Although idiopathic thrombocytopenic purpura (ITP) is the most common autoimmune hematologic disorder, little is known about the associated autoantibodies on a molecular level. Consequently, diagnostic assays and therapy for ITP lack specificity. To avoid technical limitations imposed by B-cell immortalization methods, we used repertoire cloning (Fab/phage display) to clone platelet autoantibodies and examine the relation between immunoglobulin (Ig) gene usage, clonality, and antigen specificity. Phage display libraries were constructed from splenocytes from 2 patients with chronic ITP, and competitive cell-surface selection was used to isolate several dozen unique IgG platelet-specific autoantibodies. Platelet-reactive Fabs in both patients were associated almost exclusively with rearrangements of a single Ig heavy-chain variable-region gene (VH3-30), despite an apparent diversity of antigen specificities. Comparative analysis of platelet-reactive Fab Ig gene rearrangements from each patient suggested that they evolved from a restricted number of B-cell clones through somatic mutation with high replacement-to-silent mutation ratios. Although VH3-30–encoded heavy chains were found with light chains encoded by several different Ig genes, molecular repairing experiments showed exquisite restriction on the specific heavy- and light-chain pairings that permitted platelet reactivity. Together, these data suggest that the development of platelet-reactive antibodies associated with ITP is driven by an encounter with diverse platelet antigens through the clonal expansion of B cells using genetically restricted and highly specific combinations of heavy- and light-chain gene products. The extraordinarily high usage of the VH3-30 heavy-chain gene in these patients has implications for the pathogenesis, diagnosis, and management of chronic ITP.
Introduction
Idiopathic thrombocytopenic purpura (ITP) is a common immunohematologic disorder caused by platelet-reactive autoantibodies.1 The clearance of antibody-coated platelets by tissue macrophages is accelerated, and in some cases, the antibodies also impair platelet production. Childhood-type ITP is self-limiting in about 80% of cases and may be associated with a previous viral infection. Adult-onset ITP is a chronic illness in more than 70% of cases and may occur in association with other disorders, including systemic lupus erythematosus (SLE), lymphoproliferative diseases, common variable immunodeficiency (CVID) disease, and human immunodeficiency virus (HIV) infection. The decision to treat patients with ITP takes into account the patient's age and disease severity and the anticipated natural history of the disorder. Therapy is initially directed toward impeding the clearance of antibody-coated platelets by using glucocorticoids, splenectomy, anti–blood group D [(anti-Rh(D)] immunoglobulin (Ig), intravenous γ-globulin (IVIG), and other treatments. Immunosuppressive therapy is nonspecific, often toxic, and typically reserved for patients with refractory disease.
Numerous studies have been performed to characterize the pathogenic autoantibodies responsible for platelet destruction and thereby provide a reliable way to diagnose ITP, understand its pathogenesis, and predict responsiveness to therapy. IgG antibodies that react with platelet glycoprotein (GP) IIb/IIIa and GPIb/IX have been identified in some patient serum samples and platelet eluates2-5; however, other platelet antigens also appear to be targeted,5-13 and in many cases, the antibody specificity cannot be determined or even detected.1 Furthermore, there is no formal proof that any single subset of antibodies—for example, those directed at GPIIb/IIIa—are responsible for platelet destruction. Consequently, the clinical utility of measuring serum or platelet-elutable Ig is unknown and does not have a definitive role in the diagnosis or treatment of ITP or in distinguishing between the adult-onset and childhood-onset forms of the disease.14 As a result, the diagnosis of ITP remains one of exclusion and the usefulness of available platelet-antibody tests to confirm or exclude the diagnosis independent of other criteria has not been established.1
This situation illustrates the difficulty involved in characterizing a pathologic autoimmune response by analyzing polyclonal serum. To understand clonality, genetic origin, somatic mutation, and the molecular basis of pathogenicity, repertoires of IgG antiplatelet autoantibodies, eg, those produced in vitro from the B cells of affected patients, must be studied. Conventional B-cell immortalization approaches for cloning human monoclonal antibodies result in low transformation frequencies and have a propensity for generating IgM-producing clones, thus causing a sampling bias.15,16Consequently, all but one17 of the reported human antiplatelet autoantibodies isolated from patients with ITP have been of the IgM class and no more than 2 or 3 unique antibodies have been isolated from a given patient.11,12 18-20 As a result, it has been difficult to assess the genetic diversity among ITP-associated autoantibodies within an individual patient, among patients, and in different clinical settings.
To address this problem, we combined antibody phage display, a molecular approach for cloning human immune repertoires,21with a novel competitive cell-surface–selection scheme22to isolate and study repertoires of IgG antiplatelet autoantibodies from 2 unrelated patients with chronic ITP. Using this strategy, we isolated dozens of IgG platelet-reactive autoantibodies from each patient, thus permitting a comprehensive analysis of their genetic origin, extent of somatic mutation, and clonal relatedness. Our results suggest that the development of platelet-reactive autoantibodies is driven by an encounter with diverse platelet antigens through the clonal expansion of B cells using genetically restricted and highly specific combinations of heavy- and light-chain gene products.
Materials and methods
Platelet preparations
Platelet-rich plasma (PRP) was prepared by centrifuging (500g) freshly isolated whole blood collected in sodium citrate (final concentration, 10.5 mM/L) containing 3 μM/L prostaglandin E1 (PGE1; Sigma Chemicals, St Louis, MO) at room temperature for 15 minutes. For some experiments, platelets were obtained from fresh banked platelet concentrates derived from CP2D-anticoagulated whole blood (Fenwall; Baxter Healthcare, Deerfield, IL). PRP from both sources was washed 3 times in acid-citrate-dextrose (ACD; 145 mM/L sodium chloride, 5 mM/L citric acid, 9 mM/L sodium citrate, and 17 mM/L dextrose [pH 6.5]) supplemented with 1% wt/vol bovine serum albumin (BSA).
Patients
Fab/phage display libraries were constructed from splenic mononuclear cells from 2 unrelated adults with chronic ITP (ITP patient A and ITP patient B) and one control patient with thrombocytopenia but not ITP. Both patient A (a 56-year-old man) and patient B (a 43-year-old woman) had ITP refractory to prednisone and IVIG for at least 8 months. After splenectomy, platelet counts rose to the normal range. Patient A subsequently died of unrelated causes, and patient B has been in clinical remission for more than 4 years. Splenocytes from the control patient, a 65-year-old man with nonimmune, multifactorial thrombocytopenia, were harvested at autopsy after he died of respiratory failure.
Construction of Fab/phage display libraries
Using previously described methods for cloning IgG1κ and λ immune repertoires,23 we prepared total RNA from about 108 splenocytes, amplified heavy- and light-chain–rearranged Ig gene segments by reverse transcriptase–polymerase chain reaction, and cloned the DNA into a phagemid expression vector (pComb3H; Scripps Institute, La Jolla, CA). After electroporation into XL1-Blue bacteria (Stratagene, La Jolla) and coinfection with VCSM13 helper phage (Stratagene), Ig DNA was packaged into filamentous phage particles that expressed the human Fab molecules fused to the pIII bacteriophage coat protein.
Panning Fab/phage display libraries
Fab/phage display libraries were enriched for platelet-reactive Fabs by a modification of a previously described method using competitive cell-surface selection and magnetically activated cell sorting.22 Platelets were washed free of BSA in phosphate-buffered saline (PBS) and PGE1, resuspended to a concentration of 5 × 108/mL, and surface biotinylated by adding sulfo-N-hydroxysuccinimide biotin (Pierce, Rockford, IL) to 400 μg/mL. After 2 washes with ACD and BSA, 2 × 108biotinylated platelets were incubated with 20 μL streptavidin-coated paramagnetic microbeads (Miltenyi Biotec, Sunnyvale, CA) for 10 minutes at room temperature in a total volume of 100 μL ACD, BSA, and PGE1. ACD-BSA buffer (1 mL) containing about 5-fold excess (by surface area) human red blood cells (RBCs; 1 × 107) was added. The cell admixture was centrifuged and resuspended in 50 μL ACD, BSA, and PGE1 containing about 3 × 1011 colony-forming units of Fab/phage display library. After a 2-hour incubation at room temperature with intermittent mixing, the suspension of platelets, RBCs, and phage was loaded on a MiniMACS column (Mitenyi Biotec, Germany) pre-equilibrated with ACD and BSA. Column washes (to remove RBCs and irrelevant Fab-phage), elution of platelet-bound Fab-phage, and amplification of panned libraries were performed as described previously.22
Production of soluble antiplatelet Fab Ig
To screen, isolate, and characterize individual monoclonal platelet-binding Fabs, randomly picked bacterial colonies derived from phage titering plates were grown to an optical density600of 0.5, isopropyl-β-d-thiogalactopyranoside (1 mM/L) was added, and cultures were shaken overnight at 30°C. Soluble Fabs were isolated from bacterial pellets by osmotic shock24 and used in flow cytometric experiments and enzyme-linked immunosorbent assays (ELISAs) without further purification. Where indicated, soluble Fabs were purified by nickel-chelation chromatography.23Aliquots of bacterial pellets were used to prepare plasmid DNA (Qiawell Plus; Qiagen, Valencia, CA) for nucleotide sequencing or antibody chain shuffling. Heavy- and light-chain DNA was sequenced and analyzed as described previously.24 Because of the large number of sequences (> 60), only alignments of the predicted amino acid sequences for a subset of antibodies are shown here; the rest of the sequence data are provided on the Blood website; see the Supplemental Data Set link at the top of the online article.
Characterization of antibody binding by flow cytometry
Platelets were stained by using 5 μL PRP (∼ 5 × 106 platelets) and 50 μL Fab. After a 30-minute incubation, platelets were washed with ACD and BSA, and bound antibody was detected by using a phycoerythrin (PE)-conjugated F(ab′)2 fragment of goat antihuman F(ab′)2-specific Ig (Jackson ImmunoResearch, West Grove, PA) diluted 1:25 in wash buffer. Samples were analyzed by using a microfluorometer (FACScan; Becton Dickinson, Mountain View, CA). Forward- and side-scatter gates for platelet populations were determined by using murine antihuman GPIIIa (SSA6; Dr J. Bennett, University of Pennsylvania) counterstained with PE-conjugated goat antimouse reagent (Southern Biotechnology, Birmingham, AL). Platelets from 3 unrelated donors with type I Glanzmann thrombasthenia were provided by Dr M. Poncz (University of Pennsylvania). A stable K562 cell line expressing GPIa/IIa was provided by Dr M. Zutter (Washington University, St Louis, MO).
Blocking experiments were conducted to compare the repertoires of recombinant platelet-reactive autoantibodies from ITP patients A and B with those in the serum of other patients with chronic ITP. Platelet aliquots were preincubated with each of 19 different ITP serum samples or a pool of normal serum, then mixed with antibodies from ITP patient A or B expressed as phage displayed Fabs. Blocking of recombinant patient autoantibodies by ITP serum was then detected with biotinylated anti-M13 antibody and PE-streptavidin.24 Binding of recombinant autoantibodies in the presence of normal serum was defined as 100%, and inhibition in the presence of ITP serum was normalized to that value. Administration of IVIG to ITP patients A and B just before splenectomy precluded use of their serum in competition assays.
Characterization of antibody binding by ELISA and immunofluorescence
Antibodies to platelet GPIIb/IIIa, GPIb/IX, or GPIa/IIa were measured by using a PakAuto kit (GTI, Brookfield, WI); those to cardiolipin were assessed with a QuantaLite kit (Inova, San Diego, CA). Binding to cytoplasmic or nuclear determinants was assessed by immunofluorescence with HEp-2 cells (ANA Kit, Antibodies Incorporated, Davis, CA).
Immunoprecipitation of platelet-Fab immune complexes
Immunoprecipitation of biotinylated platelet membrane proteins was performed as described previously25 except that Protein L (Pierce) was used instead of Protein A to capture immune complexes. Precipitated material was electrophoresed on 4% to 12% polyacrylamide gels under nonreducing and reducing conditions and electrophoretically blotted on nitrocellulose membranes. Precipitated, biotinylated platelet membrane proteins were detected with biotinylated horseradish peroxidase–avidin complexes (ABC Staining Kit, Pierce).
Light-chain–library shuffling
To randomly pair the H44 heavy chain with a library of light chains, 10 μg plasmid DNA from clone H44L4 (a GPIIb/IIIa-specific Fab isolated from ITP patient A) was digested for 6 hours at 37°C withSacI and XbaI (Roche Molecular Biochemicals, Indianapolis, IN) to remove the endogenous light-chain L4, and the heavy-chain–containing vector fragment was gel purified. A preparation of κ and λ light-chain segments from the original, unpanned ITP patient A library was obtained by digesting an equivalent amount of plasmid DNA purified from the bacterial pellet obtained during library preparation with SacI/XbaI and gel purifying the excised light chains. Vector containing heavy-chain H44 was then ligated to the library of light chains and electroporated into XL1-Blue bacteria. Transformants were plated on carbenicillin-containing Luria-Bertani plates from which antibody clones were randomly selected, produced as soluble Fab preparations, and assayed for platelet binding by flow cytometry. Plasmid minipreparations were performed on the bacterial pellets derived from the expression experiments, and nucleotide sequencing was done to verify the presence of heavy-chain H44 and to determine the sequence of the light chain to which it randomly paired.
Exchanging heavy and light chains among platelet-binding clones
Light-chain gene segments from clones H36/L76, H44/L4, and H47/L64 were freed from their respective plasmid DNAs bySacI/XbaI digestion, and the restriction products from the 3 clones (ie, 3 heavy-chain–containing plasmids and 3 free light chains) were combined. Religation regenerated the 3 original Fabs and created 6 novel heavy-chain–light-chain pairs. After bacterial transformation, several dozen bacterial clones were randomly selected to produce Fabs for platelet-binding assays and to isolate plasmid DNA to determine heavy- and light-chain composition.
Fab binding to modified staphylococcal protein A
Binding of Fabs to the superantigen domain of staphylococcal protein A (SpA) was measured by ELISA using SpA that had been chemically modified with iodine monochloride to destroy its native Fc-binding domain (designated mod-SpA).26 Mod-SpA (1 μg in 50 μL) was coated on the wells of a 96-well microplate and incubated overnight at 4°C. After a rinse with distilled water, wells were blocked for 1 hour at 37°C with PBS and 1% BSA, and Fab samples were added (50 μL/well). After a 2-hour incubation at 37°C, the wells were washed 3 times with PBS, and a mixture of alkaline phosphatase–conjugated goat antihuman κ (1:10 000) and λ (1:5000) light-chain reagents was added (Sigma Chemical). Wells were incubated at 37°C for an additional hour, washed again with PBS, and developed with P-nitrophenyl phosphate.
Results
Isolation of monoclonal human platelet autoantibodies
The purpose of this study was to characterize on a genetic level the repertoires of platelet autoantibodies in chronic ITP. To isolate the repertoires, Fab/phage display technology was used to avoid the technical limitations inherent in experimental approaches that rely on B-cell immortalization to produce human monoclonal antibodies. IgG κ and λ libraries were constructed from splenic lymphocytes from 2 patients with chronic ITP and a control patient with multifactorial thrombocytopenia not due to ITP. The libraries (each comprising > 2 × 108 independent transformants) were panned against intact platelets (as opposed to isolated platelet membrane GPs) to present the libraries with all possible autoantigenic determinants and to do so in a physiologically relevant manner that would preserve native antigen structure and optimize capture. By employing a magnetically activated competitive cell-surface panning strategy in which selection of platelet binders was done in the presence of an irrelevant cell type (RBCs), we prevented the capture of panreactive or nonspecific Fab-phage.
Individual Fab clones were randomly selected from platelet-selected libraries and assessed for platelet binding by flow cytometry. For the 2 patients with ITP, 78 of 294 clones were positive, of which 39 were determined to be unique antibodies on the basis of the heavy- and light-chain DNA sequence. In contrast, only 1 of 77 additional clones randomly selected from the unpanned ITP libraries and none of 59 clones isolated from the control libraries (16 from the original unpanned and 43 from the platelet-selected libraries) showed platelet reactivity.
We then asked whether the panned ITP Fab libraries would bind to a cohort of antigens recognized by polyclonal antibodies in serum from patients with ITP. The capacity of 19 ITP serum samples to block the binding of phage displayed Fabs was assessed by flow cytometry using fluorescently labeled anti-M13 (phage) antibody relative to normal control serum samples. Fab-phage from ITP patient A was inhibited 25% ± 15% (range, 0%-41%) on average; that from ITP patient B was inhibited 41% ± 17% (range, 14%-74%). Analogous studies with serum from these patients were precluded by administration of IVIG immediately before sample collection.
Sequence analysis of platelet autoantibodies
The heavy- and light-chain nucleotide sequences from the 39 unique platelet autoantibodies were aligned with the V Base Directory of Human V Gene Segments27 to examine their genetic origins and possible genetic interrelatedness. As shown in Figure1 (red boxes), all heavy chains from ITP patient A (6 of 6) and all but 4 heavy chains from ITP patient B (29 of 33) used VH3-30. Usage of light-chain variable-region genes was less restricted but comprised a limited set of VLgenes, including the Vκ genes A19/A3, A27, and L6.
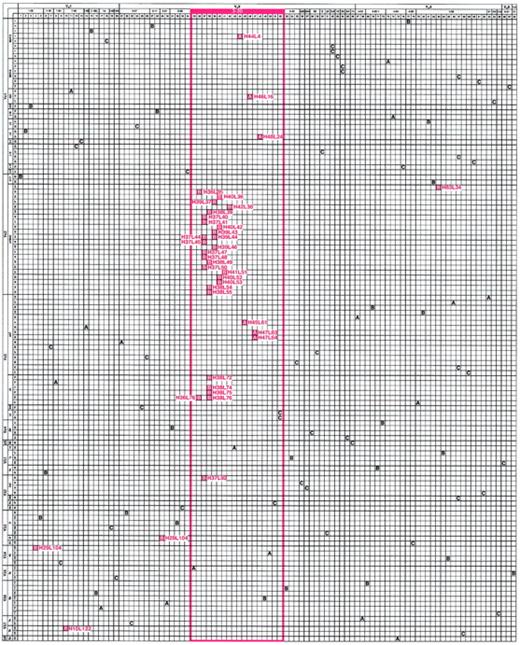
Matrix illustrating the genetic composition of platelet autoantibodies.
The horizontal axis represents the unique γ heavy chains (H01 through H98) and the vertical axis represents the unique κ and λ light chains (L01 through L124) used by antibodies cloned and sequenced from the patients with ITP and the control patient. The letter at the intersection of a heavy-chain–light-chain pair indicates the composition of a platelet-reactive (red box) or platelet-unreactive (clear box) antibody isolated from ITP patient A or B or control patient C. For positive clones, H and L designations are indicated. The order of heavy chains (left to right) and light chains (top to bottom) was determined by multiple alignments based on amino acid similarity and then grouped by putative Ig variable-region germline gene and germline gene family. Note the marked predominant use of the VH3-30 germline gene to encode platelet-binding antibodies in both patient repertoires (red-framed area).
Matrix illustrating the genetic composition of platelet autoantibodies.
The horizontal axis represents the unique γ heavy chains (H01 through H98) and the vertical axis represents the unique κ and λ light chains (L01 through L124) used by antibodies cloned and sequenced from the patients with ITP and the control patient. The letter at the intersection of a heavy-chain–light-chain pair indicates the composition of a platelet-reactive (red box) or platelet-unreactive (clear box) antibody isolated from ITP patient A or B or control patient C. For positive clones, H and L designations are indicated. The order of heavy chains (left to right) and light chains (top to bottom) was determined by multiple alignments based on amino acid similarity and then grouped by putative Ig variable-region germline gene and germline gene family. Note the marked predominant use of the VH3-30 germline gene to encode platelet-binding antibodies in both patient repertoires (red-framed area).
Selective usage of a particular heavy- or light-chain gene in a cohort of antibodies may occur because of in vivo or in vitro preselection factors (eg, greater gene usage by the pre-existing pool of B cells or cloning artifacts) or if an encounter with antigen drives clonal expansion and somatic mutation of a restricted population of B cells that use that particular gene. To address the first possibility, we assessed the diversity of the unpanned ITP patient A and patient B libraries. Analysis of the heavy and light chains of a random cohort of 43 of the 76 non–platelet-binding clones from the original libraries found no duplicate sequences and marked heterogeneity in V gene representation before selection for platelet binding (Figure 1, A and B clear boxes). Specifically, 20 different VH genes and 20 different VL genes were represented and their distribution was similar to that typically found for IgG-secreting lymphocytes in the repertoire of adults.28 The absence of platelet reactivity of recombinant antibodies from the control library was not due to inefficient library construction or lack of VH3-30 heavy-chain representation (Figure 1, C boxes), since 26 different VH genes and 25 different VL genes were used, including 3 antibodies encoded by VH3-30. Therefore, the highly restricted, near-total use of VH3-30 by the 39 platelet-binding ITP patient autoantibodies did not reflect a skewed representation of genes within the original pool of splenic lymphocytes, nor was it the result of a cloning artifact introduced during construction of the Fab/phage display libraries.
We next addressed the possibility that the increased usage of a given V gene results from clonal expansion of restricted B-cell populations. To do this, we exploited the fact that rearranged Ig genes have extensive diversity; ie, there is only a remote probability that 2 B cells will not only randomly select an identical combination of VH, D, and JH (for heavy chain) or VL and JL (for light chain) gene segments but will also splice the genes together to create identical junctional regions. Alignments of the heavy- and light-chain variable-region amino acid sequences of a cohort of 39 platelet autoantibodies from ITP patients A and B were performed. Examination of the complete set of heavy-chain sequences (see the Blood website's Supplemental Data Set) revealed evidence of clonal expansion for a subset of B cells using VH3-30 in both patients (Figure2A). The members of each clone appear to have resulted from recombination of VH3-30, D1-26, and JH4b gene segments, and within each clone, they showed identical junctional regions. The fact that the CDR3 regions of clone A and clone B were quite distinct indicates that neither resulted from an interlibrary contaminant.
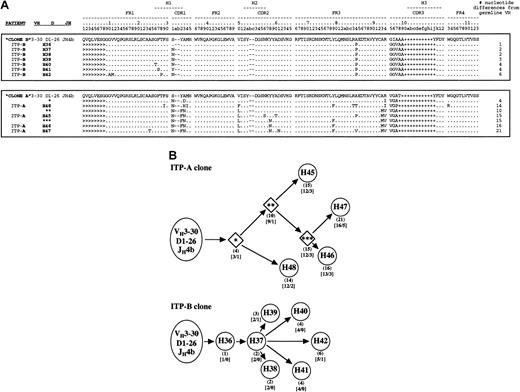
Alignment of clonally related platelet autoantibody heavy-chain amino acid sequences and their putative ontogenic trees.
The H and L nomenclature is the same as in Figure 1. (A) Groups of related sequences comprising expanded heavy-chain clones in each patient library (clone A and clone B) are enclosed in boxes. The coalignment with the rest of the 16 unique heavy chains is available on the Blood website; see the Supplemental Data Set link at the top of the online article. For clone A, the putative intermediate heavy-chain sequences are also shown (1, 2, and 3 asterisks). The number of nucleotide differences from germline VH is tabulated to the right of each sequence. Because D segments showed poor homology with known D genes, mutations were not scored in these regions. Replacement mutations are indicated by letters, identities as “.”, and insertions as −, and + to maintain spacing due to variability in CDR3 length. Sequences derived from the 5′ V-region primers used for library construction22 are marked as >. CDR-region designations are according to the system of Kabat et al29; numbering and hypervariable loop designations are according to the system of Chothia et al.30 (B) Analysis of nucleotide data in each patient revealed a distinct set of somatically mutated heavy chains sharing common VHDJH rearrangements of VH3-30, D1-26, and JH4b gene segments. Circles represent isolated and sequenced clones (Figures 1 and 2A); diamonds (for ITP patient A only) represent putative intermediates. For each member of a patient's clone, the number of nucleotide mutations from its germline VH gene is shown in parentheses, and the resulting number of replacement (R) or silent mutations (S) is shown in brackets. For each patient clone ontogenic tree, the distance in the horizontal direction represents the extent of mutation from the proposed germline origin within the constraints of the diagram.
Alignment of clonally related platelet autoantibody heavy-chain amino acid sequences and their putative ontogenic trees.
The H and L nomenclature is the same as in Figure 1. (A) Groups of related sequences comprising expanded heavy-chain clones in each patient library (clone A and clone B) are enclosed in boxes. The coalignment with the rest of the 16 unique heavy chains is available on the Blood website; see the Supplemental Data Set link at the top of the online article. For clone A, the putative intermediate heavy-chain sequences are also shown (1, 2, and 3 asterisks). The number of nucleotide differences from germline VH is tabulated to the right of each sequence. Because D segments showed poor homology with known D genes, mutations were not scored in these regions. Replacement mutations are indicated by letters, identities as “.”, and insertions as −, and + to maintain spacing due to variability in CDR3 length. Sequences derived from the 5′ V-region primers used for library construction22 are marked as >. CDR-region designations are according to the system of Kabat et al29; numbering and hypervariable loop designations are according to the system of Chothia et al.30 (B) Analysis of nucleotide data in each patient revealed a distinct set of somatically mutated heavy chains sharing common VHDJH rearrangements of VH3-30, D1-26, and JH4b gene segments. Circles represent isolated and sequenced clones (Figures 1 and 2A); diamonds (for ITP patient A only) represent putative intermediates. For each member of a patient's clone, the number of nucleotide mutations from its germline VH gene is shown in parentheses, and the resulting number of replacement (R) or silent mutations (S) is shown in brackets. For each patient clone ontogenic tree, the distance in the horizontal direction represents the extent of mutation from the proposed germline origin within the constraints of the diagram.
By examining nucleotide alignments with germline genes (data not shown), we constructed ontogeny trees for the 2 putative clones to illustrate how the patterns of somatic mutation in the respective heavy chains may have evolved in vivo (Figure 2B). For the ITP patient A clone in particular, a parsimonious mutation scheme (ie, postulating the minimum number of mutations) was used to derive putative intermediate heavy chains (Figure 2B, 1, 2, and 3 asterisks). The members of this clone appear to have undergone a marked degree of somatic mutation (from 4 to 21 nucleotide changes in the VHsegment alone) that resulted in high replacement-to-silent (R:S) ratios, both hallmarks of an immune response characterized by antigen-driven selection.31,32 For the ITP patient B clone, there were fewer mutations overall, but almost every mutation resulted in an amino acid replacement and clonal expansion was apparent. Therefore, the marked usage of VH3-30 in these cohorts of platelet-binding antibodies resulted at least partly from a restricted number of autoreactive B cells undergoing clonal expansion. The use of VH3-30 may also be important in conferring platelet binding because it encodes H44, a clonally unrelated heavy chain, and at least one IgM platelet autoantibody generated by conventional tissue-culture techniques.33
Determination of platelet autoantibody specificity by ELISA and flow cytometry.
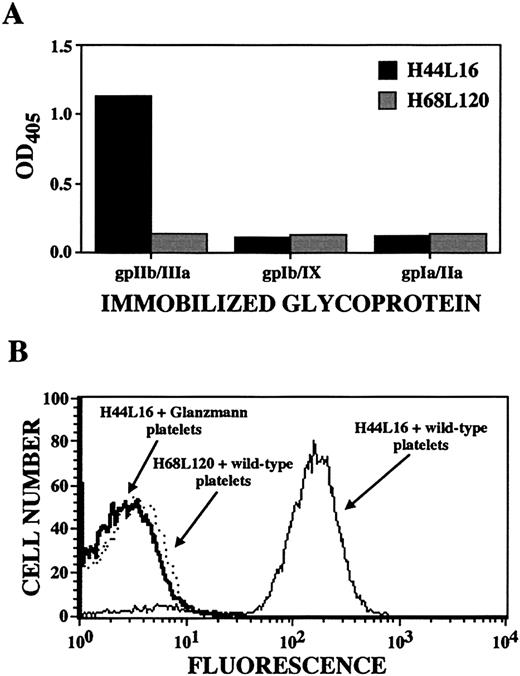
Shown are results for ITP patient A antibody H44L4, which was judged to be specific for platelet GPIIb/IIIa because of its binding to immobilized GPIIb/IIIa (but not GPIb/IX or GPIa/IIa; panel A), its binding to wild-type platelets but not GPIIb/IIIa-deficient platelets from 3 patients with Glanzmann thrombasthenia (one of 3 examples is shown in the flow cytogram; panel B), and its immunoprecipitation of GPIIb/IIIa molecules. Antibody H68L120, an anti–blood group B antibody isolated from the same original ITP patient A library,38was used as a negative control as indicated.
Determination of platelet autoantibody specificity by ELISA and flow cytometry.
Shown are results for ITP patient A antibody H44L4, which was judged to be specific for platelet GPIIb/IIIa because of its binding to immobilized GPIIb/IIIa (but not GPIb/IX or GPIa/IIa; panel A), its binding to wild-type platelets but not GPIIb/IIIa-deficient platelets from 3 patients with Glanzmann thrombasthenia (one of 3 examples is shown in the flow cytogram; panel B), and its immunoprecipitation of GPIIb/IIIa molecules. Antibody H68L120, an anti–blood group B antibody isolated from the same original ITP patient A library,38was used as a negative control as indicated.
H44 and the remaining heavy chains (H4, H10, H29, and H83) each had its own unique VHDJH recombination, and except for H29, somatic mutation occurred in their VH segments as well. Light chains also underwent somatic mutation (their aligned sequences are available in the online Blood supplement). Some κ light chains in the cohort may be clonally related (eg, L43, L44, and L45), but because the VLJL junction is not as diverse as the junctional regions for the heavy chain, clonal relatedness among light chains is more difficult to prove. Only a few λ light chains were present in platelet-reactive Fabs, none of which appeared to be clonally related.
Identification of recombinant platelet-autoantibody specificity
Autoantibodies from patients with ITP often recognize complexes composed of platelet glycoproteins GPIIb/IIIa or GPIb/IX,2,4,5,7,17,33-37 although autoantibodies against other identified and unidentified antigens have been described.5-13 Panning on intact platelets ensures that all relevant antigens were present during the selection process and that their native conformation was preserved.
Each of the 39 unique platelet-reactive antibodies showed specificity for this cell type. None bound to Chinese hamster ovary cells, K562 cells, erythrocytes, or leukocytes on flow cytometric analysis (not shown). In addition, none showed surface, cytoplasmic, or nuclear binding to HEp-2 cells on immunofluorescence analysis and none bound to cardiolipin. However, only the antigen specificity of H44L4 could be determined with relative unambiguity. In an ELISA, H44L4 reacted with purified, immobilized GPIIb/IIIa but not with GPIb/IX or GPIa/IIa (Figure 3A). H44L4 did not recognize platelets from 3 unrelated donors with type I Glanzmann thrombasthenia (Figure 3B), whereas all other platelet-reactive Fabs bound comparably to wild-type platelets and Glanzmann platelets. Furthermore, H44L4-immunoprecipitated polypeptides migrated in accordance with the behavior of GPIIb/IIIa under reducing and nonreducing conditions (Figure 4).
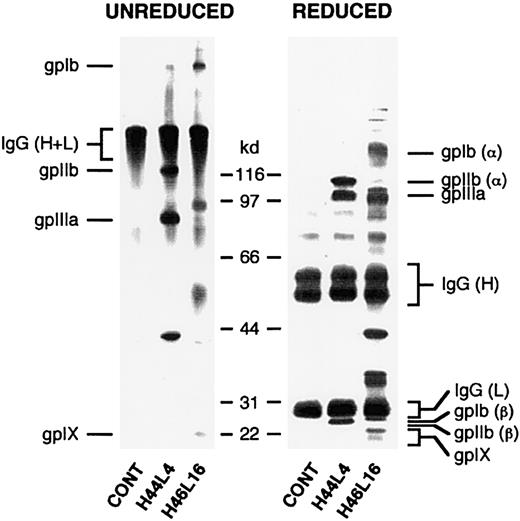
Determination of platelet autoantibody specificity by immunoprecipitation.
Biotinylated platelets solubilized after incubation with recombinant Fabs and antigen-Fab complexes were captured on Protein L dextran beads. Immunoprecipitated material was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis under nonreducing (left) or reducing (right) conditions, transferred to nitrocellulose, and detected with enzyme-labeled avidin-biotin complexes. Shown in this figure are results with ITP patient A–derived antibodies H44L4 and H46L16. Note that the presence of polypeptide bands with a relative molecular weight of about 150 kd (unreduced) and about 50 kd and 25 kd (reduced) represent platelet-bound autologous IgG that was biotinylated during the platelet-labeling procedure and coprecipitated by Protein L.
Determination of platelet autoantibody specificity by immunoprecipitation.
Biotinylated platelets solubilized after incubation with recombinant Fabs and antigen-Fab complexes were captured on Protein L dextran beads. Immunoprecipitated material was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis under nonreducing (left) or reducing (right) conditions, transferred to nitrocellulose, and detected with enzyme-labeled avidin-biotin complexes. Shown in this figure are results with ITP patient A–derived antibodies H44L4 and H46L16. Note that the presence of polypeptide bands with a relative molecular weight of about 150 kd (unreduced) and about 50 kd and 25 kd (reduced) represent platelet-bound autologous IgG that was biotinylated during the platelet-labeling procedure and coprecipitated by Protein L.
None of the other Fabs immunoprecipitated polypeptides in a manner consistent with the behavior of GPIIb/IIIa, a finding in agreement with the results of the ELISA and flow cytometry analysis using Glanzmann platelets; nor did any of them react with a stable K562 cell line expressing GPIa/IIa. However, autoantibodies H46L16, H47L64, and H48L24, 3 Fabs with clonally related heavy chains, immunoprecipitated polypeptides with molecular weights consistent with those for GPIb/IX. On ELISA, this set of Fabs did not bind significantly above background levels to purified immobilized GPIb/IX, but blocking of the relevant epitope by the mouse monoclonal capturing antibody could not be excluded. Until further analyses are performed, assignment of GPIb/IX as the specificity for clone A will remain tentative. Neither 2 antibodies from clone B (H37L50 and H42L38) nor the 2 non–VH3-30–encoded antibodies (H4L106 and H83L34) specifically immunoprecipitated labeled protein, perhaps because their target polypeptides were not biotinylated sufficiently or lost conformation during solubilization or because their targets are not proteins.
Contribution of the heavy and light chains of H44L4 to GPIIb/IIIa specificity
For certain antibodies, antigen specificity is determined primarily by one or the other component chain.39-42Identification of the platelet GPIIb/IIIa complex as the antigenic target of Fab H44L4 allowed us to examine the contribution of its constituent heavy and light chains to antigen recognition. If the VH3-30 heavy chain of H44L4 is solely responsible for GPIIb/IIIa binding, then the specific light chain that is used might be of little relevance, as long as it is permissive. Alternatively, the fine specificity of the VH3-30 heavy chain might be modified or actually determined by the paired light chain.24 The amenability of phage display–derived antibodies to molecular manipulation allowed us to examine this issue in some detail.
We first paired the H44 heavy chain with a panel of light chains and surveyed the resultant combinatorial Fabs for their capacity to bind platelets. To do this, we created a new library in which heavy-chain H44 was recombined with the entire light-chain repertoire from the original ITP patient A library. Only one of 101 Fabs expressing the H44 heavy chain paired with random light chains reacted with platelets. Like the original H44L4 antibody, this recombinant Fab recognized GPIIb/IIIa on ELISA (data not shown). Sequence analysis confirmed that H44 was used to encode this Fab. The presence of H44 in 20 randomly selected nonreactive Fabs was also confirmed. Thus, mere usage of the H44 heavy chain alone was insufficient to confer GPIIb/IIIa reactivity on a Fab molecule. This finding suggests that specific VH-VL pairing is required to impart this binding specificity.
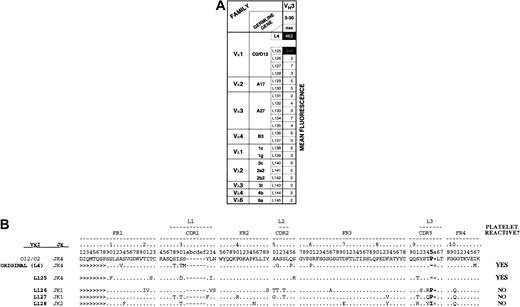
To examine this idea further, we sequenced the light-chain gene segments of the platelet-reactive Fab and those encoding the reference set of 20 H44-expressing Fabs that lacked platelet reactivity. Interestingly, the single positive Fab (H44L125) employed an O12/O2 κ variable light-chain gene and Jκ4 J-segment gene, as did the original H44L4 Fab (Figure 5). Indeed, light-chains L4 and L125 appear to have derived from the same B-cell clone, because they shared an especially distinctive VJ junction in which 3 nucleotides had been lost, resulting in deletion of the germline-encoded proline usually found at amino acid position 95 (Figure 5B). Because this residue lies in the CDR3 region of the light chain, deletion of 95P may confer or at least contribute to GPIIb/IIIa specificity. This idea is supported further by the observation that none of the 3 sampled non–platelet-reactive Fabs that use an O12/O2 light chain (Figure 5B; clones H44L126, H44L127, and H44L128) had a deletion at position 95.
Platelet binding of randomized light chains paired with platelet autoantibody heavy-chain H44.
The heavy chain of GPIIb/IIIa-specific H44L4 was paired again with a library of more than 106 light chains derived from the original, unselected ITP patient A library, and 101 resorted clones were screened for platelet binding by flow cytometry. (A) Matrix illustrating the genetic composition of the single retrieved positive resorted clone (designated H44L125). For comparison, 20 (of the 100) randomly chosen negative clones (designated H44L126 through H44L145) and the original H44L4 antibody are tabulated. Numbers in shaded boxes represent mean fluorescent intensities. Note that the single positive platelet-binding clone comprises a light chain derived from the same Ig light-chain gene as the original L4 light chain (012/02), yet no other 012/02-encoded light chain (eg, L125-L128) conferred binding when paired with H44. (B) Sequence analysis of cohort of 012/02-encoded light chains retrieved in resorting experiment shows that light-chain L125, which reconstitutes platelet binding, may be clonally related to the original L4 light chain because of a distinctive VJ junction characterized by loss of an entire amino acid residue at position 95 (boldface region).
Platelet binding of randomized light chains paired with platelet autoantibody heavy-chain H44.
The heavy chain of GPIIb/IIIa-specific H44L4 was paired again with a library of more than 106 light chains derived from the original, unselected ITP patient A library, and 101 resorted clones were screened for platelet binding by flow cytometry. (A) Matrix illustrating the genetic composition of the single retrieved positive resorted clone (designated H44L125). For comparison, 20 (of the 100) randomly chosen negative clones (designated H44L126 through H44L145) and the original H44L4 antibody are tabulated. Numbers in shaded boxes represent mean fluorescent intensities. Note that the single positive platelet-binding clone comprises a light chain derived from the same Ig light-chain gene as the original L4 light chain (012/02), yet no other 012/02-encoded light chain (eg, L125-L128) conferred binding when paired with H44. (B) Sequence analysis of cohort of 012/02-encoded light chains retrieved in resorting experiment shows that light-chain L125, which reconstitutes platelet binding, may be clonally related to the original L4 light chain because of a distinctive VJ junction characterized by loss of an entire amino acid residue at position 95 (boldface region).
These results indicate that only a limited set of light chains impart or are permissive for GPIIb/IIIa specificity when paired with a given VH3-30 gene product. This finding led us to investigate the limitations in heavy- and light-chain pairings required to generate platelet-reactive Fabs. As a corollary, we asked whether the specific light chain actually determines antigen specificity, in view of the finding that VH3-30 heavy-chain gene usage is so prevalent among platelet-reactive antibodies.
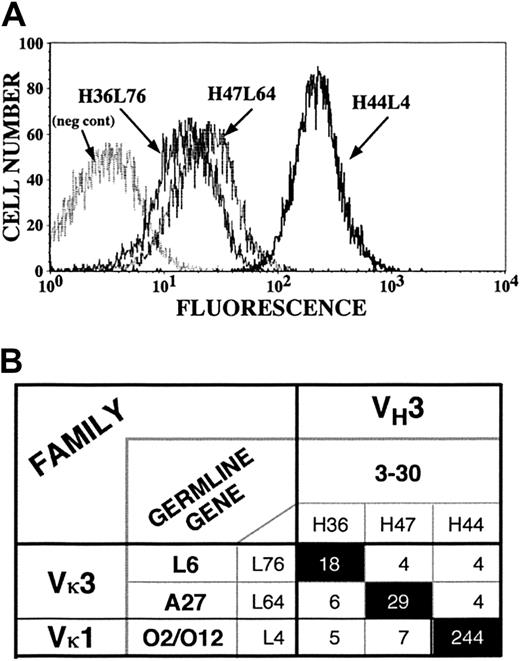
To address these issues, we exploited the distinctive flow cytometric (Figure 6A) and immunoprecipitation patterns of H44L4 (a GPIIb/IIIa-specific Fab that uses Vκ-O12/O2), H47L64 (a putative GPIb/IX-specific Fab and clone A member that uses Vκ-A27), and H36L76 (a clone B member that uses Vκ-L6), each of which uses VH3-30–encoded heavy chains. By mixing their plasmid DNAs, restriction digesting each light chain away from its originally associated heavy chain, and religating the resultant admixture of heavy- and light-chain gene segments, we generated all 9 possible combinations of the 3 heavy chains and 3 light chains. Forty-three randomly selected clones, which included several examples of each combination, were assessed for platelet binding. Only the heavy- and light-chain combinations that reconstituted the 3 original Fabs bound to platelets (Figure 6B), and their flow cytometric patterns were indistinguishable from those of the parental molecules (data not shown). Thus, although VH3-30 is used frequently by autoantibodies that bind to platelets, it is not only the specific light chain but also the particular heavy- and light-chain pairing that imparts platelet reactivity and specificity.
Exchange of light chains among platelet autoantibody clones.
Heavy and light chains for 3 platelet-binding clones (H44L4, H47L64, and H36L76) were interchanged to generate 9 possible combinations (6 novel and 3 reconstituted originals). (A) Flow cytograms comparing the fluorescent intensities of the 3 index antibodies. (B) Matrix showing that only reconstituted original heavy-chain–light-chain pairs conferred platelet binding. Numbers in boxes represent mean fluorescent intensities.
Exchange of light chains among platelet autoantibody clones.
Heavy and light chains for 3 platelet-binding clones (H44L4, H47L64, and H36L76) were interchanged to generate 9 possible combinations (6 novel and 3 reconstituted originals). (A) Flow cytograms comparing the fluorescent intensities of the 3 index antibodies. (B) Matrix showing that only reconstituted original heavy-chain–light-chain pairs conferred platelet binding. Numbers in boxes represent mean fluorescent intensities.
Binding of platelet autoantibodies to the superantigen domain of SpA
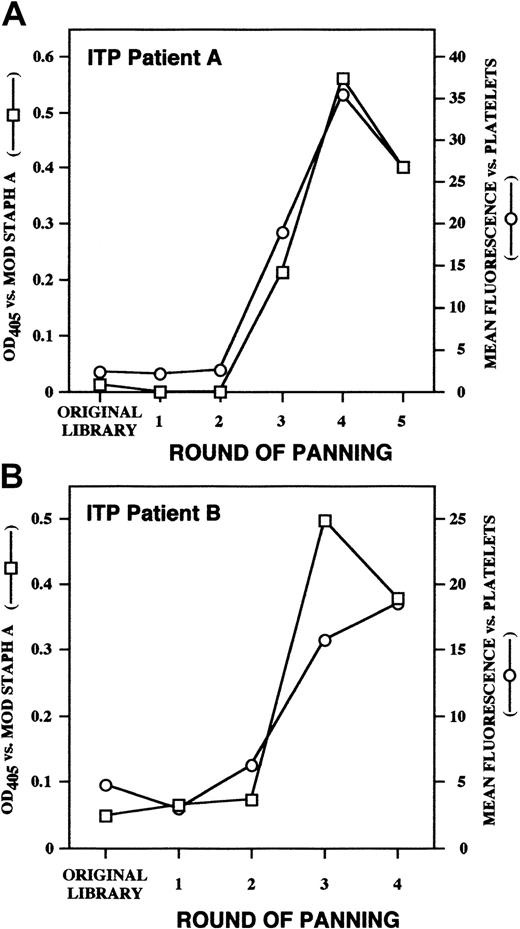
The mechanism by which extracorporeal absorption of plasma from ITP patients with affinity columns containing SpA is sometimes efficacious is unknown, given that the amount of IgG removed is only about 2% of that removed during plasmapheresis,1,43 a treatment that is rarely effective in chronic ITP.44,45 A B-cell superantigen site on SpA has been described that is independent of its well-characterized Fc binding site and that interacts with variable regions of antibodies encoded by certain members of the VH3 family, notably VH3-30.46,47Modification of SpA by iodination completely destroys Fc-binding activity, whereas Fab-binding activity is retained.26 We asked whether this modified SpA would bind our platelet autoantibodies by virtue of their genetic restriction. We found that as we selected platelet-binding autoantibodies from polyclonal, polyspecific Fab libraries through sequential rounds of panning, there was concurrent selection for binding activity to the superantigen domain of SpA (Figure 7).
Binding of platelet-selected Fabs to mod SpA.
Polyclonal Fab preparations derived from the original unselected ITP patient A and patient B Fab/phage display libraries (panel A and B, respectively) and from the libraries after each round of platelet panning were assayed for platelet binding by flow cytometry (circles, right set of axes) and for binding to mod-SpA by ELISA (squares, left set of axes).
Binding of platelet-selected Fabs to mod SpA.
Polyclonal Fab preparations derived from the original unselected ITP patient A and patient B Fab/phage display libraries (panel A and B, respectively) and from the libraries after each round of platelet panning were assayed for platelet binding by flow cytometry (circles, right set of axes) and for binding to mod-SpA by ELISA (squares, left set of axes).
Discussion
Platelet autoantibodies are encoded by a restricted set of VH genes
Use of the VH3-30 heavy-chain gene was found to be highly represented among platelet-reactive Fabs from both patients with ITP compared with its prevalence in the general library and despite differences in antigen specificity (P < 10−13 by Fisher exact test; Figures 1 and 4). Interestingly, this same heavy-chain gene was found to encode an IgM anti-GPIIb autoantibody derived by hybridoma technology from another patient with ITP,20,33 as well as several platelet-reactive, IgG phage display–derived antibodies from ITP patients selected because of their ability to bind to IVIG.48,49 This marked genetic restriction to the VH3-30 heavy-chain gene for antiplatelet autoantibodies may provide an explanation for the association of ITP with seemingly unrelated disorders, such as autoimmune hemolytic anemia, SLE, chronic lymphocytic leukemia, CVID, and HIV infection, in which VH3-30 and related gene products are expanded or involved in disease pathogenesis.50-56
Why use of the VH3-30 heavy chain is overrepresented among antibodies that show platelet reactivity is a pivotal question. One possibility is that antibodies encoded by most other VHgene products are less able to bind to platelets. Such a restriction on antigen recognition could explain why we did not identify any VH3-23 heavy-chain gene product among platelet-reactive Fabs, even though it is the most frequently used VH gene in the repertoire.57-61 However, several antibodies in our cohort of platelet binders were encoded by VH genes other than VH3-30, including VH1-02, VH1-46, VH3-21, and VH4-59 (Figure1 and Blood online supplement). Remarkably, this identical group of VH genes was found by Boucher et al62to encode all but one antibody in a large number of human anti-Rh(D) RBC alloantibodies. As noted by these investigators, products of these germline genes are among the most cationic in the human VHrepertoire. The resulting constitutive net positive charge may allow the antibodies to effectively permeate the highly negative RBC ζ potential, thus permitting contact with antigen.63 Because platelets have an even greater density of cell-surface negative charges as a result of their thick glycocalyx rich in acidic mucopolysaccharides,64 65 platelet-surface charge may play a similar role in biasing the use of cationic germline VHsegments. Therefore, use of cationic VH genes may facilitate access to the membrane surface, but specificity for a particular antigen may be determined by heavy-chain CDR3 and light chain.
We investigated this role for light chain by pairing the VH3-30–encoded H44 heavy-chain product found in our platelet GPIIb/IIIa-specific Fab with all members of the entire light-chain repertoire from the same library (∼108 light chains). Only one other platelet-reactive, GPIIb/IIIa-specific Fab was retrieved (Figure 5). Remarkably, the light chain in this Fab was not only very similar in sequence to the light chain found in the original antibody, but it appeared, on the basis of CDR3 analysis, to have derived from the same B-cell clone in vivo. Furthermore, light chains from platelet-reactive Fabs that use VH3-30 heavy-chain genes were not interchangeable. Indeed, when the genes from a set of platelet-reactive Fabs with differing specificity were permitted to recombine randomly, only the original combinations of heavy and light chains led to detectable platelet binding (Figure 6). These observations suggest that platelet antigen specificity cannot result from simple pairing of an array of permissive heavy- and light-chain gene products but requires precise interactions between particular heavy chains and their light-chain companions.
Role of autoantigen and clonal expansion in chronic ITP
The study of human autoimmune disease is greatly facilitated by focusing on disorders such as ITP, in which it is clear that the associated autoantibodies are unequivocally involved in pathogenesis. However, the role played by self-antigens in the evolution of autoreactive antibodies and the clonality of the autoimmune response are not well understood. On the basis of light-chain restriction, previous reports suggested that platelet autoantibodies in chronic ITP are clonally restricted.66-70 Consistent with these findings, several features of the platelet-reactive autoantibodies described in the current study indicate that they arose as part of an antigen-driven clonal expansion rather than being the result of polyclonal B-cell activation triggered by nonspecific stimuli. First, most antibodies isolated from each patient shared a single heavy-chain VHDJH rearrangement indicating their derivation from a single B cell (Figure 2B). Second, somatic mutation with high R:S ratios was evident in heavy- and light-chain variable regions (Figure 2 and Blood online supplement). Third, each of the platelet-reactive Fabs was derived from an IgG library, indicating that isotype switching had occurred, another hallmark of a T-cell–dependent, antigen-driven immune response. Finally, the requirement for precise heavy- and light-chain pairing to generate antigen specificity (Figures 5 and 6) also typifies antigen-driven immune responses.40,41,71 72
It may be these characteristics that distinguish pathogenic antiplatelet autoantibodies from “benign” ones cloned from samples from unaffected donors. Such naturally occurring platelet-binding antibodies, in contradistinction to those observed in the current study, are nearly always IgM, are often polyreactive, have little or no somatic mutation of their variable regions, or show a combination of these characteristics.73-75 These differences are analogous to those used to distinguish pathogenic from benign autoantibodies in murine models of autoimmunity.31,33Whether the B cells that produce benign antiplatelet autoantibodies are the clones that go on to lose self-tolerance, switch isotypes, somatically mutate their variable-region genes, and secrete pathogenic autoantibodies is not clear. In fact, it may be this clonally unrelated pool of natural, nonpathologic antiplatelet autoantibodies that normally functions to keep production of pathologic autoantibodies in check through a mechanism of competitive tolerance, as has been proposed for murine rheumatoid factors.76
Clinical and therapeutic implications of VH gene restriction
Current treatments for chronic ITP are characterized by relatively nonspecific immune intervention. If restriction of platelet autoantibodies to the VH3-30 heavy-chain gene is confirmed by studies of additional immune repertoires, exploitation of this restriction may facilitate the design of more targeted forms of immunotherapy. For example, it is known that SpA has a B-cell superantigen site—distinct from its well-characterized Fc-binding domain—that is specific for the gene products of certain VH3-encoded Igs, notably VH3-30.46,47 Consistent with this activity, we found that panning of ITP patient A and B phage display libraries on platelets resulted in concomitant enrichment for both platelet and mod-SpA binders (Figure 7). In studies in mice, targeted deletion of VH3-30 homologs by apoptotic cell death occurred on in vivo administration of recombinant mod-SpA superantigen,77,78 suggesting that infusion of small amounts of mod-SpA might likewise down-regulate production of platelet autoantibodies in ITP. In this regard, and in light of recent studies demonstrating shedding of up to 200 μg SpA from SpA-silica columns during extracorporeal immunoabsorption procedures,79 the long-term remissions may be a consequence of infused SpA and not the removal of antibody by the columns per se. Future studies testing the therapeutic effectiveness of this or other VH3-30–targeted reagents, such as anti-idiotypic antibodies derived from mice80,81 or humans,49may provide novel approaches for regulating immune-repertoire composition. Furthermore, development of reagents for rapid identification of the genetic origins of platelet autoantibodies may help predict responsiveness to such novel molecular therapies in individual patients.
We thank Tylis Chang for his contribution to the early phase of this study and Stephen Kacir for excellent technical assistance.
Supported by grant R01-HL61844 from the National Institutes of Health.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Don L. Siegel, Blood Bank/Transfusion Medicine Section, Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Room 510, Stellar-Chance Building, 422 Curie Blvd, Philadelphia, PA 19104; e-mail:siegeld@mail.med.upenn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal