Male DNA, of presumed fetal origin, can be detected in the maternal circulation decades after delivery and is referred to as fetal microchimerism (FM). We previously found quantitatively greater FM in the circulation of women with the autoimmune disease scleroderma (SSc) than of healthy women. However, it is unknown whether this difference is due to intact circulating cells or free DNA released from breakdown in disease-affected tissues. To distinguish the origin of FM, we developed a real-time quantitative polymerase chain reaction (PCR) assay for the Y-chromosome–specific sequenceDYS14, and tested 114 women in peripheral blood mononuclear cells (PBMCs) and/or plasma. Fifty-seven controls and 57 SSc patients were studied, 48 and 43 of whom, respectively, had given birth to at least one son. Circulating FM was quantitatively greater in PBMCs from SSc patients (n = 39; range, 0.0-12.5 male genome–equivalent cells per million maternal cells), compared with healthy women (n = 39; range, 0.0-4.4; P = .03). In contrast, there was no difference between patients (n = 25) and controls (n = 22) in plasma, and no evidence of free DNA. FM was enriched among T lymphocytes compared with PBMCs (P = .01) in controls (n = 14), but not in SSc patients (n = 14); the latter finding was most likely due to immunosuppressive medications. In conclusion, this real-time quantitative assay showed that quantitative differences in the circulation of women with SSc are due to cells and not to free DNA. As FM was not uncommon in healthy women, including among T cells, and because graft-versus-host disease has similarities to SSc, these results also suggest that FM merits investigation in pheresis products used for stem cell transplantation.
Introduction
During normal human pregnancy, fetal DNA and cells pass into the maternal circulation.1 The term microchimerism refers to the presence of a small amount of DNA or cells from one individual in another individual. Persistent microchimerism of presumptive fetal origin (fetal microchimerism [FM]) has recently been described in the circulation of women with systemic sclerosis (SSc), also called scleroderma, and in healthy women.2,3 SSc is an autoimmune disease characterized by an excessive deposition of collagen in the skin and internal organs and by vascular and immunologic abnormalities. Studies of FM in SSc were initiated on the basis of a constellation of observations,4 including the clinical similarities of SSc to chronic graft-versus-host-disease (cGvHD),5 the increased incidence of SSc observed in women during the postreproductive years,6 and the long-term persistence of FM after pregnancy.2 In prior studies, we tested this hypothesis using an assay that had been developed for use in prenatal diagnosis.7 The assay tested multiple aliquots of whole blood for a Y-chromosome–specific sequence by means of a direct quantitative polymerase chain reaction (PCR) test. Using this assay to test whole peripheral blood from women who had given birth to at least one son, we found quantitatively greater levels of FM in women with SSc compared with healthy controls.3 Recently, Lo et al8 reported that during pregnancy, plasma affords a better source for measuring FM than does the cellular fraction of maternal blood. Plasma fetal DNA is rapidly cleared following delivery.9 Thus, the increase of FM observed in whole blood samples from women with SSc compared with controls could be derived from circulating cells or from fetal DNA released into the plasma from disease-affected tissues. The latter possibility would be analogous to increased levels of FM seen in the serum of patients with pre-eclampsia thought to reflect DNA liberated from fetal nucleated cells.10 We therefore developed a sensitive real-time quantitative PCR assay and employed it to study FM in peripheral blood mononuclear cells (PBMCs), plasma, and also T lymphocytes of parous healthy women and women with SSc.
Patients, materials, and methods
Study subjects
All patients satisfied the American College of Rheumatology criteria for SSc.11 A total of 114 women were studied, including 57 healthy controls and 57 patients with SSc. It was found that 68% (n = 39) of patients had diffuse SSc and 32% (n = 19) had limited SSc. Among the 57 healthy women, 44 had given birth to at least one son; 11 had given birth only to daughters, and 2 were nulliparous. Among the 57 SSc patients, 43 had given birth to at least one son; 9 had given birth only to daughters; and 5 were nulliparous. Seven patients of 57 had received blood transfusions, 4 after and 3 before disease onset. The majority of women were white, representing 75% of patients and 93% of controls. Five percent and 2%, respectively, were African American; 11% and 4% were Asian; 2% and 0% were Native American; 4% and 2% were of Hispanic origin; and 4% and 0% of patients were of “mixed” ethnic origin. Informed consent was obtained for all study participants.
Procurement of blood specimens and preparation of PBMCs and plasma
Whole blood samples were drawn into acid citrate dextrose solution A-vacutainer tubes. The tubes were centrifuged at 400g, and plasma was carefully removed. In a subset of studies, some plasma samples were subsequently filtered by means of 0.45-μm filters. The remaining blood was processed to isolate PBMCs by Ficoll Hypaque (Pharmacia Biotech, Uppsula, Sweden) with density gradient centrifugation at 1.077g/mL.
DNA extraction
To prepare samples for the quantitative PCR assay, DNA was extracted from PBMCs by means of an Isoquick Nucleic Acid Extraction Kit (ORCA Research, Bothell, WA) according to the manufacturer's instructions and resuspended in 10 mM Tris-HCl Tris (tris(hydroxymethyl)aminomethane–Hcl), (pH 9.0) or was extracted from plasma by means of a High Pure PCR template Preparation Kit (Roche, Basel, Switzerland), following the isolation procedures recommended by the manufacturer. For DNA extraction, 500 μL plasma sample was used. An elution volume of 100 μL was used.
Real-time quantitative PCR
The theoretical and practical aspects of real-time quantitative PCR have previously been described.12 The amplification and product-reporting system used was based on the 5′ nuclease assay.13 In this system, in addition to the 2 amplification primers used in conventional PCR, a dual-labeled fluorogenic probe is included. Two fluorescent dyes, a reporter and a quencher, are attached to the probe. With both dyes attached to the probe, reporter dye emission is quenched. During each extension cycle, the 5′–3′ nuclease activity of the Taq DNA polymerase cleaves the reporter dye from the probe. Once separated from the quencher, the reporter dye emits its characteristic fluorescence. Amplification data were collected by a Perkin-Elmer Applied Biosystems 7700 sequence detector, stored in a Macintosh computer and analyzed by means of Sequence Detection System software (PE Applied Biosystems, Foster City, CA).
The DYS14 sequence (GenBank sequence accession number, X06325) was selected as the Y-chromosome–specific sequence for use in the quantitative PCR (QPCR) assay. The reason for this choice was that, while single-copy genes, such as the SRY,can be reliably used in developing a QPCR assay, the use of a single-copy gene results in less sensitivity than a multiple-copy gene. On the other hand, some multiple-copy genes, for example,DYZ1, have been postulated to exhibit autosomal cross-reactivity.14,15 The DYS14, on the other hand, is believed to be strictly Y specific.16
QPCR amplification primers were designed and used with the following sequences: DYS14 forward primer, 5′-CATCCAGAGCGTCCCTGG-3′; and DYS14 reverse primer, 5′-TTCCCCTTTGTTCCCCAAA-3′. The dual-labeled fluorescent DYS14-probe sequence was 5′ (FAM)-CGAAGCCGAGCTGCCCATCA (TAMRA)-3′. Simultaneously, the same amount of target DNA was tested for theβ-globin gene to confirm the quality (ability to amplify) of the extracted DNA. The quantitative results were used to calculate the fractional concentration of FM (by DYS14 PCR) among total DNA (by β-globin PCR). The amplification primers and fluorescent probe used for amplifying the β-globin gene were as previously described.12
Real-time quantitative PCR reactions were set up in a reaction volume of 50 μL. The target consisted of either 66 ng DNA (in 5 μL) from PBMCs or 5 μL resuspended DNA after extraction from plasma. A conversion factor of 6.6 pg DNA per cell was used for expressing the results as male genome–equivalent cells per milliliter of maternal plasma or per million maternal cells, with 66 ng DNA corresponding to 10 000 male genome–equivalent cells.17 Each PCR reaction consisted of 5 μL 10 × platinum buffer (Gibco BRL, Burlington, ON, Canada), 300 nM each amplification primer, 100 nM dual-labeled probe, 200 nM each deoxynucleoside triphosphate (dNTP) (Gibco BRL), 3.5 mM MgCl2 (Gibco BRL), 1.5 U platinumTaq (Gibco BRL), and 60 nM the standard reference dye ROX (Synthegen, Houston, TX). The amplification conditions consisted of an initial incubation at 50°C for 2 minutes, followed by incubation at 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 56°C for 1 minute. For quantitative measurement of fetal DNA, a calibration curve was constructed with the use of DNA from a healthy male individual diluted in DNA from a healthy female individual. Fourteen aliquots of 10 000 male genome–equivalent cells (66 ng) were tested for each patient and control, including a duplicate for β-globin and 12 aliquots for the Y-specific PCR. Standard curves for β-globin andDYS14 were run simultaneously. Average numbers of male genome–equivalent cells per million maternal cells was calculated for each subject, combining information across aliquots of PBMCs. For plasma samples, each patient was tested in duplicate, and the mean quantity of each duplicate was expressed in copies per milliliter of maternal plasma as previously described.9
T-lymphocyte isolation by fluorescence-activated cell sorting (FACS)
PBMCs were filtered on nylon wool (Du Pont Biotechnologies, Boston, MA) to avoid cell aggregation and were resuspended in phosphate-buffered saline (PBS)/1% fetal calf serum. Staining was performed as previously described18 on 10 to 20 × 106 cells with anti-CD3–cychrome and/or anti-CD4–phycoerythrin (PE) and/or anti-CD8–fluorescein isothiocyanate (FITC) (Becton Dickinson, Mountain View, CA) at 3 μL per antibody per million cells. After staining incubation, DNase (Boehringer-Mannheim, Indianapolis, IN, or GIBCO BRL) was added (30 U per million cells) after the last wash to avoid cell aggregation during the FACS process. Cells were sorted by a single laser on a FACS (Becton Dickinson), on the basis of the markers described above. An aliquot of the sorted cells was run, and the percentage of sorted and correctly gated cells were calculated to assess the purity. Purity consistently exceeded 99%. Cells were collected in RPMI. Cell sorting was immediately followed by centrifugation and 2 washes to remove all remaining DNase. Cell aliquots were stored at −80°C in cell lysis buffer until DNA extractions were performed.
Statistical analysis
We investigated whether the number of male genome–equivalent cells per million maternal cells in PBMCs, or per milliliter in maternal plasma, was higher in women with SSc compared with healthy women. The analysis considered women who had given birth to at least one son. P was calculated by means of a regression model; robust variances were estimated by means of the method of generalized estimating equations. For multiple measurements based on blood drawn on the same day, the DNA-equivalent fetal cells per million maternal cells values were averaged. Multiple measurements based on repeated blood draws per woman were entered into the analysis as repeated measures, with adjustment for possible correlation between values within a subject.
Results
Quantitative assessment of male DNA in plasma of healthy women and women with SSc
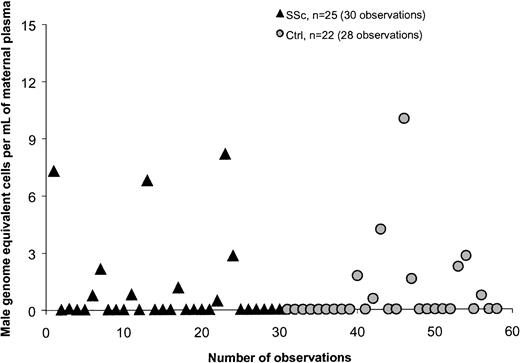
Plasma samples from 66 women were investigated for quantitative assessment of male DNA. Study subjects included 29 healthy women and 37 women with SSc. Twenty-two healthy women and 25 women with SSc had given birth to at least one son. The majority of women who had given birth to a son (healthy women as well as women with SSc) had no detectable male DNA in the plasma, 73% and 64%, respectively (Figure1). The range of male genome–equivalent cells was 0 to 10 per million maternal cells among controls and 0 to 8.2 per million maternal cells among patients, both with median values of 0 (Table 1). There was no significant difference between healthy women and women with SSc. Of the 6 healthy women with positive results (6 women, 8 observations; Figure 1), all had given birth to a son. Among the 11 SSc patients with positive results, 9 had given birth to a son and 1 had a history of blood transfusion. For one SSc patient with a low positive result, the source of male DNA was unclear as she had daughters but had not received a blood transfusion. In addition, she was not a twin and did not have a history of known miscarriage. All women with SSc had given birth to sons 10 or more years previously whereas 6 healthy women had given birth to sons fewer than 10 years from the time of testing. However, results did not differ if women with more recent births were excluded from the analysis.
Quantification of male DNA in maternal plasma from healthy women (Ctrls) and women with systemic sclerosis (SSc).
Quantification of male DNA in maternal plasma from healthy women (Ctrls) and women with systemic sclerosis (SSc).
Male DNA in plasma from healthy women and women with SSc
| . | Subjects, no. . | Observations, no. . | Male GenEq cells* . | P† . | |
|---|---|---|---|---|---|
| Median . | Range . | ||||
| All subjects | |||||
| Controls | 22 | 28 | 0 | 0 -10.0 | .61 |
| Cases | 25 | 30 | 0 | 0 -8.17 | |
| With patients with transfusions excluded | |||||
| Controls | 22 | 28 | 0 | 0 -10.0 | .48 |
| Cases | 22 | 27 | 0 | 0 -8.17 | |
| . | Subjects, no. . | Observations, no. . | Male GenEq cells* . | P† . | |
|---|---|---|---|---|---|
| Median . | Range . | ||||
| All subjects | |||||
| Controls | 22 | 28 | 0 | 0 -10.0 | .61 |
| Cases | 25 | 30 | 0 | 0 -8.17 | |
| With patients with transfusions excluded | |||||
| Controls | 22 | 28 | 0 | 0 -10.0 | .48 |
| Cases | 22 | 27 | 0 | 0 -8.17 | |
GenEq indicates genome equivalent.
Per milliliter plasma.
Results from a regression model with robust variance estimated by means of the method of generalized estimating equations, adjusted for the total number of cells tested.
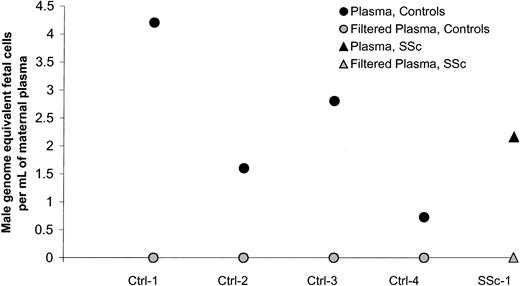
A positive result in plasma could represent free DNA or could be the result of apoptotic fetal cells in plasma, as has been described by others.19 Our method of centrifugation was gentle, using 400g, as compared with other methods that have been described as using consecutive rounds of centrifugation or centrifugation followed by filtration.20 To explore the explanation that positive results derived from apoptotic cells that were not removed with the gentle centrifugation method, we conducted additional experiments in women with positive results. Samples from 5 women (4 healthy controls and 1 woman with SSc) with high levels of plasma male DNA were studied according to 2 protocols: a 10-minute centrifugation at 400g, as initially done, and a 10-minute centrifugation followed by filtration using 0.45-μm filters to remove apoptotic cells but to leave any floating DNA in the plasma. All women had a negative result after filtration (Figure2), indicating that the positive results observed in the plasma samples were due to apoptotic cells and not to free circulating DNA.
Male DNA in maternal plasma from healthy women and women with SSc before and after filtration of the plasma.
Male DNA in maternal plasma from healthy women and women with SSc before and after filtration of the plasma.
Quantitatively greater levels of male DNA in PBMCs in women with SSc compared with healthy women
PBMCs were quantitatively assessed for male DNA for 93 women, in 46 healthy women, and 47 women with SSc. In each group, healthy women and women with SSc, 39 women had given birth to at least one son. The frequency of any detectable male DNA in PBMCs was somewhat higher in SSc patients (51%) compared with controls (31%) but was not significant (P = .07). A significant difference in FM levels between patients and controls was observed (P = .03). The range of male DNA among controls was 0 to 4.4 and among patients 0 to 12.5. Table 2summarizes a comparison between healthy women and women with SSc who had given birth to at least one son. Two patients had outlying values. When we excluded these values to see if they were influencing the estimates, the results remained significant (P = .047). Six SSc patients gave a history of blood transfusion; if these patients were excluded, the difference was not statistically significant. All statistical analyses were adjusted for some subjects who were tested on more than one occasion. Three women in the control group and 2 SSc patients were tested at 2 time points: 1 control and 1 SSc patient at 3 time points, and 1 control at 4 time points.
Male DNA in PBMCs from healthy women and women with SSc
| . | Subjects, no. . | Observations, no. . | Male GenEq cells* . | P† . | AdjustedP‡ . | |
|---|---|---|---|---|---|---|
| Median . | Range . | |||||
| All subjects | ||||||
| Controls | 39 | 47 | 0 | 0 -4.37 | .14 | .032 |
| Cases | 39 | 47 | 0 | 0 -12.5 | ||
| With large values excluded2-153 | ||||||
| Controls | 39 | 47 | 0 | 0 -4.37 | .31 | .047 |
| Cases | 38 | 46 | 0 | 0 -5.49 | ||
| With patients with transfusions excluded | ||||||
| Controls | 39 | 47 | 0 | 0 -4.37 | .56 | .11 |
| Cases | 33 | 39 | 0 | 0 -6.33 | ||
| . | Subjects, no. . | Observations, no. . | Male GenEq cells* . | P† . | AdjustedP‡ . | |
|---|---|---|---|---|---|---|
| Median . | Range . | |||||
| All subjects | ||||||
| Controls | 39 | 47 | 0 | 0 -4.37 | .14 | .032 |
| Cases | 39 | 47 | 0 | 0 -12.5 | ||
| With large values excluded2-153 | ||||||
| Controls | 39 | 47 | 0 | 0 -4.37 | .31 | .047 |
| Cases | 38 | 46 | 0 | 0 -5.49 | ||
| With patients with transfusions excluded | ||||||
| Controls | 39 | 47 | 0 | 0 -4.37 | .56 | .11 |
| Cases | 33 | 39 | 0 | 0 -6.33 | ||
GenEq indicates genome equivalent.
Per million maternal PBMCs.
Results from a regression model with robust variance estimated by means of the method of generalized estimating equations, weighted by the total number of cells tested.
Adjusted for age of last son at time of mother's blood draw.
One patient had 3 measurements at the same draw date; instead of averaging the 3 values, we took the lowest value. Another patient had a large value that we excluded to see if these outlying values were influencing the estimates.
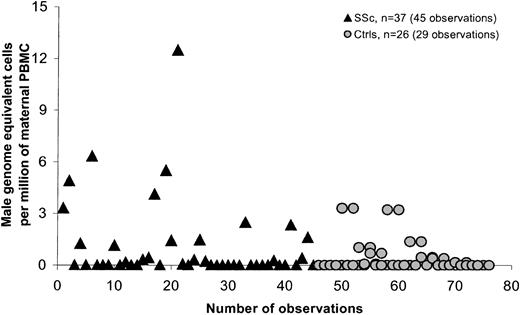
SSc is a disease that is most often diagnosed in women in the late 40s or even early 50s. Most of the SSc patients we studied had sons who were older than 10 years at the time of the patient's blood draw (95% of cases), whereas controls not infrequently had given birth fewer than 10 years previously. We therefore conducted a second analysis comparing only women for whom the son was born more than 10 years previously (Table 3; Figure3). The difference between patients and controls was significant both overall (P = .021) and after adjustment for time from birth of the most recent son (P = .006). Results were also significant when large outlying values were excluded (P = .018). The difference between patients and controls was also significant when patients who had received a blood transfusion were excluded from the analysis (P = .035). Statistical analysis was adjusted for 2 control women and 6 women with SSc who were tested on 2 different dates, and 1 SSc patient who was tested at 3 points.
Male DNA from PBMCs of healthy women and women with SSc with sons older than 10 years at time of blood draw
| . | Subjects, no. . | Observations, no. . | Male GenEq cells3-150 . | P3-151 . | AdjustedP3-152 . | |
|---|---|---|---|---|---|---|
| Median . | Range . | |||||
| All subjects | ||||||
| Controls | 26 | 29 | 0 | 0 -3.31 | .021 | .006 |
| Cases | 37 | 45 | 0 | 0 -12.5 | ||
| With large values excluded3-153 | ||||||
| Controls | 26 | 29 | 0 | 0 -3.31 | .054 | .018 |
| Cases | 36 | 44 | 0 | 0 -5.49 | ||
| With patients with transfusions excluded | ||||||
| Controls | 26 | 29 | 0 | 0 -3.31 | .135 | .035 |
| Cases | 31 | 37 | 0 | 0 -6.33 | ||
| . | Subjects, no. . | Observations, no. . | Male GenEq cells3-150 . | P3-151 . | AdjustedP3-152 . | |
|---|---|---|---|---|---|---|
| Median . | Range . | |||||
| All subjects | ||||||
| Controls | 26 | 29 | 0 | 0 -3.31 | .021 | .006 |
| Cases | 37 | 45 | 0 | 0 -12.5 | ||
| With large values excluded3-153 | ||||||
| Controls | 26 | 29 | 0 | 0 -3.31 | .054 | .018 |
| Cases | 36 | 44 | 0 | 0 -5.49 | ||
| With patients with transfusions excluded | ||||||
| Controls | 26 | 29 | 0 | 0 -3.31 | .135 | .035 |
| Cases | 31 | 37 | 0 | 0 -6.33 | ||
GenEq indicates genome equivalent.
Per million maternal PBMCs.
Results from a regression model with robust variance estimated using the method of generalized estimating equations, weighted by the total number of cells tested.
Adjusted for age of last son at time of mother's blood draw.
One patient had 3 measurements at the same draw date; instead of averaging the 3 values, we took the lowest value. Another patient had a large value that we excluded to see if these outlying values were influencing the estimates.
Male DNA in PBMCs in women with SSc and healthy women for whom the youngest son was older than 10.
Male DNA in PBMCs in women with SSc and healthy women for whom the youngest son was older than 10.
Some patients and controls who were parous but had no history of a male birth or who were nulligravid had positive results for male DNA in PBMCs. Among the 8 patients tested, 5 had positive results, all but 1 at a very low level (0.07 to 0.6 male genome–equivalent fetal cells per million maternal cells); 1 with a high result (26 male genome–equivalent fetal cells) had given birth only to a daughter but reported 3 pregnancies. Among the 4 patients who had a low level of FM, 1 reported an incomplete pregnancy, but the 3 other patients with low levels did not report any pregnancies. Among healthy controls who did not give birth to a son, 2 of 7 also had a very low-level positive result (equivalent to 0.08 and 0.2 male fetal cells per million maternal cells). One had given birth to a daughter, but the pregnancy history, in terms of the possibility of miscarriage, was unknown. The second was a nulliparous woman. Although not specifically investigated, FM is presumed to occur after an incomplete pregnancy, and it is known that large amounts of FM are found in the maternal circulation after an induced abortion.21 Thus, potential explanations include an early unrecognized miscarriage, an unreported induced abortion, an unrecognized loss of a male twin in utero, or possibly cells from sexual intercourse or transfer, via the maternal circulation, from an older male sibling.
Male DNA among T lymphocytes of healthy women and women with SSc
As FM was cellular in origin, and because T lymphocytes are implicated in a number of autoimmune diseases including SSc, we also employed the QPCR assay to test T lymphocytes isolated to greater than 99% purity by flow cytometry. Forty-one women, including 22 healthy controls and 19 SSc patients, were studied after isolation of cells with antibodies to CD3 (T lymphocytes) and/or CD3/CD4 (T-helper cells) and/or CD3/CD8 markers (T-cytotoxic cells).
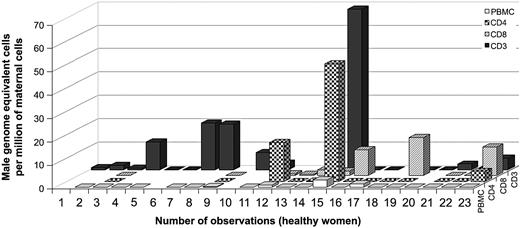
Among the healthy women, 67% (12 of 18) had FM in the T-cell compartment (CD3 marker only) (Figure 4). The range of positive results extended from 0.3 to 68.7 male genome–equivalent fetal cells per million maternal T cells. The levels of FM were greater in T lymphocytes (only CD3 markers) than in PBMCs for 71% of the healthy women for whom quantitative testing was done from the same draw date (n = 14). This result indicated FM was significantly enriched in T lymphocytes compared with PBMCs (P = .01 by Wilcoxon signed rank test). T-helper (CD3/CD4) and T-cytotoxic (CD3/CD8) cells were studied from some healthy controls (n = 13 and n = 11, respectively). Positive results were found in both populations. Thirty-one percent (4 of 13) healthy women had positive results for CD4 T cells, ranging from 0.2 to 50.3 male genome–equivalent cells per million maternal T cells. Sixty-four percent (7 of 11) had positive results in CD8 T cells, ranging from 0.6 to 16.3 male genome–equivalent cells per million maternal T cells.
Male DNA in T cells from healthy controls (n = 22).
Numbers 11 and 12 correspond to 2 observations at different draw dates from the same woman.
Male DNA in T cells from healthy controls (n = 22).
Numbers 11 and 12 correspond to 2 observations at different draw dates from the same woman.
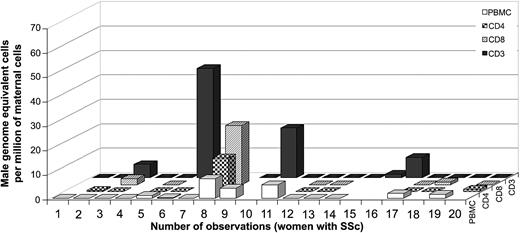
Among women with SSc, 26% (5 of 19) had a positive result for FM in the T cells isolated by antibodies to CD3 only, with a range from 1.6 to 44.8 male genome–equivalent cells per million maternal T cells (Figure 5). Thirty percent (3 of 10) had FM in T cells isolated by antibodies to CD3 and CD4 (T helper cells), with a range from 0.5 to 13.7 male genome–equivalent cells per million maternal T cells. Thirty-eight percent (3 of 8) had FM in T cells isolated by antibodies to CD3 and CD8 (T-cytotoxic cells), with a range from 1.1 to 24.2 male genome–equivalent cells per million maternal T cells. These results suggest a lower frequency and generally lower levels of FM among T cells in SSc patients than in healthy women. Additionally, no significant difference was observed in the levels of FM among T cells compared with PBMCs from the same draw date in women with SSc (n = 14). However, interpretation of results from SSc patients is confounded by the fact that the majority of patients studied were taking immunosuppressive medications. Consistent with modulation of results from immunosuppressive medication use, the total lymphocyte cell count was within the normal range (15% to 45%) for 4 SSc patients who had high levels of FM in T cells (patients no. 3, 6, 8, and 10), in contrast to the majority of patients who had low total lymphocyte counts (4% to 10%) and were negative for FM. One patient (no. 6, Figure 5) was tested on 2 draw dates with high levels in FM in T cells while taking low-dose methotrexate and undetectable FM in T cells while taking cyclophosphamide.
Male DNA in T cells from women with SSc (n = 19).
Numbers 6 and 7 correspond to 2 observations at different draw dates from the same woman.
Male DNA in T cells from women with SSc (n = 19).
Numbers 6 and 7 correspond to 2 observations at different draw dates from the same woman.
Discussion
Recent studies indicate that fetal DNA and cells routinely pass into the maternal circulation during normal pregnancy and that persistent FM can be found in the maternal circulation for decades after pregnancy completion.1,2 Persistent FM, when considered in concert with other observations, including the female predilection to autoimmunity and clinical similarities of cGvHD and some autoimmune diseases, led to the hypothesis that microchimerism is involved in autoimmune diseases such as SSc.4 In a prior study, we found that women with SSc had quantitatively greater levels of FM in peripheral blood than healthy women.3 The assay used in this study was developed for use in prenatal diagnosis and was employed in testing of multiple aliquots of whole blood targeting the Y-chromosome–specific sequence 49a, a moderately repetitive Y segment with no autosomal homology.22 This study was unable to determine the origin of fetal DNA in the maternal circulation. In women who are currently pregnant, fetal DNA has been found in almost 1000-fold higher amounts in plasma than in the cellular compartment of maternal peripheral blood,8 with levels rapidly dropping off within hours of delivery.9 These observations raise the question as to whether the increased FM that we previously found among women with SSc compared with controls is due to circulating fetal cells or to free fetal DNA released into the circulation from cellular breakdown in disease-affected tissues.
To elucidate the origin of long-term persistent FM from pregnancy in patients with SSc and healthy women, we developed a quantitative assay using real-time PCR targeting DYS14, a pseudogene of a multigene family,16,23,24 and employed the QPCR assay to test plasma and PBMC compartments of peripheral blood. In a large study of more than 100 women, we found that the majority of healthy women and women with SSc had no detectable male circulating DNA in plasma, and no difference was observed between women with SSc and healthy women. Studies from other investigators have shown that not all fetal DNA in plasma is soluble and cell-free, with some deriving from “plasmatic cells” in the process of apoptosis.19 Therefore additional studies were conducted for some women who had positive results in the plasma samples. All plasma samples became negative after being subjected to filtration, a process that removes cells, but keeps free circulating DNA. Thus, it is likely that the few positive results derived not from free DNA but from DNA associated with cells in the process of apoptosis.
In contrast to results for plasma, FM was quantitatively greater in the cellular component, in PBMCs, in women with SSc compared with healthy women. The difference between SSc patients and controls was statistically significant before and after adjusting for time since the last birth of a son. Results were also statistically significant when restricted to the study of subjects for whom sons had been born more than 10 years previously (examined because sons of some healthy women were born more recently than sons of women with SSc). Similarly, results were significant when women with SSc who had received a blood transfusion25 were excluded from the analysis.
To our knowledge, this is the first study to employ a quantitative assay to test the origin of long-term, persistent FM in maternal peripheral blood. Following our initial description of quantitatively greater levels of FM in whole blood samples,3 other studies were reported that tested for FM in peripheral blood from women with SSc and controls.26-30 The results of these studies have been variable. Most studies have used qualitative rather than quantitative methods and asked whether the frequency of any detectable FM differs in SSc patients compared with controls. Some studies have used a nested PCR test (nonquantitative) and described an increased frequency of detectable FM in women with SSc compared with controls,26 while others have used the same method and reported no significant difference.27 Variability of results may also arise owing to the particularY-chromosome–specific sequence used as a measure of FM, as these studies used a repetitive DNA family, DYZ1, that has homology with autosomal sequences, raising concern with respect to false-positive results.14,15 Other studies described a semiquantitative PCR method for the Y-chromosome–specific sequence 49a with no significant difference in SSc patients compared with controls.28 We found a marginal difference in the frequency with which FM could be detected in SSc patients compared with controls using a nonquantitative technique targeting the DYS14 sequence.29 In the current study, using the same Y-chromosome sequence, but using a real-time assay to quantify FM, we similarly observed an increased frequency of FM in women with SSc compared with controls (although prior results were just above the point of statistical significance, whereas current results were just short of significance). However, the quantitative assay clearly demonstrated a statistically significant difference in FM levels in PBMCs between patients and controls.
Another source of variability in studies of peripheral blood for FM is the compartment of blood that is investigated. Most studies have used DNA extracted from PBMCs, although some have tested DNA from whole blood, and others have tested DNA white blood cells. Thus, both differences in the techniques and differences in the source of sample from peripheral blood are likely, at least in part, to explain variability in reports. Differences in technique include whether an assay was qualitative or quantitative, the level of sensitivity of the assay, and the specific Y-chromosome sequence targeted. Further variability may derive from the fact that most studies use male DNA as the measure of FM, but some studies summarize results for women with sons while others have summarized results without providing pregnancy history for the subjects tested.
The use of male DNA as a measure of FM can be confounded by other potential sources of microchimerism. Cells are known to engraft and persist between twins, so that a woman with a fraternal male twin may have positive result.31 Twinning is also not uncommon, so that a woman could even potentially have microchimerism from an unrecognized twin that was lost early in gestation. Blood transfusion is another source of persistent microchimerism, as has been described in the circulation of trauma patients who have received multiple transfusions.25 Although FM has not yet been formally studied, it is presumed to also occur after a miscarriage or an induced abortion. Consistent with this likelihood, FM has been detected as early as 5 weeks of gestation,32 and large amounts of fetal DNA have been reported in the blood of women undergoing elective pregnancy termination.21Similar to other reports, our study found that some women who did not have a history of a male birth had a positive test for male DNA. Some of the women with positive results had either had a previous blood transfusion or reported a previous miscarriage. The origin of male DNA in others is not known, although the most likely source may be an unrecognized miscarriage. It is at least theoretically possible that male cells could be transferred from an older male sibling via the maternal circulation, or possibly even from sexual intercourse. Results are not likely to be due to contamination, since the QPCR assay used is a homogeneous assay, which did not require the opening of the amplification wells following PCR.
We also found that FM was enriched among T lymphocytes as compared with PBMCs among healthy women. Few SSc patients had FM among T lymphocytes; however, most were taking immunosuppressive drugs, thus limiting the ability to draw conclusions. The finding of FM among T cells in healthy women, in any case, indicates that the presence of FM among T lymphocytes per se is not likely to be detrimental to the host. Factors that could be important in determining whether FM has a neutral or a detrimental (or possibly even a beneficial) effect on the host include the particular human leukocyte antigen (HLA) genotype of the host, the HLA genotype of the microchimeric cell population, and the HLA-relationship between host and nonhost cells. In prior studies, we found that HLA compatibility forDRB1 of a previously born child was associated with almost a 9-fold increased risk of SSc in the mother.3Additionally, although donor CD4 and CD8 T cells are implicated in cGvHD,33 which has similarities to SSc, the mechanism by which FM putatively contributes to SSc would not necessarily expected to be similar. There are clinical and pathological differences in the 2 disorders, and most importantly, quantitative levels of chimerism differ dramatically. In cGvHD, donor cells essentially replace circulating host cells, whereas, in SSc, circulating fetal cells at most are estimated at less than 1 in a million host cells.34 Other ways by which FM could contribute to SSc include through the disruption of host-to-host interactions, by providing, for example, a source of peptides that are presented by host cells to other host cells, referred to as the “indirect” pathway of allorecognition.35
The real-time QPCR assay described in the current studies provides a tool for future studies in which other specific cellular subsets can be quantitatively assessed, thereby helping to clarify the direction for functional studies directed at elucidating the role of FM in the healthy host and in patients with SSc. Careful quantitative assessment of FM will be useful in resolving variability among initial reports investigating FM in SSc and other autoimmune diseases, including primary biliary cirrhosis and Sjögren syndrome.36Additionally, because FM was also found among healthy women, including among T lymphocytes, and because SSc resembles graft-versus-host disease for which a parous donor increases risk,37 38results of the current study also indicate that FM merits investigation in pheresis products used for stem cell transplantation.
We would like to thank the Scleroderma Registry, Jennifer Brackensick, and Gretchen Agee for their contributions to this study. We gratefully acknowledge Jennifer Pang and Heidi Hermes for technical support and Wendy Leisenring for statistical advice.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2002-01-0295.
Supported by the Scleroderma Foundation (Grant no. 007/01) and National Institutes of Health grants AI41721 and AR48084.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Nathalie Lambert, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D2-100, Seattle, WA 98109-1024; e-mail:nlambert@fhcrc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal