughout the course of the disease, additional genetic abnormalities may be acquired and further studies are necessary to determine the clinical value of sequential genetic analysis.
We express our gratitude to Mo Tiller for excellent technical assistance. We thank Dr S. Johnson (Taunton), Dr P. Stross (Chichester), Dr M. Hilali (Dorchester), and Dr J. Rees (Cambridge) for referring patients.
Supported by research grants from the Bournemouth Leukaemia Fund, Tenovus, the Leukaemia Research Fund, and the National Health Service Executive South and West.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David G. Oscier, Department of Haematology, Royal Bournemouth Hospital, Castle Lane East, Bournemouth, BH7 7DW, United Kingdom; e-mail: david.oscier@lineone.net.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal