Abstract
Specific cytogenetic abnormalities predict prognosis in childhood acute myeloid leukemia (AML). However, it is unknown why they are predictive and whether this is related to drug resistance. We previously reported that Down syndrome (DS) AML was associated with favorable resistance profiles. Here, we successfully analyzed drug resistance and (cyto-) genetic abnormalities of 109 untreated childhood AML samples using the 4-day total cell-kill methyl-thiazol tetrazolium (MTT) assay. Patients were classified according to the genetic abnormalities in the leukemic cells: t(8;21), inv(16), t(15;17), t(9;11), other 11q23 translocations, abnormalities of chromosome 5/7, trisomy 8 alone, normal karyotype, single random, and multiple (defined as 2 or more) abnormalities. The DS AML samples were excluded from the subgroup analysis. Samples with chromosome 5/7 abnormalities were median 3.9-fold (P = .01) more resistant to cytarabine than other AML samples. The t(9;11) samples were more sensitive to cytarabine (median 2.9-fold, P = .002), etoposide (13.1-fold, P = .001), the anthracyclines (2.9- to 8.0-fold, P < .01), and 2-chlorodeoxyadenosine (10.0-fold, P = .002) than other AML samples. The trisomy 8 and t(15;17) groups were too small for meaningful analysis. All other genetic subgroups did not show specific resistance profiles. Overall, we found no differences in drug resistance in samples taken at diagnosis between patients remaining in continuous complete remission (CCR) versus the refractory/relapsed patients. Within several genetic subgroups, however, relapsed/refractory patients were more cytarabine resistant when compared with patients remaining in CCR, but numbers were small and the results were not significant. We conclude that some, but not all, cytogenetic subgroups in childhood AML display specific drug-resistance profiles.
Introduction
Genetic alterations can be detected in approximately 80% of de novo childhood acute myeloid leukemia (AML) cases and can be divided into random and nonrandom abnormalities.1 These alterations are not only recognized as important initiating events in the process of leukemogenesis but also as indicators of clinical outcome.1-5 Consequently, their presence or absence may determine the choice of therapy, for instance, by stratification into risk groups.2 For example, children with Down syndrome and AML (DS AML) often receive less intensive therapy, as a consequence of the unique biology of their disease, and its apparent sensitivity to chemotherapy, both in vivo and in vitro.6-9 Patients with acute promyelocytic leukemia with PML-RARα transcripts show striking responses to differentiation therapy with all-trans-retinoic acid (ATRA), whereas other AML cases do not.10,11 In adult AML, high-dose cytarabine was shown to be especially beneficial to patients with t(8;21) leukemias.12 However, in general, it is unknown why certain genetic subgroups respond better to chemotherapy than others and whether this is related to differences in cellular drug resistance.
There is no firm consensus considering the risk assignment of specific genetic subgroups in childhood AML. The t(8;21) and inv(16) are usually classified as good-risk abnormalities, but the favorable outcome of t(15;17) patients is mainly achieved by the addition of ATRA to regular chemotherapy.10,11 With chemotherapy alone, the clinical outcome of t(15;17) leukemias is less good (30%-50% survival), but this is also caused by a high early death rate and is not necessarily related to chemotherapy resistance.4 13
In general, in childhood AML, 11q23 translocations and abnormalities in chromosome 5/7 (including del(5q)/−5; del(7q)/−7) are considered to be poor risk.13,14 However, the group with 11q23 translocations can be further subdivided according to the particular translocation partner involved. In certain studies, the t(9;11) has been associated with a favorable outcome5,15,16 also in adult AML,17 whereas one other study did not confirm this.18 In one report, the t(10;11) showed a poor clinical outcome, whereas t(9;11) and other 11q23 translocations did not differ in prognosis.19 Kalwinsky et al suggested the better outcome of t(9;11) to be related to increased drug sensitivity, especially to etoposide.20 In addition, some groups classify multiple cytogenetic abnormalities as unfavorable.2
Differences in prognosis may reflect differences in treatment, cellular drug resistance, pharmacokinetics, and relapse potential of residual disease. We and others have already demonstrated the prognostic importance of cellular resistance in childhood leukemias.21-24 We previously reported childhood AML to be relatively resistant to a large number of drugs when compared with childhood acute lymphoblastic leukemia (ALL).25 Also, specific genetic ALL and AML subgroups may show different resistance profiles. For example, hyperdiploid (> 50) ALL is particularly sensitive to antimetabolites andl-asparaginase26; ALL with TEL-AML gene fusion is relatively sensitive to l-asparaginase27; Down syndrome AML cases show a sensitive profile to a large number of drugs9; and AML with inv(16) shows increased cytarabine sensitivity, as well as higher cytarabine incorporation in blasts, when compared with other AML cells.28
In this study we aimed at identifying differences in drug resistance profiles of separate genetic subgroups in childhood AML to determine whether these could explain their prognostic relevance. Although not the primary aim of this study, we also assessed the relation between the clinical outcome of the different cytogenetic subgroups and in vitro drug resistance for the clinically most relevant drugs.
Patients, materials, and methods
Patient samples
We tested either bone marrow or peripheral blood samples from children (0 to ≤ 18 years of age) diagnosed with de novo AML. Samples for drug resistance testing were taken with institutional review board approval from the VU University Medical Center Amsterdam and only after informed consent had been obtained. Two collaborative groups participated: the AML-BFM (Berlin-Frankfurt-Münster) Study Group (Münster, Germany) and the Dutch Childhood Leukemia Study Group (DCLSG, The Hague, The Netherlands). Both groups provided us with patient samples and performed central review of the clinical and cell biologic data, including the (cyto-) genetics, the French-Amercian-British (FAB)–type classification, and treatment outcome. Most patients were treated according to 5 different BFM-like protocols, which included the AML-BFM studies 87 (n = 5 of 204), 93 (n = 95 of 204), and 98 (n = 19 of 204) as well as the Dutch DCLSG acute nonlymphoblastic leukemia (ANLL) 87 (n = 29 of 204) and 94 (n = 43 of 204) studies.29-31 The other 13 patients were treated according to other protocols. Because the results on children with DS AML have been reported elsewhere, we did not analyze this particular subgroup in this study.9
Cytogenetic and molecular genetic analyses
Cytogenetic analysis was performed in 7 cytogenetic laboratories in The Netherlands and centrally reviewed by one of the coauthors (R.M.S.). In Germany all samples were sent to Giessen, and karyotyping was performed at the Oncogenetic Laboratory of the Children's Hospital by the coauthor J.H. and coworkers.
Cytogenetic studies on either bone marrow or peripheral blood samples, taken prior to induction therapy, were performed using standard banding techniques.32 Routinely, at least 20 banded metaphases were analyzed and the results described according to the International System for Human Cytogenetic Nomenclature 1995.33Patients with fewer than 5 metaphases assessable for cytogenetic analysis and showing an apparently normal karyotype were excluded.
In a number of samples, additional molecular genetic studies were performed to confirm or exclude the presence of specific genetic alterations, either by dual-colored fluorescent in situ hybridization (FISH) using a probe for the MLL gene at 11q23 34 and a cosmid contig for detecting inv(16)(p13q22) 35 or by reverse transcriptase–polymerase chain reaction (RT-PCR) (AML1-ETO, PML-RARα, CBFβ-MYH11, MLL-AF4, and MLL-AF9).36
The patients were assigned to different commonly accepted categories based on the cytogenetic and molecular genetic abnormalities at diagnosis: t(8;21), inv(16), t(15;17), t(9;11), other 11q23 translocations (ie, non-t(9;11)), trisomy 8 alone, abnormalities of chromosome 5/7 (including monosomy 5, del(5q), add(5), and monosomy 7), random single chromosomal abnormalities, multiple chromosomal abnormalities (defined as 2 or more abnormalities), normal karyotype, and DS AML.
Drug-resistance testing
The cellular drug resistance studies were performed at the Research Laboratory of Pediatric Oncology, VU University Medical Center, Amsterdam. Mononuclear cells were isolated by density gradient centrifugation with Ficoll Isopaque. In case of a low blast percentage (< 80%), we determined the presence of contaminating mature granulocytes and/or lymphocytes. To eliminate granulocytes, the samples were frozen in liquid nitrogen and thawed at a later stage to perform cellular resistance testing.25 Lymphocytes were removed using immunomagnetic beads.37
Cellular resistance was measured using a 4-day total cell kill assay, as described before.38 Briefly, cells were seeded in 96-well microculture plates (80 μL of 0.8 × 106/mL to 1.0 × 106/mL) and cultured for 4 days. Drugs were added in 6 concentrations and always in duplicate. Six wells contained culture medium only, and 6 other wells contained culture medium with cells to determine the control cell survival (CCS). After 4 days methyl-thiazol tetrazolium (MTT) was added, which can be converted by viable cells into a colored formazan product that can be measured spectrophotometrically at 562 nm.38 The optical density (OD) is linearly related to the number of viable cells. Cytotoxicity was calculated at each drug concentration by the following equation: (OD treated well/mean OD control wells) × 100%, after correction for the background OD of the wells with culture medium only. Results were considered evaluable only if the control wells contained at least 70% leukemic cells (determined by morphology after May-Grünwald Giemsa [MGG] staining) after 4 days of culture and if the mean control OD, after correction for the background, at day 4, exceeded 0.05 arbitrary units. The LC50 value, which is the drug concentration that kills 50% of the leukemic cells, was used as a measure of resistance. Sample source (bone marrow or peripheral blood) and cryopreservation do not influence the results obtained by cellular resistance testing.39,40 There is no statistically significant relationship between OD and cellular drug resistance data, or between OD and CCS, as we reported previously.25
The following drugs were tested (minimal and maximal concentration): amsacrine (0.006-20 μg/mL; Amsidine, Parke-Davis, Hoofdorp, The Netherlands), busulfan (1.23-300 μg/mL; Myleran, Wellcome, Zeist, The Netherlands), 2-chlorodeoxyadenosine (0.0004-40 μg/mL; Leustatin, Ortho-Biotech, Raritan, NJ), cytarabine (0.002-2.5 μg/mL; Cytosar, Upjohn, Woerden, The Netherlands), daunorubicin (0.002-2 μg/mL; Cerubidine, Rhône Poulenc, Amstelveen, The Netherlands), doxorubicin (0.008-8 μg/mL; Adriblastina, Pharmacia and Upjohn, Woerden, The Netherlands), etoposide (0.05-50 μg/mL; VePesid, Bristol-Myers, Woerden, The Netherlands), idarubicin (0.002-2 μg/mL; Zavedos, Pharmacia and Upjohn), 4-hydroperoxy-ifosfamide (0.1-100 μg/mL; releases 4-hydroxy ifosfamide in solution, which is the active metabolite of ifosfamide; Asta-Medica, Diemen, The Netherlands),l-asparaginase (0.003-10 IU/mL; Paronal, Christiaens, Breda, The Netherlands), mitoxantrone (0.001-1 μg/mL; Novantrone, AHP Pharma Wyeth Lederle, Hoofdorp, The Netherlands), prednisolone disodiumphosphate (0.008-250 μg/mL; Bufa Pharmaceutical Products, Uitgeest, The Netherlands), 6-thioguanine (1.56-50 μg/mL; Lanvis, Wellcome), and vincristine (0.05-50 μg/mL; Oncovin, Eli Lilly, Nieuwegein, The Netherlands).
Part of the data on drug resistance in AML have been published before in the context of other study questions but not in relation to leukemia-specific genetic alterations.25
Statistical analysis
Differences in the distribution of LC50 values were analyzed using the Mann-Whitney U test. For statistical comparisons of categorical variables, the χ2 test was used, or the Fisher exact test was used in case of small numbers. Correlations were assessed using the Spearman rank correlation coefficient (ρ). The duration of event-free survival (EFS) was defined as the time from diagnosis until the date of the first adverse event (relapse, death from any reason or the development of a second malignancy) or, if no such event occurred, until the date of last contact. Patients who did not attain a complete remission were considered failures at time zero. Probabilities of EFS were estimated by the methods of Kaplan and Meier with standard errors according to Greenwood and were compared using the log-rank test.
If the number of samples in a specific subgroup was fewer than 5, statistical analyses were not performed. P ≤ .01 was considered statistically significant (2-tailed test). In using the term “complementary group” in statistical comparisons, we defined “complementary” by all other AML samples (included in the study population) than those with the particular genetic abnormality.
Results
Patient characteristics
From February 1990 to February 2000, 204 de novo AML samples were sent to our laboratory for drug resistance testing. We received 119 samples from the 1245 de novo childhood AML patients included in the AML-BFM Study Group (SG) trials as well as 85 samples from 171 patients included in the DCLSG trials over that time period.
Of these 204 samples, 142 (70%) were successfully tested for drug resistance. For 109 of these patients, both genetic and drug resistance data were available, which is the (selected) study population described here. This included the subgroup of DS AML patients (n = 7), which are not analyzed as a separate subgroup in this study but were reported previously.9
Children with a successful MTT assay (n = 142) had a significantly higher white blood cell count (WBC) than children without a successful assay (WBC: median 30.7 × 109/L vs 8.0 × 109/L, P < 0.001). However, age (median 7.9 vs 5.1 years, P = .02) and sex (P = .61) were not significantly different.
Children for whom genetic data were available (n = 159) did not differ significantly from children without such data, considering age (P = .13), WBC (P = .97), or sex (P = .62).
The 109 children with data on both genetic alterations and drug resistance presented with a significantly higher WBC (median 35.2 × 109/L vs 22.0 × 109/L,P < .001) but did not differ in age (P = .41) or sex (P = .92) compared with the 95 children without these data. Patient characteristics of the total study population and the cytogenetic subgroups (see below) are presented in Table 1.
Patient characteristics of childhood AML patients (n = 109) for whom data on karyotyping or molecular genetics and cellular drug resistance were available
| . | All patients . | t(8;21) . | inv(16) . | t(15;17) . | Normal karyotype . | Abnormalities 5/7 . | t(9;11) . | Other 11q23 translocations . | Single random abnormality . | Multiple abnormalities . | Trisomy 8 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 109 | 11 | 10 | 3 | 27 | 8 | 9 | 8* | 12 | 8 | 4 |
| Median age, y (P25-75) | 7.2 (2.8-12.1) | 10.9 (7.0-14.0) | 7.6 (4.6-11.7) | 13.3 (10.1-14.7)† | 10.5 (5.8-13.0) | 9.9 (5.2-10.5) | 3.7 (2.0-6.1) | 7.1 (5.6-10.2) | 3.5 (2.2-6.2) | 11.9 (3.5-14.0) | 13.5 (9.3-15.3)† |
| Sex; M, F | 60, 49 | 6, 5 | 5, 5 | 2, 1 | 10, 17 | 5, 3 | 3, 6 | 6, 2 | 9, 3 | 5, 3 | 3, 1 |
| Median WBC, ×109/L (P25-75) | 35.2 (12.5-81.5) | 20.2 (9.0-29.9) | 53.0 (36.7-67.9) | 6.8 (3.4-22.5)† | 24.9 (15.5-70.0) | 62.7 (18.7-88.5) | 14.4 (6.6-170.4) | 130.2 (59.7-173.2) | 41.4 (24.8-122.9) | 33.5 (11.5-92.6) | 82.5 (56.7-123.1)† |
| FAB | |||||||||||
| M0 | 5 | — | — | — | 1 | 1 | — | 1 | — | 1 | — |
| M1 | 12 | 1 | — | — | 4 | 2 | — | — | 2 | 1 | 2 |
| M2 | 27 | 9 | — | — | 9 | 3 | — | — | 2 | 4 | — |
| M3 | 4 | — | — | 3 | 1 | — | — | — | — | — | — |
| M4 (M4Eo) | 31 (13) | 1 (0) | 10 (8) | — | 8 (4) | 2 (0) | 1 (0) | 2 (0) | 5 (1) | — | 1 (0) |
| M5 | 21 | — | — | — | 2 | — | 8 | 5 | 2 | 2 | 1 |
| M6 | 1 | — | — | — | 1 | — | — | — | — | — | — |
| M7 | 6 | — | — | — | — | — | — | — | — | — | — |
| RAEB-t | 2 | — | — | — | 1 | — | — | — | 1 | — | — |
| Median CCS (P25-75) | 101 (80-128) | 94 (80-120) | 104 (88-118) | 107 (92-138)† | 100 (71-134) | 89 (71-117) | 83 (69-114) | 117 (86-125) | 100 (86-120) | 115 (70-158) | 94 (79-113)† |
| Median OD per 105 cells (P25-75) | 0.50 (0.37-0.65) | 0.46 (0.30-0.51) | 0.71 (0.61-0.81) | 0.47 (0.43-0.52)† | 0.54 (0.35-0.70) | 0.48 (0.29-0.49) | 0.45 (0.41-0.56) | 0.55 (0.32-0.86) | 0.45 (0.42-0.63) | 0.36 (0.35-0.39) | 0.26 (0.16-0.37)† |
| . | All patients . | t(8;21) . | inv(16) . | t(15;17) . | Normal karyotype . | Abnormalities 5/7 . | t(9;11) . | Other 11q23 translocations . | Single random abnormality . | Multiple abnormalities . | Trisomy 8 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 109 | 11 | 10 | 3 | 27 | 8 | 9 | 8* | 12 | 8 | 4 |
| Median age, y (P25-75) | 7.2 (2.8-12.1) | 10.9 (7.0-14.0) | 7.6 (4.6-11.7) | 13.3 (10.1-14.7)† | 10.5 (5.8-13.0) | 9.9 (5.2-10.5) | 3.7 (2.0-6.1) | 7.1 (5.6-10.2) | 3.5 (2.2-6.2) | 11.9 (3.5-14.0) | 13.5 (9.3-15.3)† |
| Sex; M, F | 60, 49 | 6, 5 | 5, 5 | 2, 1 | 10, 17 | 5, 3 | 3, 6 | 6, 2 | 9, 3 | 5, 3 | 3, 1 |
| Median WBC, ×109/L (P25-75) | 35.2 (12.5-81.5) | 20.2 (9.0-29.9) | 53.0 (36.7-67.9) | 6.8 (3.4-22.5)† | 24.9 (15.5-70.0) | 62.7 (18.7-88.5) | 14.4 (6.6-170.4) | 130.2 (59.7-173.2) | 41.4 (24.8-122.9) | 33.5 (11.5-92.6) | 82.5 (56.7-123.1)† |
| FAB | |||||||||||
| M0 | 5 | — | — | — | 1 | 1 | — | 1 | — | 1 | — |
| M1 | 12 | 1 | — | — | 4 | 2 | — | — | 2 | 1 | 2 |
| M2 | 27 | 9 | — | — | 9 | 3 | — | — | 2 | 4 | — |
| M3 | 4 | — | — | 3 | 1 | — | — | — | — | — | — |
| M4 (M4Eo) | 31 (13) | 1 (0) | 10 (8) | — | 8 (4) | 2 (0) | 1 (0) | 2 (0) | 5 (1) | — | 1 (0) |
| M5 | 21 | — | — | — | 2 | — | 8 | 5 | 2 | 2 | 1 |
| M6 | 1 | — | — | — | 1 | — | — | — | — | — | — |
| M7 | 6 | — | — | — | — | — | — | — | — | — | — |
| RAEB-t | 2 | — | — | — | 1 | — | — | — | 1 | — | — |
| Median CCS (P25-75) | 101 (80-128) | 94 (80-120) | 104 (88-118) | 107 (92-138)† | 100 (71-134) | 89 (71-117) | 83 (69-114) | 117 (86-125) | 100 (86-120) | 115 (70-158) | 94 (79-113)† |
| Median OD per 105 cells (P25-75) | 0.50 (0.37-0.65) | 0.46 (0.30-0.51) | 0.71 (0.61-0.81) | 0.47 (0.43-0.52)† | 0.54 (0.35-0.70) | 0.48 (0.29-0.49) | 0.45 (0.41-0.56) | 0.55 (0.32-0.86) | 0.45 (0.42-0.63) | 0.36 (0.35-0.39) | 0.26 (0.16-0.37)† |
The clinical data of specific genetic subgroups are given. Two patients with MLL rearrangements, but without identified translocation partner chromosome, were excluded in the subgroup of other 11q23 abnormalities, while 7 Down syndrome AML patients have been reported elsewhere.
Characteristics of the MTT assay are shown for each genetic subgroup—that is, CCS after 4 days of culture and the OD per 105 cells. For statistical analysis, see “Results.”
P25-75, 25th and 75th percentile; WBC indicates white blood cell count; FAB, French-American-British morphology classification; RAEB-t, refractory anemia in excess of blasts in transformation; CCS, control cell survival after 4 days of culture; OD, optical density.
Translocation partners were t(4;11), n = 2; t(6;11), n = 3; t(10;11), n = 2; t(11;17), n = 1.
The range is given instead of P25-75.
Cytogenetic and molecular genetic abnormalities
The patients were assigned to different genetic subgroups according to the genetic abnormalities identified in the leukemic cells. Patient characteristics for the specific genetic subgroups, excluding those with Down syndrome, are shown in Table 1. The one patient with FAB M3 and normal cytogenetics was negative for the PML-RARα transcript by PCR. Of the 4 patients with normal karyotype and FAB M4Eo, 2 were negative for inv(16) with FISH and the other 2 patients were not studied with FISH due to lack of material. The one patient with FAB M4Eo and a single random chromosomal abnormality was also negative for inv(16) with FISH. Of 2 patients with 11q23 abnormalities, the partner chromosome was unknown.
For the patients for whom these genetic data were available, we compared patient characteristics at diagnosis of each specific genetic subgroup with those of individuals without this specific abnormality. The only significant difference was that children with t(9;11) were younger than the complementary AML patients (3.7 vs 8.5 years,P = .01). The subgroups with t(15;17) and trisomy 8 were not analyzed because the sample sizes were too small.
The incidence of the different specific genetic abnormalities in our study is only slightly different from those reported in the literature. However, patients with t(15;17) were underreported in our group,4,41 but the frequency of this translocation is known to differ among different geographic areas.14 The frequency of abnormalities of chromosome 5/7 was 5.7% (9 of 158) in our study, which is higher than in 2 United States series of de novo AML (2%)4,41 but similar in frequency to one other European study.14
Treatment results for cytogenetic subgroups
In Table 2 the treatment results (3-year probability of event-free survival) for each of the cytogenetic subgroups included in this study are given. This concerned 99 patients of the total group of 109 patients for whom data on genetic alterations and drug resistance were available. The 7 DS AML patients and the 2 patients with 11q23 abnormalities without known translocation partner, as well as 1 patient from the “normal karyotype” subgroup, were excluded. The latter patient was not treated with curative attempt due to comorbidity (severe neurologic disease). Confidence intervals were very large due to the small number of patients in each particular subgroup. Only the subgroup of patients with other 11q23 abnormalities did significantly worse than AML patients without these abnormalities (3-year EFS 25.0% vs 54.6%, P = 0.01, log-rank).
Treatment outcome data from the different cytogenetic subgroups included in this study, given as 3-year event-free survival rates and 95% confidence intervals
| Cytogenetic subgroup . | n . | pEFS, 3 y, % . | 95% CI . | P, log-rank . |
|---|---|---|---|---|
| All patients | 99 | 52.1 | 41.8–62.4 | |
| t(8;21) | 11 | 54.5 | 25.1–84.0 | .53 |
| Inv(16) | 10 | 66.7 | 35.9–97.5 | .39 |
| Normal karyotype | 26 | 68.4 | 50.2–86.5 | .09 |
| Abnormalities 5/7 | 8 | 50.0 | 15.4–84.6 | .56 |
| t(9;11) | 9 | 55.6 | 23.1–88.0 | .84 |
| Other 11q23 translocations | 8 | 25.0 | 0.0–55.0 | .01 |
| Single random abnormalities | 12 | 27.8 | 0.0–58.8 | .64 |
| Multiple abnormalities | 8 | 62.5 | 29.0–69.0 | .70 |
| Cytogenetic subgroup . | n . | pEFS, 3 y, % . | 95% CI . | P, log-rank . |
|---|---|---|---|---|
| All patients | 99 | 52.1 | 41.8–62.4 | |
| t(8;21) | 11 | 54.5 | 25.1–84.0 | .53 |
| Inv(16) | 10 | 66.7 | 35.9–97.5 | .39 |
| Normal karyotype | 26 | 68.4 | 50.2–86.5 | .09 |
| Abnormalities 5/7 | 8 | 50.0 | 15.4–84.6 | .56 |
| t(9;11) | 9 | 55.6 | 23.1–88.0 | .84 |
| Other 11q23 translocations | 8 | 25.0 | 0.0–55.0 | .01 |
| Single random abnormalities | 12 | 27.8 | 0.0–58.8 | .64 |
| Multiple abnormalities | 8 | 62.5 | 29.0–69.0 | .70 |
The 2 patients with 11q23 translocations without known karyotype and 1 patient in the “normal cytogenetics” subgroup (not treated with curative attempt), as well as the Down syndrome AML patients, were excluded. The subgroups with less than 5 patients were not analyzed for treatment outcome (ie, t(15;17) and trisomy 8).
P, log-rank compares the 3-year probability of event-free survival (pEFS) of that particular cytogenetic subgroup versus the complementary AML group.
n indicates number of patients included in the analysis; CI, confidence interval.
Cellular drug resistance
MTT assay.
The MTT assay could be successfully performed (for at least 1 drug) in 70% of patients. The sample source for the 109 children with data on both drug resistance and genetic abnormalities was peripheral blood in 31 and bone marrow in 78 cases. This does not influence the results of drug resistance testing.39
The success rate of the MTT assay in relation to genetic subgroups showed a relatively low success rate of 55% for samples with t(8;21). On the other hand, 100% of samples from patients with inv(16), t(15;17), or trisomy 8 were tested successfully, and samples with chromosome 5/7 abnormalities could be successfully tested in 89%.
The reasons for failure of the MTT assay included (1) assay not being performed due to lack of cells or too few blasts at the start of the culture (n = 26 samples) and (2) true assay failures: too low optical density (n = 6), insufficient blast percentage after 4 days of culture (n = 25), and 5 other failures (such as infected culture).
The control cell survival (CCS) after 4 days of culture was median 101% (range 24%-217%). There were no statistically significant differences in CCS when specific genetic subgroups were compared with the complementary group of AML samples (data shown in Table 1).
The median optical density per 105 cells (the metabolic capacity of viable cells to convert MTT into formazan) was 0.50 (range 0.12-1.16). Samples with inv(16) had a significantly higher OD per 105 cells (median 0.71) than the complementary group of AML samples (median 0.48, P = .003) (Table 1).
Dose response curves could be obtained for most drugs. However, even the highest concentration of prednisolone (250 μg/mL) was unable to kill more than 50% of the leukemic cells in most cases, resulting in LC50 values of more than 250 μg/mL.
There were no significant differences in drug resistance between AML samples that lacked data on karyotyping or molecular genetics and samples for which data on genetic alterations were available (data not shown).
Cellular drug resistance and specific genetic subgroups.
Data on in vitro drug resistance specified per genetic subgroup are given in Tables 3 and4. We compared each particular genetic subgroup with the complementary group of AML samples for which results on drug resistance testing were available.
Cellular drug resistance of the total group of AML patients as well as 5 specific genetic subgroups in childhood AML
| Drug . | All patients3-150 . | Patients with data on genetics only3-151 . | t(8;21) . | inv (16) . | Normal keryotype . | Single random abnormalities . | Multiple abnormalities . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | |
| Ara-C | 134 | 0.44 (0.22-1.27) | 105 | 0.43 (0.19-1.09) | 11 | 0.56 (0.36-1.53) | 10 | 0.47 (0.31-1.30) | 25 | 0.43 (0.28-0.61) | 10 | 0.34 (0.19-0.66) | 8 | 1.29 (0.26-1.73) |
| DNR | 122 | 0.22 (0.10-0.38) | 93 | 0.21 (0.10-0.38) | 11 | 0.33 (0.11-0.40) | 8 | 0.30 (0.27-0.40) | 23 | 0.23 (0.10-0.37) | 10 | 0.25 (0.09-0.37) | 5 | 0.43 (0.06-0.79) |
| Dox | 88 | 0.48 (0.27-0.85) | 65 | 0.48 (0.28-0.83) | 9 | 0.70 (0.49-0.80) | 3 | 0.84 (0.82-0.86)3-153 | 15 | 0.49 (0.35-0.72) | 10 | 0.56 (0.33-0.76) | 4 | 0.71 (0.12-1.34) |
| Ida | 91 | 0.13 (0.04-0.26) | 70 | 0.11 (0.03-0.25) | 6 | 0.41 (0.17-0.72) | 6 | 0.25 (0.15-0.28) | 16 | 0.11 (0.03-0.23) | 9 | 0.11 (0.09-0.25) | 3 | 0.02 (0.02-0.37)3-153 |
| VP16 | 131 | 7.42 (1.90-19.35) | 101 | 6.64 (1.68-18.18) | 10 | 6.41 (2.93-30.31) | 10 | 13.90 (10.30-20.75) | 25 | 7.45 (1.95-12.50) | 9 | 9.38 (5.18-25.00) | 8 | 3.05 (0.61-21.58) |
| 6-TG | 134 | 5.91 (3.81-10.51) | 107 | 5.99 (3.75-10.57) | 11 | 7.74 (4.86-12.81) | 10 | 7.41 (6.94-10.62) | 25 | 5.86 (3.40-7.32) | 12 | 5.46 (3.66-7.46) | 8 | 7.17 (3.43-11.26) |
| Mitox | 113 | 0.08 (0.02-0.34) | 86 | 0.06 (0.01-0.29) | 10 | 0.07 (0.05-0.42) | 8 | 0.30 (0.10-0.45) | 20 | 0.16 (0.03-0.27) | 11 | 0.13 (0.03-0.29) | 6 | 0.04 (0.00-0.18) |
| Amsa | 108 | 0.51 (0.14-1.30) | 84 | 0.41 (0.13-1.10) | 8 | 0.50 (0.16-2.15) | 10 | 0.63 (0.57-1.28) | 19 | 0.70 (0.15-1.19) | 8 | 0.36 (0.13-0.76) | 6 | 0.55 (0.10-1.30) |
| CdA | 97 | 0.020 (0.005-0.031) | 74 | 0.018 (0.004-0.029) | 6 | 0.021 (0.006-0.07) | 10 | 0.023 (0.012-0.03) | 17 | 0.020 (0.013-0.028) | 6 | 0.010 (0.003-0.024) | 6 | 0.012 (0.007-0.023) |
| Bus | 96 | 36.1 (25.2-58.6) | 73 | 35.1 (22.5-55.6) | 7 | 47.9 (27.7-65.1) | 8 | 28.6 (21.7-36.7) | 15 | 32.1 (26.2-47.2) | 7 | 37.8 (14.0-41.7) | 5 | 41.7 (25.6-76.5) |
| Ifos | 110 | 11.85 (5.68-14.79) | 85 | 11.50 (5.50-14.48) | 8 | 12.25 (9.20-14.45) | 9 | 14.48 (11.36-20.31) | 22 | 10.06 (5.27-12.69) | 6 | 8.55 (3.35-14.60) | 7 | 7.55 (5.09-13.41) |
| Pred | 137 | >250 (>250->250) | 104 | >250 (>250->250) | 10 | >250 (65->250) | 10 | >250 (207->250) | 24 | >250 (237->250) | 12 | >250 (>250->250) | 8 | >250 (215->250) |
| VCR | 129 | 2.99 (0.71-23.61) | 97 | 2.64 (0.71-23.41) | 11 | 2.26 (1.61-4.95) | 10 | 6.52 (2.34-31.62) | 21 | 10.16 (2.64-21.55) | 11 | 1.59 (0.32-11.27) | 8 | 1.96 (0.42-2.62) |
| ASP3-152 | 113 | 0.88 (0.29-1.45) | 85 | 0.80 (0.24-1.46) | 9 | 1.68 (0.99-2.00) | 9 | 0.90 (0.57-1.46) | 18 | 0.70 (0.12-1.23) | 11 | 0.96 (0.33-1.21) | 7 | 0.32 (0.15-0.55) |
| Drug . | All patients3-150 . | Patients with data on genetics only3-151 . | t(8;21) . | inv (16) . | Normal keryotype . | Single random abnormalities . | Multiple abnormalities . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | |
| Ara-C | 134 | 0.44 (0.22-1.27) | 105 | 0.43 (0.19-1.09) | 11 | 0.56 (0.36-1.53) | 10 | 0.47 (0.31-1.30) | 25 | 0.43 (0.28-0.61) | 10 | 0.34 (0.19-0.66) | 8 | 1.29 (0.26-1.73) |
| DNR | 122 | 0.22 (0.10-0.38) | 93 | 0.21 (0.10-0.38) | 11 | 0.33 (0.11-0.40) | 8 | 0.30 (0.27-0.40) | 23 | 0.23 (0.10-0.37) | 10 | 0.25 (0.09-0.37) | 5 | 0.43 (0.06-0.79) |
| Dox | 88 | 0.48 (0.27-0.85) | 65 | 0.48 (0.28-0.83) | 9 | 0.70 (0.49-0.80) | 3 | 0.84 (0.82-0.86)3-153 | 15 | 0.49 (0.35-0.72) | 10 | 0.56 (0.33-0.76) | 4 | 0.71 (0.12-1.34) |
| Ida | 91 | 0.13 (0.04-0.26) | 70 | 0.11 (0.03-0.25) | 6 | 0.41 (0.17-0.72) | 6 | 0.25 (0.15-0.28) | 16 | 0.11 (0.03-0.23) | 9 | 0.11 (0.09-0.25) | 3 | 0.02 (0.02-0.37)3-153 |
| VP16 | 131 | 7.42 (1.90-19.35) | 101 | 6.64 (1.68-18.18) | 10 | 6.41 (2.93-30.31) | 10 | 13.90 (10.30-20.75) | 25 | 7.45 (1.95-12.50) | 9 | 9.38 (5.18-25.00) | 8 | 3.05 (0.61-21.58) |
| 6-TG | 134 | 5.91 (3.81-10.51) | 107 | 5.99 (3.75-10.57) | 11 | 7.74 (4.86-12.81) | 10 | 7.41 (6.94-10.62) | 25 | 5.86 (3.40-7.32) | 12 | 5.46 (3.66-7.46) | 8 | 7.17 (3.43-11.26) |
| Mitox | 113 | 0.08 (0.02-0.34) | 86 | 0.06 (0.01-0.29) | 10 | 0.07 (0.05-0.42) | 8 | 0.30 (0.10-0.45) | 20 | 0.16 (0.03-0.27) | 11 | 0.13 (0.03-0.29) | 6 | 0.04 (0.00-0.18) |
| Amsa | 108 | 0.51 (0.14-1.30) | 84 | 0.41 (0.13-1.10) | 8 | 0.50 (0.16-2.15) | 10 | 0.63 (0.57-1.28) | 19 | 0.70 (0.15-1.19) | 8 | 0.36 (0.13-0.76) | 6 | 0.55 (0.10-1.30) |
| CdA | 97 | 0.020 (0.005-0.031) | 74 | 0.018 (0.004-0.029) | 6 | 0.021 (0.006-0.07) | 10 | 0.023 (0.012-0.03) | 17 | 0.020 (0.013-0.028) | 6 | 0.010 (0.003-0.024) | 6 | 0.012 (0.007-0.023) |
| Bus | 96 | 36.1 (25.2-58.6) | 73 | 35.1 (22.5-55.6) | 7 | 47.9 (27.7-65.1) | 8 | 28.6 (21.7-36.7) | 15 | 32.1 (26.2-47.2) | 7 | 37.8 (14.0-41.7) | 5 | 41.7 (25.6-76.5) |
| Ifos | 110 | 11.85 (5.68-14.79) | 85 | 11.50 (5.50-14.48) | 8 | 12.25 (9.20-14.45) | 9 | 14.48 (11.36-20.31) | 22 | 10.06 (5.27-12.69) | 6 | 8.55 (3.35-14.60) | 7 | 7.55 (5.09-13.41) |
| Pred | 137 | >250 (>250->250) | 104 | >250 (>250->250) | 10 | >250 (65->250) | 10 | >250 (207->250) | 24 | >250 (237->250) | 12 | >250 (>250->250) | 8 | >250 (215->250) |
| VCR | 129 | 2.99 (0.71-23.61) | 97 | 2.64 (0.71-23.41) | 11 | 2.26 (1.61-4.95) | 10 | 6.52 (2.34-31.62) | 21 | 10.16 (2.64-21.55) | 11 | 1.59 (0.32-11.27) | 8 | 1.96 (0.42-2.62) |
| ASP3-152 | 113 | 0.88 (0.29-1.45) | 85 | 0.80 (0.24-1.46) | 9 | 1.68 (0.99-2.00) | 9 | 0.90 (0.57-1.46) | 18 | 0.70 (0.12-1.23) | 11 | 0.96 (0.33-1.21) | 7 | 0.32 (0.15-0.55) |
Data on the other specific genetic subgroups are given in Table4.
Data on statistical comparisons are given in “Results.” Results are expressed as median LC50 values in μg/mL. In parentheses, the 25th-75th percentile (P25-P75) is given. n indicates the number of samples tested for that particular drug; Ara-C, cytarabine; DNR, daunorubicin; Dox, doxorubicin; Ida, idarubicin; VP16, etoposide; 6-TG, 6-thioguanine; Mitox, mitoxantrone; Amsa, amsacrine; CdA, 2-chlorodeoxyadenosine; Bus, busulfan; Ifos, 4-HOO-ifosfamide; Pred, prednisolone; VCR, vincristine; ASP, l-asparaginase.
Includes all patients for whom drug resistance data were available (n = 142), with or without cytogenetic data.
Includes patients for whom both data on drug resistance and genetic abnormalities were available (n = 109).
L-ASP in IU/mL, not in μg/mL.
Ranges are given instead of the P25-75.
Cellular drug resistance of 5 specific genetic subgroups in childhood AML
| Drug . | Abnormalities 5/7 . | 11q23 with t(9;11) . | 11q23; non-t(9;11) . | t(15;17) . | Trisomy 8 . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | |
| Ara-C | 8 | 1.56 (0.56-1.96) | 9 | 0.16 (0.11-0.25) | 8 | 0.57 (0.42-0.78) | 3 | 1.60 (1.18-1.74)4-153 | 4 | 0.28 (0.20-0.88) |
| DNR | 6 | 0.29 (0.22-0.63) | 8 | 0.08 (0.03-0.11) | 8 | 0.13 (0.10-0.41) | 3 | 0.34 (0.11-1.32)4-153 | 2 | 0.05 (0.04-0.05)4-153 |
| Dox | 4 | 0.42 (0.37-0.70) | 6 | 0.11 (0.08-0.18) | 6 | 0.68 (0.43-0.97) | 3 | 0.48 (0.39-1.71)4-153 | 3 | 0.43 (0.11-0.46)4-153 |
| Ida | 6 | 0.16 (0.04-0.27) | 6 | 0.06 (0.01-0.10) | 6 | 0.10 (0.05-0.26) | 2 | 0.62 (0.20-1.04)4-153 | 3 | 0.05 (0.03-0.10)4-153 |
| VP16 | 7 | 18.42 (8.57-27.09) | 9 | 0.68 (0.63-1.60) | 8 | 4.72 (2.35-23.98) | 3 | 12.50 (3.59-33.15)4-153 | 4 | 1.83 (1.28-4.01) |
| 6-TG | 8 | 5.89 (3.92-13.76) | 9 | 4.88 (3.13-8.80) | 8 | 9.58 (6.88-23.50) | 3 | 17.61 (4.50-43.33)4-153 | 4 | 6.10 (3.58-6.14) |
| Mitox | 8 | 0.08 (0.03-0.27) | 8 | 0.01 (0.00-0.03) | 5 | 0.05 (0.01-0.61) | 2 | 0.57 (0.04-1.10)4-153 | 2 | 0.09 (0.04-0.14)4-153 |
| Amsa | 7 | 1.08 (0.18-1.20) | 8 | 0.10 (0.05-0.29) | 7 | 0.21 (0.15-1.13) | 3 | 0.14 (0.13-2.07)4-153 | 3 | 0.07 (0.06-0.31)4-153 |
| CdA | 5 | 0.025 (0.020-0.036) | 8 | 0.002 (0.002-0.004) | 6 | 0.008 (0.001-0.026) | 2 | 7.15 (0.02-14.29)4-153 | 1 | 0.028 |
| Bus | 6 | 42.4 (28.6-67.7) | 8 | 9.6 (7.1-35.8) | 6 | 51.5 (34.9-59.4) | 3 | 49.2 (48.3-72.6)4-153 | 3 | 67.6 (55.6-180)4-153 |
| Ifos | 6 | 12.17 (11.29-14.10) | 9 | 5.97 (4.92-12.04) | 7 | 13.85 (13.82-14.50) | 2 | 17.21 (16.1-18.3)4-153 | 1 | 22.62 |
| Pred | 7 | >250 (>250->250) | 9 | >250 (>250->250) | 8 | >250 (>250->250) | 3 | >250 (>250->250)4-153 | 4 | >250 (194->250) |
| VCR | 7 | 12.50 (1.95-28.52) | 7 | 0.60 (0.24-1.18) | 7 | 18.36 (1.33-38.00) | 3 | 0.50 (0.14-9.15)4-153 | 4 | 20.19 (1.82-41.60) |
| ASP‡ | 6 | 1.45 (0.49-1.77) | 8 | 0.14 (0.01-0.59) | 5 | 1.36 (0.03-10.10) | 2 | >10.0 (>10.0->10.0)4-153 | 2 | 0.66 (0.51-0.80)4-153 |
| Drug . | Abnormalities 5/7 . | 11q23 with t(9;11) . | 11q23; non-t(9;11) . | t(15;17) . | Trisomy 8 . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | n . | Median (P25-P75) . | |
| Ara-C | 8 | 1.56 (0.56-1.96) | 9 | 0.16 (0.11-0.25) | 8 | 0.57 (0.42-0.78) | 3 | 1.60 (1.18-1.74)4-153 | 4 | 0.28 (0.20-0.88) |
| DNR | 6 | 0.29 (0.22-0.63) | 8 | 0.08 (0.03-0.11) | 8 | 0.13 (0.10-0.41) | 3 | 0.34 (0.11-1.32)4-153 | 2 | 0.05 (0.04-0.05)4-153 |
| Dox | 4 | 0.42 (0.37-0.70) | 6 | 0.11 (0.08-0.18) | 6 | 0.68 (0.43-0.97) | 3 | 0.48 (0.39-1.71)4-153 | 3 | 0.43 (0.11-0.46)4-153 |
| Ida | 6 | 0.16 (0.04-0.27) | 6 | 0.06 (0.01-0.10) | 6 | 0.10 (0.05-0.26) | 2 | 0.62 (0.20-1.04)4-153 | 3 | 0.05 (0.03-0.10)4-153 |
| VP16 | 7 | 18.42 (8.57-27.09) | 9 | 0.68 (0.63-1.60) | 8 | 4.72 (2.35-23.98) | 3 | 12.50 (3.59-33.15)4-153 | 4 | 1.83 (1.28-4.01) |
| 6-TG | 8 | 5.89 (3.92-13.76) | 9 | 4.88 (3.13-8.80) | 8 | 9.58 (6.88-23.50) | 3 | 17.61 (4.50-43.33)4-153 | 4 | 6.10 (3.58-6.14) |
| Mitox | 8 | 0.08 (0.03-0.27) | 8 | 0.01 (0.00-0.03) | 5 | 0.05 (0.01-0.61) | 2 | 0.57 (0.04-1.10)4-153 | 2 | 0.09 (0.04-0.14)4-153 |
| Amsa | 7 | 1.08 (0.18-1.20) | 8 | 0.10 (0.05-0.29) | 7 | 0.21 (0.15-1.13) | 3 | 0.14 (0.13-2.07)4-153 | 3 | 0.07 (0.06-0.31)4-153 |
| CdA | 5 | 0.025 (0.020-0.036) | 8 | 0.002 (0.002-0.004) | 6 | 0.008 (0.001-0.026) | 2 | 7.15 (0.02-14.29)4-153 | 1 | 0.028 |
| Bus | 6 | 42.4 (28.6-67.7) | 8 | 9.6 (7.1-35.8) | 6 | 51.5 (34.9-59.4) | 3 | 49.2 (48.3-72.6)4-153 | 3 | 67.6 (55.6-180)4-153 |
| Ifos | 6 | 12.17 (11.29-14.10) | 9 | 5.97 (4.92-12.04) | 7 | 13.85 (13.82-14.50) | 2 | 17.21 (16.1-18.3)4-153 | 1 | 22.62 |
| Pred | 7 | >250 (>250->250) | 9 | >250 (>250->250) | 8 | >250 (>250->250) | 3 | >250 (>250->250)4-153 | 4 | >250 (194->250) |
| VCR | 7 | 12.50 (1.95-28.52) | 7 | 0.60 (0.24-1.18) | 7 | 18.36 (1.33-38.00) | 3 | 0.50 (0.14-9.15)4-153 | 4 | 20.19 (1.82-41.60) |
| ASP‡ | 6 | 1.45 (0.49-1.77) | 8 | 0.14 (0.01-0.59) | 5 | 1.36 (0.03-10.10) | 2 | >10.0 (>10.0->10.0)4-153 | 2 | 0.66 (0.51-0.80)4-153 |
For comparison with other subgroups and the total group of AML patients, please see Table 3.
Data on statistical comparisons are given in “Results.” Results are expressed as median LC50 values in μg/mL. In parentheses, the 25th-75th percentile (P25-P75) is given. n indicates the number of samples tested for that particular drug; Ara-C, cytarabine; DNR, daunorubicin; Dox, doxorubicin; Ida, idarubicin; VP16, etoposide; 6-TG, 6-thioguanine; Mitox, mitoxantrone; Amsa, amsacrine; CdA, 2-chlorodeoxyadenosine; Bus, busulfan; Ifos, 4-HOO-ifosfamide; Pred, prednisolone; VCR, vincristine; ASP, l-asparaginase.
L-ASP in IU/mL, not in μg/mL.
Ranges are given instead of the P25-75.
For samples with t(8;21) there was a trend to be resistant to idarubicin (P = .014, median 4.1-fold). For the other drugs, including the anthracyclines daunorubicin and doxorubicin, no significant differences were found.
For samples with inv(16) normal cytogenetics, single, or multiple chromosomal abnormalities, there were no significant differences in drug resistance between the specific subgroup and the complementary AML samples (data not shown).
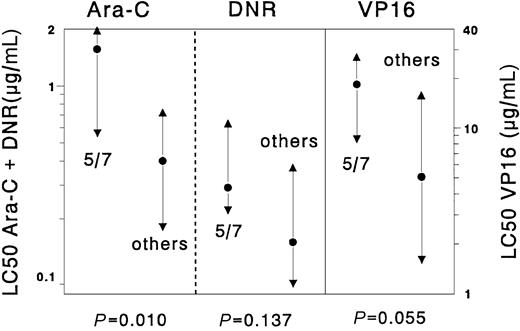
Samples with chromosome 5/7 abnormalities were significantly more resistant to cytarabine (median 3.9-fold, P = .01) than the complementary samples. The 3.6-fold difference for etoposide between samples with chromosome 5/7 abnormalities and the complementary AML samples was not significant (P = .055). For the other drugs no significant differences were found. Results are depicted in Figure 1.
Differences in cellular resistance to cytarabine (Ara-C), daunorubicin (DNR), and etoposide (VP16), comparing samples with abnormalities in chromosome 5/7 versus all other de novo childhood AML cases.
Results are expressed as LC50 values (μg/mL). Dots represent the median LC50 values, the triangles the 25th and 75th percentiles. Cases with chromosome 5/7 abnormalities (n = 8) were significantly more resistant to cytarabine (median 3.9-fold) but not to etoposide and daunorubicin when compared with the other AML samples (n = 101) without chromosome 5/7 abnormalities.
Differences in cellular resistance to cytarabine (Ara-C), daunorubicin (DNR), and etoposide (VP16), comparing samples with abnormalities in chromosome 5/7 versus all other de novo childhood AML cases.
Results are expressed as LC50 values (μg/mL). Dots represent the median LC50 values, the triangles the 25th and 75th percentiles. Cases with chromosome 5/7 abnormalities (n = 8) were significantly more resistant to cytarabine (median 3.9-fold) but not to etoposide and daunorubicin when compared with the other AML samples (n = 101) without chromosome 5/7 abnormalities.
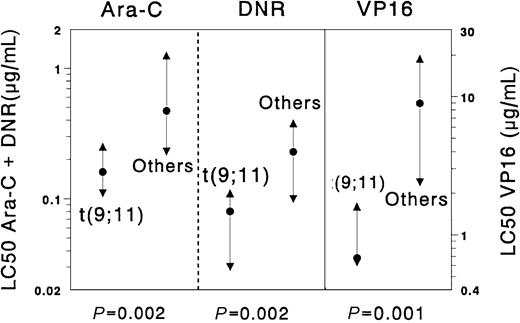
The t(9;11) subgroup appeared to be significantly more sensitive than other AML samples for the following drugs: cytarabine (median 2.9-fold,P = .002), etoposide (13.1-fold, P = .001), daunorubicin (2.9-fold, P = .002), doxorubicin (4.5-fold,P = .001), mitoxantrone (8.0-fold, P = .004), 2-chlorodeoxyadenosine (10.0-fold, P = .002), and amsacrine (5.1-fold, P = .011). Moreover, we found large differences between t(9;11) samples and the complementary AML samples for vincristine (t(9;11) were median 4.9-fold more sensitive) andl-asparaginase (t(9;11) were median 6.4-fold more sensitive), although these differences were not statistically significant (P = .035 and .051, respectively). Results are depicted in Figure 2. We also compared the samples with non-t(9;11) 11q23 translocations with all other AML samples, which revealed no significant differences.
Differences in cellular resistance to cytarabine (Ara-C), daunorubicin (DNR), and etoposide (VP16) when the subgroup of childhood AML samples with t(9;11) was compared with all other de novo childhood AML cases.
Results are expressed as LC50 values (μg/mL). Dots represent the median LC50 values, the triangles the 25th and 75th percentiles. Samples with t(9;11) (n = 9) were significantly more sensitive to cytarabine (Ara-C, median 2.9-fold), daunorubicin (DNR, 2.9-fold), and etoposide (VP16, 13.1-fold) than the complementary group of AML samples (n = 100).
Differences in cellular resistance to cytarabine (Ara-C), daunorubicin (DNR), and etoposide (VP16) when the subgroup of childhood AML samples with t(9;11) was compared with all other de novo childhood AML cases.
Results are expressed as LC50 values (μg/mL). Dots represent the median LC50 values, the triangles the 25th and 75th percentiles. Samples with t(9;11) (n = 9) were significantly more sensitive to cytarabine (Ara-C, median 2.9-fold), daunorubicin (DNR, 2.9-fold), and etoposide (VP16, 13.1-fold) than the complementary group of AML samples (n = 100).
Within the group of 11q23 translocations, the samples with t(9;11) were significantly more sensitive than samples without t(9;11) for cytarabine (median 3.6-fold, P = .005) and doxorubicin (6.2-fold, P = .004) The 6.9-fold difference for etoposide was only borderline significant (P = .012). Differences for the other drugs were not significant. There were no significant differences in age, WBC, sex, CCS, or OD per 105 cells between these 2 groups. There were no significant correlations between resistance to each of the major AML drugs and age for the t(9;11) samples.
Numbers in the t(15;17) and the trisomy 8 subgroups were too small for meaningful statistical analysis of drug resistance profiles. The median LC50 values of the 3 samples with t(15;17) were generally higher than those of the complementary AML group, but there was a large variation in LC50 values between these 3 samples. Samples with trisomy 8 showed lower median LC50 samples for a number of drugs, such as etoposide, cytarabine, daunorubicin, idarubicin, and amsacrine; for the other drugs, results were either similar or more resistant to the complementary AML group. Again, individual variations in LC50 values were large.
Cellular drug resistance and treatment outcome.
Although this study was not designed to assess the relationship between cellular drug resistance and treatment outcome, we analyzed whether patients who suffered from refractory disease or relapse differed in resistance (on cells taken at initial diagnoses) from patients remaining in continuous complete remission (CCR) for the following drugs: cytarabine, daunorubicin, 6-thioguanine, and etoposide. Overall, we could not find such differences for these 2 groups of patients. However, when we performed the same analysis within the specific cytogenetic subgroups, we found that refractory/relapsed patients in the inv(16) subgroup were 4-fold more resistant to cytarabine than the patients in CCR (P = .08). In the subgroup of patients with single random abnormalities, refractory/relapsed (n = 7) patients were 2.3-fold more resistant to cytarabine than patients in CCR (n = 5, P = .39). In the t(9;11) subgroup, relapsed/refractory patients (n = 3) were 1.8-fold more resistant to cytarabine than patients in CCR (n = 6, P = .36).
Discussion
Because of their prognostic significance, the presence of specific genetic abnormalities in childhood AML at diagnosis is currently used to determine treatment strategy.1,2,4,5,14 The underlying mechanisms responsible for these differences in treatment outcome are, however, largely unknown. We have already shown that differences in drug resistance play a role in explaining differences in prognosis in specific genetic subgroups in childhood ALL.26 27Therefore, in this study we attempted to determine whether differences in drug resistance profiles of the genetic subgroups found in childhood AML could, at least partially, explain their prognostic significance.
For 2 genetic subgroups we found a specific drug-resistance profile. Leukemic cells from patients with chromosome 5/7 abnormalities were significantly more resistant to cytarabine and showed a trend for resistance to etoposide. The second subgroup with a specific resistance profile were the t(9;11) cases, which displayed a remarkably sensitive profile, with enhanced sensitivity to cytarabine, 2-chlorodeoxyadenosine, etoposide, the anthracyclines, and amsacrine. Interestingly, t(9;11) samples were median 4.9-fold more sensitive to vincristine and 6.4-fold more sensitive to l-asparaginase than the other AML samples, although these differences only showed a trend for statistical significance. The median LC50 values for t(9;11) samples for vincristine and l-asparaginase are similar to those reported earlier for childhood ALL.25
We previously reported a remarkable sensitive drug-resistance profile for DS AML, which involved the following drugs: cytarabine, the anthracyclines, etoposide, 6-thioguanine, amsacrine, vincristine, and busulfan.9 When we compared the t(9;11) cases with the DS AML cases, we found DS AML to be significantly more sensitive to 6-thioguanine only (median 2.1-fold, P = .003, data not shown). For all other drugs there were no significant differences between these 2 groups. Apparently, both AML subgroups have similar drug sensitivity profiles.
Based on these in vitro drug-resistance profiles, we can currently distinguish 3 different cytogenetic subgroups in de novo childhood AML: on the “sensitive” side the t(9;11) and DS AML patients, on the “resistant” side AML with abnormalities of chromosome 5/7. All other AML cases can be classified as “intermediately” sensitive. In the latter group, however, we may have missed relevant differences in cellular drug resistance due to the small numbers in the specific cytogenetic subgroups and/or selection of samples. Looking at treatment outcome of patients included in this study, we found no evidence for differences in prognosis between the different cytogenetic subgroups, with the exception of the subgroup of “other 11q23 abnormalities” that did significantly worse. This may imply selection of samples; however, results have to be interpreted with caution because patients were treated on many different treatment protocols over a long time and numbers were small.
For samples with abnormalities of chromosome 5/7, our findings are in full agreement with the clinical data presented by most collaborative groups.2,6,8,13,14 However, a good prognosis for t(9;11) patients has not been reported by all collaborative groups.5,15-18,20 This translocation occurs predominantly in AML, but it is also found in myelodysplasia and ALL, although this is rare.42,43 The translocation leads to the MLL-AF9 gene fusion, which involves transcription modulation or regulation.42,44 Although no explanation is readily available why the t(9;11) cases display a relatively sensitive profile, it stresses the importance of identifying the partner chromosome in MLL rearranged leukemias. We found no significant correlations between age and resistance to the major AML drugs in t(9;11) cases, which is in agreement with reports suggesting that good prognosis for childhood t(9;11) AML is age independent.16 17
We could not demonstrate enhanced drug sensitivity in the t(8;21) or inv(16) genetic subgroups. The reported good prognosis of these subgroups may be caused by other factors, such as diminished relapse potential of minimal residual disease. PCR positivity for the AML1-ETO transcript can exist even in long-term remissions in patients with t(8;21)-positive AML.45 In addition, t(8;21) is not associated with a good prognosis in all pediatric series. In the Pediatric Oncology Group (POG) 8821 study, patients with normal cytogenetics had comparable outcome to those with t(8;21),4 while in the study by Martinez-Climent the t(8;21) subgroup had inferior outcome.15 Leblanc et al reported a very favorable CR rate in t(8;21) patients but also reported a relatively high relapse rate.5 The Medical Research Council (MRC) reported that the prognosis of good-risk patients (including the inv(16) and t(8;21) cases) and the standard-risk patients was rather similar in terms of disease-free survival.2 This might support our finding of a large group of “intermediate sensitive” cases, which included the t(8;21) and inv(16) samples. However, in our study a relatively large number of the t(8;21) samples did not survive the 4-day cell culture, suggesting that we may have analyzed a selected, relatively resistant, subset of t(8;21) samples. Finally, the numbers in the specific genetic subgroups in our study were rather small, which may hamper the detection of relevant differences in drug resistance.
Two other studies have reported a relationship between in vitro drug resistance and genetic alterations in AML. Nørgaard et al studied a combined set of childhood and adult AML samples and could not find a significant correlation between drug resistance and cytogenetic findings, although there was a trend for daunorubicin resistance in the unfavorable cytogenetics subset.46 Tosi et al, studying adult AML samples, found leukemic cells from patients with inv(16) to be significantly more sensitive to cytarabine than cells with “poor-risk” abnormal karyotypes.28 The inv(16) cells also incorporated more cytarabine into DNA. We have not been able to demonstrate increased cytarabine sensitivity in our inv(16) subset or in the t(8;21) cases. Therefore, we cannot confirm the hypothesis of enhanced cytarabine sensitivity in core binding factor leukemias.12 28
Although this study was not designed to detect a possible relationship between in vitro drug resistance of AML cells obtained at initial diagnosis and treatment outcome, we analyzed whether patients remaining in continuous complete remission (CCR) differed in cellular resistance from refractory/relapsed patients. We were not able to demonstrate such a relationship when all patients were grouped together. However, when analyzing this within the specific cytogenetic subgroups separately, relapsed/refractory patients were more cytarabine resistant than patients in CCR in several subgroups, but results were not significant and the low numbers preclude any firm conclusions. Overall, the data suggest that cellular drug resistance is not strongly related to long-term clinical outcome, and therefore other factors must have played a more important a role in determining clinical outcome in this cohort of childhood AML patients. This might involve differences in pharmacokinetics or pharmacogenomics, clonal evolution, and/or relapse potential of residual disease. Another explanation may be that our assay correctly predicts the cytotoxicity profile of the bulk of AML cells but does not reflect a small but resistant subclone of leukemic stem cells that eventually causes relapse.47However, a recent study in adult AML did show independent prognostic significance of cellular drug resistance for both early response and long-term clinical outcome.48 Moreover, in pediatric AML, associations with early response have been reported either using the MTT assay39,49 or a clonogenic assay.50 51 It must be stressed that cellular drug resistance is a treatment-independent parameter of the leukemic cells taken at diagnosis. However, prognostic significance in terms of clinical outcome is treatment dependent, and as such the absence of prognostic significance in this specific cohort of patients does not mean that differences in drug resistance are not relevant.
In conclusion, we found more or less similar cellular drug resistance patterns for most (cyto-) genetic subgroups, but the t(9;11) AML cases were found to have a distinct and relatively sensitive drug profile, which may explain their superior clinical outcome, as reported in the literature. AML cells with chromosome 5/7 abnormalities, on the other hand, were found to be relatively resistant to cytarabine and possibly also to other drugs. These results suggest the possibility for subgroup-directed chemotherapy in certain karyotypic or molecular genetic subgroups in de novo childhood AML and may be useful in the rational design of future treatment protocols.
The authors thank all hospitals and clinicians participating in the AML-BFM Study Group and the DCLSG who provided us with the patient samples. We are grateful to the reference laboratories of the AML-BFM SG, with regard to the cytogenetic studies especially to J. Bruch, S. Röttgers, A. Teigler-Schlegel and S. Viehmann, and with regard to the morphology data to J. Ritter, D. Reinhardt, and E. Kurzknabe. Board members of the AML-BFM Study Group are C. Bender-Götze, F. Berthold, J. Boos, U. Creutzig, A. Feldges, H. Gadner, N. Graf, G. Henze, J. Hermann, H. Jürgens, H. Kabisch, D. Körholz, T. Klingebiel, C. M. Niemeyer, A. Reiter, J. Ritter, and J. Stary. Participants of the Dutch Working Group on Cancer Genetics and Cytogenetics are R. M. Slater, E. Van den Berg-De Ruiter, A. Geurts van Kessel, A. Hagemeijer-Hausman, A. Hamers, S. L. Bhola, C. G. Beverstock, and M. Blij-Philipsen. We also thank Mrs A. Heus for secretarial help and the technicians of the Laboratory of Pediatric Oncology for their expertise in testing the patient samples.
Supported by the “Landelijke Vereniging van Crematoria,” Eindhoven, The Netherlands.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
C. M. Zwaan, Department of Pediatric Hematology/Oncology, VU University Medical Center, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands; e-mail:cm.zwaan@vumc.nl.