Abstract
Mycosis fungoides (MF) is a cutaneous T-cell lymphoma characterized by multifocal disease and protracted clinical course. The few studies that have assessed T-cell receptor (TCR) gene rearrangements (GRs) present at different anatomic sites in MF have generally reported a common clone. We used a previously validated 4-color polymerase chain reaction (PCR) assay to assess the size and V-family usage of TCR-γ GRs in 102 concurrent and/or sequential morphologically involved biopsy specimens (91 skin and 11 lymph nodes) from 39 MF patients. This assay detected TCR-γ clonal GRs in 89 samples (87%) from 36 patients (92%). In 24 patients (77%), an identical clonal GR was present in at least 2 skin samples. However, in one third of these patients, additional different clonal GRs were also noted. Four patients (13%) had clonal GRs that were distinct in different skin samples. In 3 patients (10%), no GR was detected in any sample. In a comparison of lymph node and skin samples, 8 patients had the identical clonal GRs at both sites, 2 patients had different clonal GRs, and 1 patient had no GR identified at either site. Independent of clinical stage, patients who had the same GR detected in multiple concurrent biopsy specimens at the time of diagnosis were more likely to have progressive disease than those who had different GRs (P = .04). Four-color TCR-γ PCR analysis can uncover multiple distinct clonal GRs in different samples consistent with multiclonal or oligoclonal disease in a significant proportion of MF patients. Demonstration of identical clonal GRs in multiple biopsy specimens at the time of diagnosis may provide prognostic information related to disease progression.
Introduction
Mycosis fungoides (MF) is typically an indolent CD4+ cutaneous T-cell lymphoma that usually presents with patch-stage disease that can be controlled with topical therapies. A subset of MF patients, however, develop rapidly progressive disease characterized by large cutaneous tumors and/or extracutaneous spread. The early identification of patients who will progress from patch-stage MF to aggressive disease is an important goal.
Although epidermotropism and atypia of lymphocytes remain the hallmarks of MF, the presence of a clonal T-cell receptor (TCR) gene rearrangement (GR) can support the diagnosis.1 2Conventional polymerase chain reaction (PCR) methods to assess clonality at the TCR-γ locus have been widely used. In these analyses, the presence of a dominant amplified band following electrophoresis of the PCR products is equated with the presence of a monoclonal T-cell population. Lack of a discrete band (ie, smear pattern) usually indicates a polyclonal T-cell population.
Only a few studies have determined the relationship between T-cell clones present at different anatomic sites in patients with MF.3-8 By comparing band size following multiplex TCR-γ PCR and denaturing gradient gel electrophoresis (DGGE), Delfau-Larue et al8 concluded that identical T-cell clones were usually present at multiple cutaneous lesions over the course of the disease. Here, we sought to assess the degree of clonal heterogeneity in multiple MF samples from the same patient using a more detailed 4-color TCR-γ PCR assay that allows accurate comparison of the size and V-family of GR(s).9 We identified some patients with common and/or stable clonal GR pattern, others with both persistent and variable clonal GRs, and others with nonshared clonal GR(s), suggesting that some MF patients have multiple T-cell clones.
Materials and methods
Patient samples
We studied 91 skin and 11 lymph node biopsy specimens from 39 patients being followed for cutaneous T-cell lymphoma in the Dermatology Clinics of the University of Texas MD Anderson Cancer Center from January 1980 to November 2001. Analyses were done on 83 skin samples from 31 patients who had multiple skin lesions biopsied simultaneously and/or sequentially (in 3 of those patients, a sequential lymph node biopsy specimen was also analyzed) and on 8 patients who had a single skin sample and a sequential or concurrent lymph node biopsy specimen.
Diagnosis of MF required the presence of an epidermotropic, cytologically atypical CD4+ T-cell infiltrate, as outlined in the World Health Organization classification of lymphoid neoplasms.10 The National Cancer Institute (NCI)–Navy classification was used to grade MF involvement in lymph nodes, with minimal numbers of identifiable tumor cells indicated as LN-1 or LN-2; partial involvement as LN-3; and complete effacement by tumor as LN-4.10,11 Staging was determined in accordance with the Mycosis Fungoides Cooperative Group criteria.11Patients were classified in 2 categories according to their clinical status during follow-up: progressive disease or nonprogressive disease. Progressive disease was defined by the following clinical criteria: (1) skin disease unresponsive to external therapies (eg, nitrogen mustard, psoralen and long-wave ultraviolet radiation) and requiring systemic therapies, and/or (2) significant tumor dissemination to extracutaneous sites such as blood or lymph nodes, and/or (3) death due to disease. Four cases of chronic dermatitis were also included. Comparisons of data were made using a one-tailed Wilcoxon-Mann-Whitney exact test. A logistic regression model was employed for multivariable analysis.
Molecular analysis of the TCR-γ locus using a 4-color PCR assay
DNA was extracted from fixed, paraffin-embedded tissue in all cases using the QIAamp tissue kit (QIAGEN GmbH, Hilden, Germany). Amplification of a 400-bp sequence from the human β-globin gene was performed in all cases to confirm that the quality of DNA was adequate. Analysis was done using a recently developed, novel 4-color TCR-γ PCR assay combined with GeneScan analysis, as previously described.9 To validate this 4-color assay, a comparison study with conventional PCR–DGGE was previously performed, analyzing 86 lymphoproliferative lesions by both methods.9
For the 4-color TCR-γ PCR assay, we used 4 sets of consensus Vγ and Jγ primers, without GC clamps, as described by Theodorou et al.12 Each Vγ family primer was end-labeled with a different fluorescent dye: tetramethyl-6-rhodamine (TAMRA, red) for VγI,-carboxyfluorescein (FAM, blue) for VγII, hexachloro-6-carboxyfluorescein (HEX, black) for VγIII, and tetrachloro-6-carboxyfluorescein (TET, green) for VγIV. Thus, if one of the segments of the VγI family is used in the GR, the PCR products will be fluorescently labeled with TAMRA dye and will show a red color on the GeneScan. If VγII, III, and IV are used, the PCR products will be fluorescently labeled with FAM, HEX, and TET and will show a blue, black, and green color, respectively, on the GeneScan. Primers were included in a multiplex 30-μL PCR reaction performed in a 9700 thermal cycler (PE/Applied Biosystems, Foster City, CA).
Following PCR, amplified products were denatured and separated by high-resolution capillary electrophoresis using the 310-Genetic Analyzer (PE/Applied Biosystems), as previously described.9 Fluorescence data were analyzed using GeneScan and Genotyper software (PE/Applied Biosystems).
The PCR assay was performed for each case in triplicate, and only those cases with reproducible peaks in at least 2 determinations were considered as clonal GRs. For this study, a monoclonal pattern was defined as 1 or 2 clonal GR(s). Three or more clonal GRs that were reproducible, distinct, and quantifiable above the polyclonal background characterized an oligoclonal pattern.
The sensitivity of this PCR technique was established by diluting DNA extracted from Jurkat (lymphoblastic lymphoma) or Hut-78 (Sezary syndrome) T-cell lines and mixing with DNA from a reactive lymph node that served as a polyclonal T-cell control.9 In this assay, clonal TCR-γ GRs could be detected above the polyclonal background in dilutions of up to 1:1000.
DNA sequencing
The amplification products in 10 cases were sequenced as previously described.9 In cases with only a single clonal GR, the PCR reaction mixture was directly sequenced. In cases with more than one clonal GR, PCR products were first separated by 7.5% polyacrylamide gel electrophoresis. Each band was then excised and DNA was extracted. Sequencing was done using the fluorescence dye terminator cycle method. NCBI Blast (National Center for Biotechnology Information Basic Local Alignment Search Tool) software was used to compare the sequence homology of the clones with TCR-γ germline sequences. Sequence Navigator (PE/Applied Biosystems) software was used to compare the sequence homology between different clones.
Results
Summary of analyzed cases
Thirty-nine patients were studied, 19 men and 20 women, ranging in age from 9 to 73 years (average, 49 years) (Table1). By TNM staging, 19 patients (54%) presented with patch or plaque-type cutaneous lesions (stages IA, IB, and IIA), whereas 20 (46%) also had cutaneous tumors and/or lymph node or visceral involvement (stages IIB and IV, respectively). The length of clinical follow-up ranged from 12 months to 32 years, with an average length of 8.6 years. The clinical outcome distribution of the patients at the last follow-up was as follows: 6 patients were in complete remission (CR), 26 were alive but not in CR (8 of those with minimal cutaneous disease, and 3 had progressive lymphoma), 6 had died of MF, and 1 had died of causes unrelated to MF.
Relation between clinical characteristics and the TCR-γ PCR results
| Patient number . | Age at diagnosis, y . | Sex . | Stage . | Type of samples . | TCR-γ GR pattern . | Follow-up, mo . | Behavior . | Status at the last follow-up . |
|---|---|---|---|---|---|---|---|---|
| 1 | 30 | M | IA | S | Stable GR | 181 | P | Alive with disease |
| 2 | 30 | F | IA | C | Common GR | 144 | P | Alive with disease |
| 3 | 68 | F | IA | C | Common GR | 96 | NP | Alive, CR |
| 4 | 44 | M | IA | C and S | No GR | 77 | NP | Alive with disease |
| 5 | 38 | M | IA | S | Stable GR | 70 | P | Alive with disease |
| 6 | 44 | F | IA | S | No GR | 66 | P | Alive, CR |
| 7 | 9 | F | IA | S | Stable GR | 64 | NP | Alive with disease |
| 8 | 39 | F | IA | C | Common GR | 43 | P | Alive with disease |
| 9 | 40 | M | IA | S | Stable GR | 39 | NP | Alive with disease |
| 10 | 66 | M | IA | C | Different GR | 38 | NP | Alive, CR |
| 11 | 59 | M | IB | C and S | Common and stable GR | 224 | P | DOD |
| 12 | 57 | M | IB | S | Stable GR | 133 | P | Alive with disease |
| 13 | 68 | F | IB | S | Stable GR | 63 | NP | Alive, CR |
| 14 | 60 | M | IIA | S | Stable GR | 159 | P | Alive with disease |
| 15 | 56 | M | IIA | S | Stable GR | 139 | P | Alive with disease |
| 16 | 62 | F | IIA | S | Stable GR | 66 | P | Alive with disease |
| 17 | 57 | M | IIA | LN | Different GR | 59 | NP | Died without disease |
| 18 | 60 | M | IIA | LN | Different GR | 36 | NP | Alive, CR |
| 19 | 50 | F | IIA | C | Common GR | 17 | P | Alive with disease |
| 20 | 37 | M | IIB | C and S | Common and stable GR | 387 | P | Alive with disease |
| 21 | 56 | M | IIB | C and S | Different and unstable GR | 240 | NP | Alive with disease |
| 22 | 17 | F | IIB | C and S | Common and stable GR | 234 | P | Alive with disease |
| 23 | 53 | M | IIB | C and S | Common and stable GR | 180 | P | DOD |
| 24 | 25 | F | IIB | C and S | Common and stable GR | 166 | P | Alive with disease |
| 25 | 61 | M | IIB | S and LN | Common and stable GR | 91 | P | Alive with disease |
| 26 | 61 | F | IIB | C and S | Different and stable GR | 87 | P | Alive with disease |
| 27 | 61 | M | IIB | C | Common GR | 46 | P | Alive with disease |
| 28 | 43 | F | IV | LN | Common GR | 100 | P | Alive, CR |
| 29 | 61 | F | IV | S | Unstable GR | 63 | P | Alive with disease |
| 30 | 73 | F | IV | C and LN | No GR | 41 | P | DOD |
| 31 | 73 | F | IV | LN | Common GR | 12 | P | DOD |
| 32 | 44 | M | IVA | C | Different GR | 146 | P | Alive with disease |
| 33 | 58 | F | IVA | S | Stable GR | 110 | P | Alive with disease |
| 34 | 66 | F | IVA | LN | Common GR | 93 | P | Alive with disease |
| 35 | 48 | M | IVA | LN | Common GR | 70 | P | DOD |
| 36 | 43 | F | IVA | S and LN | Common and stable GR | 52 | P | Alive with disease |
| 37 | 49 | M | IVA | LN | Common GR | 38 | P | Alive with disease |
| 38 | 25 | F | IVA | C | Common GR | 12 | P | DOD |
| 39 | 29 | F | IVB | LN | Common GR | 152 | P | Alive with disease |
| Patient number . | Age at diagnosis, y . | Sex . | Stage . | Type of samples . | TCR-γ GR pattern . | Follow-up, mo . | Behavior . | Status at the last follow-up . |
|---|---|---|---|---|---|---|---|---|
| 1 | 30 | M | IA | S | Stable GR | 181 | P | Alive with disease |
| 2 | 30 | F | IA | C | Common GR | 144 | P | Alive with disease |
| 3 | 68 | F | IA | C | Common GR | 96 | NP | Alive, CR |
| 4 | 44 | M | IA | C and S | No GR | 77 | NP | Alive with disease |
| 5 | 38 | M | IA | S | Stable GR | 70 | P | Alive with disease |
| 6 | 44 | F | IA | S | No GR | 66 | P | Alive, CR |
| 7 | 9 | F | IA | S | Stable GR | 64 | NP | Alive with disease |
| 8 | 39 | F | IA | C | Common GR | 43 | P | Alive with disease |
| 9 | 40 | M | IA | S | Stable GR | 39 | NP | Alive with disease |
| 10 | 66 | M | IA | C | Different GR | 38 | NP | Alive, CR |
| 11 | 59 | M | IB | C and S | Common and stable GR | 224 | P | DOD |
| 12 | 57 | M | IB | S | Stable GR | 133 | P | Alive with disease |
| 13 | 68 | F | IB | S | Stable GR | 63 | NP | Alive, CR |
| 14 | 60 | M | IIA | S | Stable GR | 159 | P | Alive with disease |
| 15 | 56 | M | IIA | S | Stable GR | 139 | P | Alive with disease |
| 16 | 62 | F | IIA | S | Stable GR | 66 | P | Alive with disease |
| 17 | 57 | M | IIA | LN | Different GR | 59 | NP | Died without disease |
| 18 | 60 | M | IIA | LN | Different GR | 36 | NP | Alive, CR |
| 19 | 50 | F | IIA | C | Common GR | 17 | P | Alive with disease |
| 20 | 37 | M | IIB | C and S | Common and stable GR | 387 | P | Alive with disease |
| 21 | 56 | M | IIB | C and S | Different and unstable GR | 240 | NP | Alive with disease |
| 22 | 17 | F | IIB | C and S | Common and stable GR | 234 | P | Alive with disease |
| 23 | 53 | M | IIB | C and S | Common and stable GR | 180 | P | DOD |
| 24 | 25 | F | IIB | C and S | Common and stable GR | 166 | P | Alive with disease |
| 25 | 61 | M | IIB | S and LN | Common and stable GR | 91 | P | Alive with disease |
| 26 | 61 | F | IIB | C and S | Different and stable GR | 87 | P | Alive with disease |
| 27 | 61 | M | IIB | C | Common GR | 46 | P | Alive with disease |
| 28 | 43 | F | IV | LN | Common GR | 100 | P | Alive, CR |
| 29 | 61 | F | IV | S | Unstable GR | 63 | P | Alive with disease |
| 30 | 73 | F | IV | C and LN | No GR | 41 | P | DOD |
| 31 | 73 | F | IV | LN | Common GR | 12 | P | DOD |
| 32 | 44 | M | IVA | C | Different GR | 146 | P | Alive with disease |
| 33 | 58 | F | IVA | S | Stable GR | 110 | P | Alive with disease |
| 34 | 66 | F | IVA | LN | Common GR | 93 | P | Alive with disease |
| 35 | 48 | M | IVA | LN | Common GR | 70 | P | DOD |
| 36 | 43 | F | IVA | S and LN | Common and stable GR | 52 | P | Alive with disease |
| 37 | 49 | M | IVA | LN | Common GR | 38 | P | Alive with disease |
| 38 | 25 | F | IVA | C | Common GR | 12 | P | DOD |
| 39 | 29 | F | IVB | LN | Common GR | 152 | P | Alive with disease |
S indicates sequential; C, concurrent; P, progressive; NP, nonprogressive; CR, complete remission; DOD, dead of disease; LN, lymph node.
GR patterns were classified as “clonal” if there were 1 or 2 dominant peaks above the polyclonal amplified background. Cases were regarded as oligoclonal if there were 3 or more discrete peaks with no dominant peak identified, and as negative when all 4 primer pairs showed an absence of discrete, reproducible peaks. We considered that a patient had a stable clonal pattern when the same size and family of TCR-γ GR was present in the initial and at least one follow-up sample. An unstable clonal pattern indicated that different GR(s) were identified in all the sequential samples analyzed from the same patient. We considered that a patient had a common GR when the same size and family of TCR-γ GR was present in multiple concurrent skin samples.
A total of 91 involved skin samples and 11 lymph node samples from 39 MF patients were analyzed. Histologic analysis of the skin samples showed low-grade patch or plaque lesions in 64, large-cell transformation (LCT) in 21, and tumor-stage disease without LCT in 6. The following patterns of lymph node involvement were observed: 1 LN-1, 1 LN-2, 2 LN-3, and 7 LN-4. The 4 skin samples analyzed from 4 patients with clinical findings of chronic dermatitis all showed small-cell lymphocytic infiltrates with no significant cytologic atypia.
Overall, 36 of 39 patients had a detectable clonal TCR-γ GR in at least one biopsy specimen, with the remaining 3 patients having no clonal TCR-γ GR detected in any sample. All 4 patients with chronic dermatitis had abundant amplified DNA with no discrete peaks identified, consistent with a polyclonal T-cell population. Of the 64 skin biopsy specimens involved by low-grade patch or plaque MF, 46 samples (72%) had a monoclonal pattern, 9 (14%) were oligoclonal, and 9 (14%) had no TCR-γ GR identified. Of the 6 skin biopsy specimens involved by tumor-stage MF without LCT, 5 (83%) had a monoclonal pattern and 1 had an oligoclonal pattern. Of the 21 LCT samples, 17 (81%) had a monoclonal pattern, 1 (5%) was oligoclonal, and 3 (14%) from one patient were polyclonal.
Because each Vγ primer used in this assay was labeled with a different fluorescent dye, we could identify the Vγ family used in each clonal TCR-γ GR. The VγI family was used most often, in 78 (48%) of a total of 164 clonal GRs. VγII, VγIII, and VγIV were used in 26 (16%), 41 (24%), and 19 (12%) clonal GRs, respectively. Thirteen clonal TCR-γ GRs from 10 skin specimens of 4 patients were directly sequenced, which confirmed the authenticity of TCR-γ GRs using the appropriate Vγ and Jγ family sequences, in all cases.
Assessing the degree of clonal heterogeneity
We first compared the TCR-γ clonal pattern in 41 MF skin samples from 17 patients who had multiple biopsy specimens taken from different involved sites on the same day. A common clonal TCR-γ GR was observed in simultaneous skin samples from 11 patients (65%) (Figure1). However, in 5 of these 11 patients, additional nonshared and reproducible TCR-γ GRs were also noted. By contrast, different clonal TCR-γ GRs were observed in simultaneous specimens from 4 patients (24%) (Figure2). In one patient, an initial set of 2 skin samples showed an identical clonal TCR-γ pattern, but a second set of 2 skin samples obtained 12 years later showed a different but reproducible clonal pattern. The biopsies from 2 MF patients did not show a clonal TCR-γ GR in any analyzed samples.
TCR-γ analysis of concurrent MF skin biopsy specimens showing a common clonal GR.
(A) A skin sample obtained from the right thigh showed a dominant TCR-γ gene rearrangement (GR) of 228 bp using the VγI primer. (B) The second specimen, obtained from left-thigh skin, showed an identical dominant T-cell GR (same Vγ family and the same size). Asterisks indicate TCR-γ GRs. The red peaks without asterisks correspond to internal size standards. Red indicates VγI; blue, VγII; black, VγIII; and green, VγIV.
TCR-γ analysis of concurrent MF skin biopsy specimens showing a common clonal GR.
(A) A skin sample obtained from the right thigh showed a dominant TCR-γ gene rearrangement (GR) of 228 bp using the VγI primer. (B) The second specimen, obtained from left-thigh skin, showed an identical dominant T-cell GR (same Vγ family and the same size). Asterisks indicate TCR-γ GRs. The red peaks without asterisks correspond to internal size standards. Red indicates VγI; blue, VγII; black, VγIII; and green, VγIV.
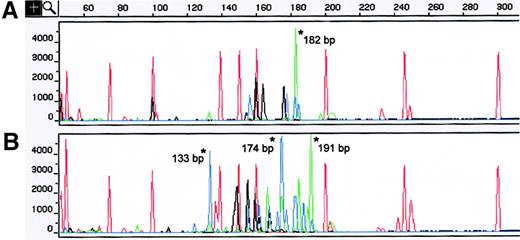
TCR-γ analysis of concurrent MF skin biopsy specimens showing different clonal GRs.
(A) A skin sample obtained from the left foot displayed a dominant clonal TCR-γ GR of 182 bp that used the VγIV primer. (B) The left-hand lesion showed 3 different TCR-γ GRs of 191, 174, and 133 bp that used the VγIV and VγII families. Asterisks indicate TCR-γ GRs. The red peaks without asterisks correspond to internal size standards. Red indicates VγI; blue, VγII; black, VγIII; and green, VγIV.
TCR-γ analysis of concurrent MF skin biopsy specimens showing different clonal GRs.
(A) A skin sample obtained from the left foot displayed a dominant clonal TCR-γ GR of 182 bp that used the VγIV primer. (B) The left-hand lesion showed 3 different TCR-γ GRs of 191, 174, and 133 bp that used the VγIV and VγII families. Asterisks indicate TCR-γ GRs. The red peaks without asterisks correspond to internal size standards. Red indicates VγI; blue, VγII; black, VγIII; and green, VγIV.
We next compared the clonal TCR-γ GR pattern in skin samples from 22 patients who had multiple involved skin biopsy specimens taken at different times over the course of disease (up to 12 years between samples). A common stable TCR-γ GR was observed in 18 patients (82%). However, in 5 (28%) of these 18 patients, additional nonshared but reproducible TCR-γ GR(s) were also noted. Different clonal TCR-γ GRs were seen in sequential samples from 2 patients (9%). Initial and sequential skin samples from 2 patients identified no TCR-γ GR.
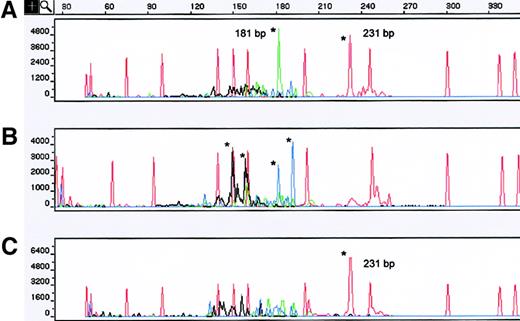
One patient had concurrent skin samples from left and right thigh lesions at the time of diagnosis displaying different clonal TCR-γ patterns (Figure 3A,B). However, analysis of a skin sample obtained from the left thigh 4 years later yielded a dominant clonal TCR-γ VγI GR of 231 bp, identical to that obtained in the earlier left-thigh lesion (Figure 3C). Sequencing confirmed that the identical clonal TCR-γ GRs obtained 4 years apart were the same T-cell clone.
TCR-γ analysis of both concurrent and sequential MF skin biopsy specimens.
(A) A sample obtained from left-thigh skin showed 2 dominant TCR-γ GRs of 231 and 181 bp that used the VγI and VγIV family primers. (B) A concurrent skin sample obtained from the right thigh showed an oligoclonal pattern with 4 dominant TCR-γ GRs that used VγII and VγIII family primers, respectively. (C) The third specimen, obtained 4 years later from the left thigh (same area as sample shown in A), displayed the same TCR-γ GR of 231 bp that used the VγI family primer. Sequencing confirmed that both GRs using VγI detected 4 years apart were the same. Asterisks indicate TCR-γ GRs. The red peaks without asterisks correspond to internal size standards. Red indicates VγI; blue, VγII; black, VγIII; and green, VγIV.
TCR-γ analysis of both concurrent and sequential MF skin biopsy specimens.
(A) A sample obtained from left-thigh skin showed 2 dominant TCR-γ GRs of 231 and 181 bp that used the VγI and VγIV family primers. (B) A concurrent skin sample obtained from the right thigh showed an oligoclonal pattern with 4 dominant TCR-γ GRs that used VγII and VγIII family primers, respectively. (C) The third specimen, obtained 4 years later from the left thigh (same area as sample shown in A), displayed the same TCR-γ GR of 231 bp that used the VγI family primer. Sequencing confirmed that both GRs using VγI detected 4 years apart were the same. Asterisks indicate TCR-γ GRs. The red peaks without asterisks correspond to internal size standards. Red indicates VγI; blue, VγII; black, VγIII; and green, VγIV.
Finally, we studied 11 patients who had lymph node and skin samples morphologically involved by MF. In 8 patients (73%), identical clonal TCR-γ GR(s) at both sites were observed, whereas in 2 patients (18%), different clonal TCR-γ GR(s) were present. One patient had no GR identified at either site.
Table 2 summarizes the overall occurrence of clonal heterogeneity and clonal persistence in all skin and lymph node samples studied. Thirty of the 36 patients (77%) with clonal GRs had a common clone noted in multiple biopsies studied. However, in 9 of these 30 patients, additional nonshared clonal GRs were also noted in one or more samples. The remaining patients either had divergent TCR-γ clonal patterns (6 patients, 17%) or had no detectable GR (3 patients, 6%).
Summary of clonal heterogeneity and clonal persistence in MF patients
| . | Common GR . | Different GR . | No GR . |
|---|---|---|---|
| Concurrent skin samples | 11 patients (65%) (additional GRs in 5/11) | 4 patients (24%) | 2 patients (11%) |
| Sequential skin samples | 18 patients (82%) (additional GRs in 5/18) | 2 patients (9%) | 2 patients (9%) |
| Skin/lymph node samples | 8 patients (73%) | 2 patients (18%) | 1 patient (9%) |
| . | Common GR . | Different GR . | No GR . |
|---|---|---|---|
| Concurrent skin samples | 11 patients (65%) (additional GRs in 5/11) | 4 patients (24%) | 2 patients (11%) |
| Sequential skin samples | 18 patients (82%) (additional GRs in 5/18) | 2 patients (9%) | 2 patients (9%) |
| Skin/lymph node samples | 8 patients (73%) | 2 patients (18%) | 1 patient (9%) |
Correlation of clonal patterns with clinical parameters
Of the 17 patients with multiple concurrent biopsies from skin taken from different involved sites on the same day, 12 had evidence of progressive disease after a mean follow-up of 8 years, and 3 showed nonprogressive disease over a similar follow-up period. Two patients had no GR identified and were omitted from analysis. In this “concurrent biopsy” group, among the 12 patients with progressive disease, 10 showed multiple samples with a common GR and 2 showed different GRs. Of the 3 nonprogressing patients, 2 showed different GRs in the multiple concurrent biopsies and one had a common TCR-γ GR. There was a statistically significant association between the finding of an identical TCR-γ GR pattern in multiple concurrentskin samples and clinical progression (P = .04). However, there was no statistically significant correlation between the TCR-γ clonal pattern of the sequential skin samples and clinical progression. Multivariate regression analysis showed no statistically significant association between the clinical variables of presenting stage, age, or duration of disease and the clonal pattern observed.
Discussion
We describe here the use of a multicolor PCR method to follow more accurately the clonal pattern in multiple MF samples from a given patient over the course of disease. Using this assay, which allows determination of both the particular Vγ family used and the exact product size, we noted a subset of MF patients who had either clonal heterogeneity (approximately 30%) or nonpersistent clones (approximately 10%) over the course of disease. Clonal heterogeneity was also seen in synchronously obtained biopsy specimens in that the dominant TCR-γ GR at one skin site was not the same as that in lymph node and/or other skin samples in 6 patients (17%). Our data support variably persistent clonal T-cell expansions in some cases of MF and suggest that serial PCR analyses using this technique can be useful to document the presence of a dominant persistent or systemically disseminated clone. In our study, patients with a common clone identified in multiple concurrent skin samples at the time of diagnosis were more likely to demonstrate clinically progressive disease.
We have previously compared this 4-color PCR technique with conventional PCR–DGGE methodology.9 In that study, we concluded that the 4-color PCR assay is at least equivalent to conventional PCR methods in its sensitivity. But more important, this assay more readily detects differences in clones because of the more reproducible size determination of GRs possible with capillary electrophoresis, complemented by using different colors for each set of Vγ family primers. The ease of quantification and database entry of results from different experiments is critical given that analysis of multiple biopsy specimens may occur over many years. It is also possible to assess the adequacy of PCR amplification and/or contamination of samples given the unique amplification pattern of background of the reactive T cells that are present in each sample. This method also indicated that “pseudoclonality” (ie, TCR-γ GRs that are not reproducibly detected on repeat assay of the same sample13) may be misleading in a small number of cases. Our sensitive technique revealed that the dominant GR in one sample could sometimes be present but only minimally amplified over the polyclonal background in some other samples.
Only a few previous studies have considered the clonal relationship between T cells present in different lesions of MF.3-8 In the most complete conventional PCR-based study, Delfau-Larue et al8 found identical clones in different lesions from the same patient. A few other studies have shown similar results; however, in all of them the sample number was small and sequence confirmation of clonal identity was not always done.3-7 An earlier study, however, using restriction fragment length analysis of the TCR-β locus, found clonal heterogeneity in 5 of 23 patients (22%) with cutaneous T-cell lymphoma.14
Several mechanisms might explain the occurrence of clonal heterogeneity in patients with MF. Given the frequent history of chronic dermatitis in these patients, it is likely that early lesions emerge from polyclonal or oligoclonal activation of T cells. The role of certain superantigens (eg, bacterial proteins) in driving this early proliferation has been suggested.15 Independent outgrowth of several clones at different sites might then occur after many years of antigen stimulation. Persistence of any given clone could be dependent on further oncogenic transformation and/or therapeutic intervention. Topical or systemic treatments might preferentially eradicate a fast-growing clone, with subsequent emergence of a minor clone. Less likely, clonal heterogeneity could also be explained by the evolution of subclones from the dominant T-cell clone by subsequent rearrangements or deletions at the TCR locus. In this study, more than 90% of clinicopathologically defined MF patients studied had identifiable TCR-γ GRs. The few cases of MF patients here who lacked identifiable GRs may be attributable to previously deleted Vγ rearrangements or mutated V or J family genes not detectable with our consensus primers, rather than true polyclonal disease.
Several groups have reported the use of simultaneous analyses of TCR-γ GRs in skin and peripheral blood samples as a method of molecular staging at the time of diagnosis.5,8,16-20However, given the low incidence of extracutaneous involvement in most MF patients, the prognostic value of detection of a shared clonal TCR-γ GR in blood remains unclear.20 We did, however, note a statistically significant correlation between the presence of identical TCR-γ GRs in multiple concurrent skin samples and evidence of clinical progression. Among the 12 patients with multiple concurrent skin samples who have had progressive disease, 10 showed a common GR and only 2 showed different GRs. Of the 3 nonprogressing patients, 2 showed different GRs and one showed a common GR.
In conclusion, the presence of a GR by PCR analysis of a single MF lesion may not be sufficient to establish the presence of a dominant and systemically disseminated clone, especially in those patients with early-stage disease. Thus, detailed clonal analysis of multiple sites shows promise as a tool for stratifying early-stage MF patients. We are currently doing prospective studies with long-term follow-up to assess the statistical power of this method for identifying those MF patients with a higher risk of disease progression.
We thank David Sanders for assistance with figure preparation.
Supported by research grant CA16672 awarded by the National Cancer Institute, Department of Health and Human Services.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dan Jones, Department of Hematopathology, Box 72, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: dajones@mdanderson.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal