Abstract
Molecular mechanisms by which the Src homology 2 domain-containing inositol 5-phosphatase (SHIP) negatively regulates phagocytosis in macrophages are unclear. We addressed the issue using bone marrow–derived macrophages from FcγR- or SHIP-deficient mice. Phagocytic activities of macrophages from FcγRII(b)−/− and SHIP−/− mice were enhanced to a similar extent, relative to those from wild type. However, calcium influx was only marginally affected in FcγRII(b)−/−, but greatly enhanced in SHIP−/− macrophages. Furthermore, SHIP was phosphorylated on tyrosine residues upon FcγR aggregation even in macrophages from FcγRII(b)−/− mice or upon clustering of a chimeric receptor containing CD8 and the immunoreceptor tyrosine-based activation motif (ITAM)–bearing γ-chain or human-restricted FcγRIIa. These findings indicate that, unlike B cells, SHIP is efficiently phosphorylated in the absence of an immunoreceptor tyrosine-based inhibition motif (ITIM)–bearing receptor. We further demonstrate that SHIP directly bound to phosphorylated peptides derived from FcγRIIa with a high affinity, comparable to that of FcγRII(b). Lastly, FcγRIIa-mediated phagocytosis was significantly enhanced in THP-1 cells overexpressing dominant-negative form of SHIP in the absence of FcγRII(b). These results indicate that SHIP negatively regulates FcγR-mediated phagocytosis through all ITAM-containing IgG receptors using a molecular mechanism distinct from that in B cells.
Introduction
Phagocytosis of IgG-coated particles is initiated by clustering of the phagocytic receptors for the Fc moiety of IgG (FcγRs). There are 3 murine forms of FcγRs, encoded by 3 distinct genes.1 Two of these forms, FcγRI and FcγRIII, consist of ligand-binding α-chain and common γ-chain, and are able to promote phagocytosis. The γ-chain contains an immunoreceptor tyrosine-based activation motif (ITAM) in its cytoplasmic region, a motif shared among all immunoreceptors. There are a total of 8 human FcγR genes: 3 for FcγRI (A-C); 3 for FcγRII (A-C); and 2 for FcγRIII (A and B). FcγRIA-C and FcγRIIIA are the respective equivalents of murine FcγRI and FcγRIII, whereas FcγRIIA and FcγRIIIB are unique to human and absent in mouse.
The other class of murine and human FcγR, FcγRII(b), is a single-chain receptor containing an immunoreceptor tyrosine-based inhibition motif (ITIM) in its cytoplasmic region and does not promote phagocytosis. The term FcγRII(b) will be used to indicate murine FcγRII and human FcγRIIb. FcγRII(b) functions as a negative regulator in B cells and mast cells by recruiting inhibitory molecules such as SH2 domain-containing inositol 5-phosphatase (SHIP) and SH2 domain-containing protein tyrosine phosphatase-1 (SHP-1).2,3 SHIP is only recruited and activated when FcγRII(b) is coclustered with B-cell receptor (BCR)4,5or FcεRI.6-8 Hence, coclustering of the ITAM-bearing BCR with the ITIM-bearing FcγRII(b) provokes efficient SHIP phosphorylation and a block in cell activation. Indeed, all of the ITIM-bearing inhibitory receptors provoke inhibitory functions only upon their coclustering with an activating receptor. The signal transduction process following such coclustering was termed coinhibition,9,10 or negative signaling.3Other examples of coinhibition include natural killer (NK) cell–mediated lysis, a process blocked when killer cell inhibitory receptor (KIR) is coclustered with the NK cell–activating receptors (reviewed in Taylor et al11). Likewise, the paired immunoglobulinlike receptor B (PIR-B) in mast cells or B cells blocks signal transduction only when coclustered with an antigen or Ig receptor.12 Lastly, the gp49 inhibitory receptor blocks mast cell secretion of mediators when coclustered with IgE receptors.13 The block of cell activation in each of these examples is completely dominant over the activation signal provided by the ITAM-bearing receptor.
Clustering of γ-chain–containing FcγRs by particles opsonized with IgG triggers intracellular events such as activation of protein tyrosine kinases of the Src family14 and Syk,15 calcium mobilization,16 and actin polymerization,17 leading to internalization of the particles. The molecular mechanisms of signal transduction for FcγR-mediated phagocytosis is similar to that of antigen receptors in lymphocytes. In addition to Src-family and Syk protein tyrosine kinases, phosphatidylinositol 3-kinase (PtdIns 3-kinase) activity is required for FcγR-mediated phagocytosis.18,19 After FcγRs are clustered by binding of immune complexes, these molecules are recruited to the phosphorylated tyrosines of the γ-chain of FcγRI/III and are sequentially activated to transduce the signal to downstream events such as actin polymerization and particle internalization.20
Recently, it was reported that FcγRII(b)-deficient macrophages show greater phagocytic and calcium mobilization responses upon FcγRIII engagement, indicating the FcγRII(b) inhibits γ-chain–containing IgG receptor function.21 Similar findings were made using macrophages of SHIP−/− mice.22 In B cells, FcγRII(b) imparts a negative signal by plasma membrane recruitment of SHIP,23 and the SH2 domain of SHIP is required for its recruitment by ITIM receptors.5 Thus, the functional experiments of macrophages from FcγRII(b)−/−21 and SHIP−/−22 mice suggest that, like the B cell, coclustering the ITAM-containing γ-chain FcγR and the ITIM-containing FcγRII(b) inhibits macrophage activation and function by recruitment and phosphorylation of SHIP. Recent experiments in human macrophages and neutrophils support the notion that coclustering FcγRs containing ITAM and ITIM sequences regulate cellular function.24
In contrast to this model, other studies suggest that SHIP is efficiently phosphorylated upon clustering of ITAM-bearing FcγR25 26 and that an ITIM receptor is not necessary. Thus, it is unclear whether SHIP is involved in the regulation of macrophage activation through ITIM-containing receptors like FcγRII(b). Likewise, it is unclear whether FcγRII(b)-mediated inhibition of macrophage activation involves SHIP.
Here, we explored the requirement for the ITIM-bearing FcγRII(b) to induce SHIP phosphorylation and to regulate phagocytosis in bone marrow–derived macrophages from wild-type or gene-targeted mice and in cell lines expressing chimeric receptors of FcγRI/III γ-chain with extracellular region of CD8. We report that, in contrast to the B-cell model, SHIP phosphorylation is efficiently induced in FcγRII(b)-deficient macrophages, and can be elicited by an ITAM-containing receptor chimera through direct binding to ITAM-containing phagocytic receptors. Furthermore, the SH2 domain of SHIP has an affinity for phosphorylated ITAM tyrosines of human FcγRIIa comparable to the affinity for phosphotyrosines of the ITIM. Thus, SHIP is able to regulate FcγR-mediated phagocytosis independently of FcγRII(b).
Materials and methods
Animals
The FcγRII(b)−/− or γ-chain−/−single-deficient and FcγRII(b)−/−/ γ-chain−/−double-deficient mice were purchased from Taconic Farms (Westminster, NY). The SHIP−/− mice were kindly provided by Dr G. Krystal, Terry Fox Laboratory, British Columbia Cancer Agency, Vancouver, BC, Canada. All gene-targeted mice were of C57Bl/6 background. C57Bl/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and used as wild-type controls.
Antibodies
We purchased 2.4G2 (anti–mouse FcγRII/III),27antigen-presenting cell (APC)–conjugated anti–Mac-1 (IgG2b), fluorescein isothiocyanate (FITC)–conjugated anti–CD8, and APC-conjugated IgG2b from Pharmingen (San Diego, CA). The monoclonal IgG2a (UPC10) was from Caltag (Burlingame, CA). The FITC-conjugated F(ab′)2 fragment of rabbit anti–mouse IgG was from Jackson ImmunoResearch (West Grove, PA). The polyclonal anti–sheep red blood cells antibody was from Sigma (St Louis, MO). The polyclonal mouse IgG was from Pierce (Rockford, IL). The rabbit anti–SHIP antibody was described previously and used for immunoprecipitation.5 Antiphosphotyrosine monoclonal antibody 4G10 was purchased from Upstate Biotechnology (Lake Placid, NY). Anti–CD8 monoclonal antibody was purified from culture supernatant of hybridoma, OKT8 (American Type Culture Collection, Manassas, VA), and used as a F(ab′)2 fragment. Horseradish peroxidase (HRP)–conjugated goat anti–mouse IgG antibody and HRP-conjugated sheep anti–rabbit Ig antibody were obtained from Kappel (West Chester, PA) and Amersham Pharmacia (Piscataway, NJ), respectively. Anti–myc (9E10) monoclonal antibody was purchased from Roche Molecular Biochemicals (Indianapolis, IN).
Cell culture and transfection
RAW264.7 and THP-1 were obtained from American Type Culture Collection. The cells were maintained in complete medium (RPMI supplemented with 10% fetal calf serum [FCS], 2 mMl-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin). Transfection of cDNA was performed by electroporation at 310 V, 975 μF by Gene Pulser (Bio-Rad, Hercules, CA). Stable transfectants were selected and maintained in complete medium containing 1 mg/mL G418 (Invitrogen, Carlsbad, CA). CD8+cells stained with FITC-conjugated anti–CD8 antibody or green fluorescence protein (GFP)–positive cells were sorted by Moflo Cytometer (Cytomation, Fort Collins, CO).
Bone marrow–derived macrophages
Bone marrow–derived macrophages (BMMs) were prepared by standard methods from gene-targeted mice. Briefly, the bone marrow cells were isolated by flushing femurs and tibias and cultured overnight in 10 cm2 dishes with complete medium containing 20% L cell–conditioned medium at 37°C in 5% CO2. Nonadherent cells were transferred to new dishes and cultured for an additional 5 days at 37°C in 5% CO2 for experiments.
Flow cytometry
BMMs and RAW264.7 were harvested from plates using Cell Dissociation Medium (Sigma). Staining and flow cytometry were performed according to standard methods and analyzed by FACSCalibur and CELLQUEST software (Becton Dickinson, San Jose, CA).
Phagocytosis assay
Phagocytic index was measured as previously described.20 Briefly, sheep red blood cells (RBCs) were labeled by fluorescent dye (PKH26; Sigma) according to manufacturer's instruction. The RBCs were opsonized by polyclonal anti–sheep RBC (IgG-RBC) and used as targets for phagocytosis. For FcγRIIa-restricted phagocytosis, RBCs were biotinylated and treated with streptavidin. The streptavidin-labeled RBCs were then coupled with biotinylated Fab fragments of IV.3 antibody. Phagocytes were plated on 24-well plates at 2 × 105 cells per well and incubated overnight at 37°C in 5% CO2. Opsonized RBCs (4 × 106) were added to the prechilled 24-well plates and incubated on ice for 10 minutes to be formed rosettes. The cells were warmed to 37°C to initiate phagocytosis. Uninternalized RBCs were removed by incubation with ammonium chloride potassium (ACK) buffer (10 mM HEPES, pH 7.3, 150 mM NH4Cl). Internalized RBCs were visualized under fluorescence microscope and counted. Phagocytic index was defined as a number of internalized RBCs per 100 phagocytes.
Calcium mobilization measurements
Heat-aggregated IgG (ΔIgG) was prepared by heating 10 mg/mL normal mouse IgG at 64°C for 30 minutes. Cells were incubated in complete medium containing 2.5 μM Indo-1 AM (Molecular Probe, Eugene, OR) for 30 minutes at 37°C. The cells were stimulated with 40 μg/mL ΔIgG and monitored by spectrofluorometry (Perkin-Elmer, Norwalk, CT). The Indo-1 fluorescence emission was converted to Ca++i according to the manufacturer's instructions.
Immunoprecipitation and immunoblot
All procedures were essentially as described earlier.20 Briefly, cells were lysed in TN-1 buffer (50 mM Tris-HCl, pH 8.0, 125 mM NaCl, 10 mM ethylenediaminetetraacetic acid [EDTA], 1% Nonidet P-40, 10 mM NaF, 3 mM Na3VO4, 10 mM Na4P2O7, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 100 μg/mL phenylmethylsulfonyl fluoride) and centrifuged at 16 000g for 10 minutes at 4°C to remove insoluble materials. The resulting supernatants were subjected to immunoprecipitation using the indicated antibodies followed by protein A– or protein G–agarose (Invitrogen). The beads were extensively washed with TN-1 and the proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were electrophoretically transferred to nitrocellulose membranes, blotted with appropriate antibodies, and visualized by enhanced chemiluminescence (ECL) system (Pierce).
Reverse transcriptase–polymerase chain reaction and construction of plasmids
Total RNAs were isolated from RAW264.7 cells and reverse-transcribed to cDNAs by standard methods. The intracellular portion of γ-chain, corresponding to amino acids 47-86, were obtained by polymerase chain reaction (PCR) and the product was fused to extracellular and transmembrane regions of human CD8. The resulting cDNA was cloned into pEF/myc/cyto (Invitrogen). The intracellular portion of human FcγRIIa, corresponding to the amino acids 285-307, were also fused to the extracellular and transmembrane portions of human CD8, and cloned into pEF/myc/cyto. The substitution of tyrosine residues within ITAM of FcγRIIa with phenylalanine was performed based on PCR technique using a CD8/IIa chimera as a template. For GFP-SH2-SHIP expression vector, the cDNA fragment of SH2-SHIP, corresponding to amino acids 1-114 of murine SHIP,28 was generated by PCR and ligated into pEGFP-N1 (Clontech). The materials were confirmed by sequencing.
In vitro peptide binding assay
Whole-cell lysates were incubated with biotinylated peptides described elsewhere.5,29 Peptides were collected with Neutravidin-Sepharose (Pierce) after 5 washes with TN1 lysis buffer. The proteins associated with the peptides were analyzed by immunoblot. The identical protocol was done in experiments using the purified, recombinant GST-SHIP SH2 domain, as earlier described.5,30 31 The purified, recombinant GST-SHIP SH2 domain fusion protein showed a single band on SDS-PAGE analysis corresponding to the fusion protein.
Affinity measurements of the SH2 domain of SHIP to phosphopeptides
Affinities of SH2 domain of SHIP to phosphopeptides were determined by BIAcore system (BIAcore, Uppsala, Sweden) according to the manufacturer's instructions. In this system, the amount of analytes (GST-SH2-SHIP) bound to the sensor chip via phosphopeptides was correlated with the response unit (RU) observed. Biotinylated peptides were immobilized to streptavidin-coated chips. No direct binding of GST-SH2-SHIP to the streptavidin-coated sensor chip was observed. The GST-SH2-SHIP in the binding buffer (phosphate-buffered saline [PBS] containing 0.05% Tween-20) was injected at a flow rate of 30 μL/min for 5 minutes at 25°C. Binding was monitored and the chip was continuously washed with the binding buffer for another 5 minutes at 25°C. The chip was regenerated by washing with PBS containing 0.05% SDS. The kinetic parameters were calculated by the BIAevaluation 3.0 software (BIAcore) according to data from at least 5 different concentrations of the analytes injected.
Results
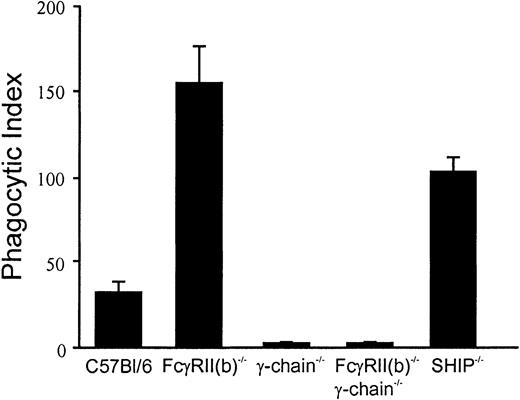
FcγR-mediated phagocytosis is enhanced in bone marrow–derived macrophages isolated from FcγRII(b)−/− and SHIP−/− knockout mice
To examine the roles of FcγRII(b) and SHIP on FcγR-mediated phagocytosis, the phagocytic abilities of the BMMs from C57Bl/6 wild-type, FcγRII(b)−/−, FcR γ-chain−/− (γ-chain−/−), FcγRII(b)−/−/ γ-chain−/−, and SHIP−/− mice were compared using IgG-opsonized sheep red blood cells (IgG-RBCs) as phagocytic targets (Figure1). BMMs from either γ-chain−/− or FcγRII(b)−/−/ γ-chain−/−double-deficient mice were incapable of phagocytosis, due to the lack of phagocytic receptors FcγRI and FcγRIII, as reported previously.32 However, phagocytic activities of BMMs from FcγRII(b)−/− and SHIP−/− were greatly enhanced, compared with that of wild-type BMMs These observations indicate that both FcγRII(b) and SHIP negatively regulate FcγR-mediated phagocytosis. The data are consistent with the possibility that, like B cells, paired coclustering an ITAM- and an ITIM-containing receptor with an IgG-coated particle blocks cell activation.
Phagocytosis assay using BMMs from gene-targeted mice.
Fluorescent IgG-opsonized RBCs were incubated with BMMs from gene-targeted mice indicated at a ratio of 20:1. Internalized IgG-RBCs were counted under a fluorescence microscope. The results were expressed as the number of the internalized IgG-RBCs per 100 BMMs (phagocytic index). Results shown are the average of duplication and are representative of 2 independent experiments.
Phagocytosis assay using BMMs from gene-targeted mice.
Fluorescent IgG-opsonized RBCs were incubated with BMMs from gene-targeted mice indicated at a ratio of 20:1. Internalized IgG-RBCs were counted under a fluorescence microscope. The results were expressed as the number of the internalized IgG-RBCs per 100 BMMs (phagocytic index). Results shown are the average of duplication and are representative of 2 independent experiments.
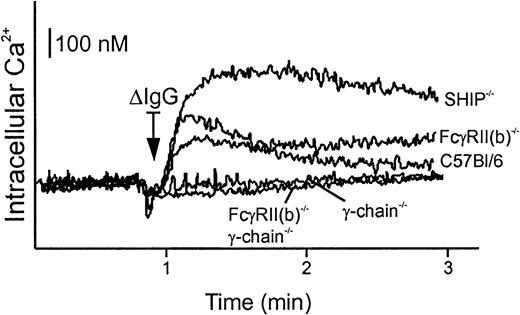
Calcium mobilization is enhanced in BMMs of SHIP−/−but not those of FcγRII(b)−/− animals
Clustering of phagocytic receptors, like all ITAM-containing receptors, is accompanied by an increase of intracellular calcium.33 In B cells, the calcium mobilization induced by clustering of B-cell receptors is inhibited by coclustering of B-cell receptor with the ITIM-bearing FcγRII(b),34,35 which promotes SHIP recruitment.5,31 To explore the possibility that the inhibitory effect of SHIP on phagocytosis is associated with FcγRII(b) like the B-cell model, we compared the intracellular calcium mobilization in BMMs from gene-targeted mice upon stimulation with ΔIgG. Stimulation of macrophages with ΔIgG engages all mouse FcγR, including phagocytic receptors FcγRI and FcγRIII, and inhibitory receptor FcγRII(b). This model enables us to measure the calcium mobilization through both activating receptors FcγRI and FcγRIII, and investigate the contribution of FcγRII(b) by using cells from gene-targeted animals. The model is an improvement over earlier studies that used 2.4G2 monoclonal antibody (mAb) to stimulate peritoneal macrophages21 because 2.4G2 does not recognize FcγRI.
Calcium mobilization upon stimulation of ΔIgG was only marginally increased in BMMs derived from FcγRII(b)-deficient mice, relative to BMMs from wild-type mice (Figure 2). However, calcium influx was greatly enhanced in BMMs from SHIP−/− compared with those of wild-type, or FcγRII(b)−/−. Minimal calcium mobilization was observed in BMMs from γ-chain−/− or from double-deficient BMMs, as reported elsewhere.21 These findings indicate that the negative signal induced by FcγRII(b) coclustering minimally affects IgG receptor–triggered intracellular calcium, whereas SHIP has a more pronounced influence. These observations suggest that SHIP functions through other receptors besides FcγRII(b), unlike the B-cell model.
Calcium mobilization upon FcγR stimulation in BMMs from gene-targeted mice.
BMMs (5 × 105) from gene-targeted mice indicated were loaded with Indo-1 AM and stimulated with 40 μg/mL ΔIgG. The intracellular Ca++ was monitored by spectrofluorometry. The bar indicates intracellular Ca++ as a reference. The arrow indicates the time when ΔIgG was added.
Calcium mobilization upon FcγR stimulation in BMMs from gene-targeted mice.
BMMs (5 × 105) from gene-targeted mice indicated were loaded with Indo-1 AM and stimulated with 40 μg/mL ΔIgG. The intracellular Ca++ was monitored by spectrofluorometry. The bar indicates intracellular Ca++ as a reference. The arrow indicates the time when ΔIgG was added.
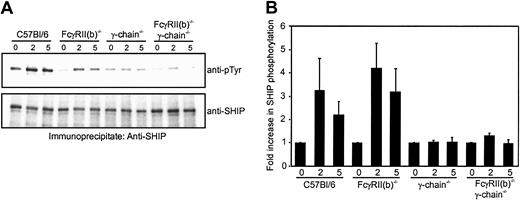
SHIP is efficiently phosphorylated by FcγR clustering in the absence of FcγRII(b)
B-cell lines show efficient SHIP phosphorylation when the cells express FcγRII(b), but less SHIP phosphorylation when FcγRII(b) is absent.5 To test whether SHIP phosphorylation in macrophages likewise requires expression of FcγRII(b), we determined the tyrosine phosphorylation of SHIP in BMMs from the various gene-targeted mice using ΔIgG as a stimulus. We found that SHIP phosphorylation was increased upon stimulation with ΔIgG in BMMs from wild-type mice, but not in BMMs from mice lacking FcR γ-chain (Figure3A). These data show that SHIP phosphorylation minimally requires clustering of ITAM-bearing receptors, FcγRI and/or FcγRIII, associated with the γ-chain. Surprisingly, SHIP was significantly phosphorylated in BMMs from FcγRII(b)−/− mice. The stoichiometry of SHIP phosphorylation was estimated from several identical experiments by quantitating the ratio of phosphorylated SHIP to total immunoprecipitated SHIP in BMMs from wild-type or the gene-targeted animals. The data are expressed as fold increase and presented in Figure 3B. These data show that SHIP phosphorylation does not require the inhibitory receptor FcγRII(b), unlike the B-cell model.5
Tyrosine phosphorylation of SHIP upon FcγR stimulation in BMMs from gene-targeted mice.
(A) BMMs (4 × 106) from gene-targeted mice indicated were stimulated with 40 μg/mL ΔIgG for indicated minutes, lysed, and immunoprecipitated with anti–SHIP antibody. The immunoprecipitates were blotted with antiphosphotyrosine antibody (upper panel; anti-pTyr) or anti–SHIP antibody (lower panel). (B) The amount of tyrosine-phosphorylated SHIP to total SHIP shown in panel A was quantified and expressed as fold increase of the ratio. The results were shown as relative values of the time-zero controls and as averages from 2 independent experiments. Bars represent standard errors of duplicate measurements.
Tyrosine phosphorylation of SHIP upon FcγR stimulation in BMMs from gene-targeted mice.
(A) BMMs (4 × 106) from gene-targeted mice indicated were stimulated with 40 μg/mL ΔIgG for indicated minutes, lysed, and immunoprecipitated with anti–SHIP antibody. The immunoprecipitates were blotted with antiphosphotyrosine antibody (upper panel; anti-pTyr) or anti–SHIP antibody (lower panel). (B) The amount of tyrosine-phosphorylated SHIP to total SHIP shown in panel A was quantified and expressed as fold increase of the ratio. The results were shown as relative values of the time-zero controls and as averages from 2 independent experiments. Bars represent standard errors of duplicate measurements.
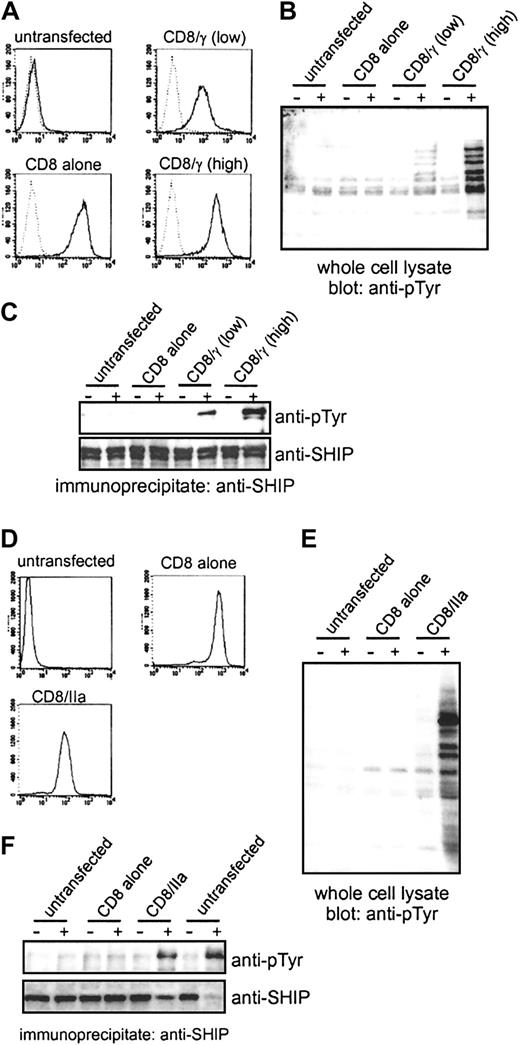
Clustering of the γ-chain or FcγRIIa is sufficient to induce SHIP phosphorylation in macrophages
The efficient phosphorylation of SHIP in the BMMs from FcγRII(b)−/− mice raises the possibility that clustering of activating, phagocytic FcγRs is sufficient for SHIP activation. To directly address this issue, and to eliminate any potential contribution from FcγRII(b), we transfected the RAW264.7 mouse macrophage cell line with a chimeric receptor containing the intracellular region of FcγR γ-chain fused to the unrelated extracellular region of human CD8 (CD8/γ). Because the ITAM sequence of the γ-chain is sufficient to trigger phagocytosis,36the receptor chimera enables us to discriminate signaling through the γ-chain associated with phagocytic FcγRI/III from that of FcγRII(b), and to examine whether SHIP is phosphorylated upon clustering of ITAM-bearing γ-chain alone. The transfected RAW264.7 macrophages were sorted based on CD8 expression levels to derive stable transfectants expressing high or low levels of CD8/γ and CD8 alone (Figure 4A). The stable transfectants were stimulated with biotinylated F(ab′)2 fragment of anti-CD8 (OKT8) and the receptor was clustered by the addition of streptavidin. This stimulation protocol using F(ab′)2fragments was applied to avoid stimulation of any endogenous IgG receptors of RAW264.7. We confirmed by flow cytometry that the F(ab′)2 fragment of OKT8 failed to recognize untransfected cells (Figure 4A, untransfected), indicating that the endogenous FcγRs are not engaged by this stimulation protocol. Stimulation with biotinylated F(ab′)2 fragment of OKT8 followed by streptavidin revealed tyrosine phosphorylation appearing in whole-cell lysates (Figure 4B) in CD8/γ transfectants, but not in untransfected RAW264.7 cells, or in the transfectants expressing CD8 alone. Likewise, we found that SHIP tyrosine phosphorylation was greatly increased after OKT8 stimulation in cells expressing either high or low levels of the chimeric receptor, depending on the expression of chimeras (Figure 4C). However, SHIP phosphorylation was absent in cells transfected with CD8 only. These data indicate that clustering of the γ-chain ITAM is sufficient for SHIP phosphorylation and that participation of FcγRII(b) is not necessary.
Clustering of FcγRIIa or the γ-chain of FcγRs is sufficient for SHIP phosphorylation.
(A) The expression of CD8 chimeras in stable RAW264.7 transfectants was examined by fluorescence-activating cell sorter (FACS) analysis. The cells were stained with biotinylated F(ab′)2 fragments of OKT8 followed by FITC-conjugated streptavidin and analyzed by FACS. Dotted lines indicate fluorescence of unstained cells. (B,C) The RAW264.7 transfectants were stimulated with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin. Whole-cell lysates (B) or SHIP immunoprecipitates (C) were separated by SDS-PAGE and blotted with antiphosphotyrosine (anti-pTyr). The filter in C was reprobed with anti–SHIP antibody (lower panel). (D) The expression of CD8 chimeras in stable THP-1 transfectants was examined by FACS analysis using biotinylated F(ab′)2 fragments of OKT8 followed by FITC-conjugated streptavidin. (E,F) THP-1 transfectants were stimulated with biotinylated F(ab′)2fragments of OKT8 followed by streptavidin. Whole-cell lysates (E) or SHIP immunoprecipitates (F) were blotted with anti-pTyr. In panel F, untransfected cells were also stimulated with Fab fragments of IV.3 antibody followed by F(ab′)2 fragments of goat anti–mouse antibody (2 lanes on the farthest right). The membrane was reprobed with anti–SHIP antibody (lower panel).
Clustering of FcγRIIa or the γ-chain of FcγRs is sufficient for SHIP phosphorylation.
(A) The expression of CD8 chimeras in stable RAW264.7 transfectants was examined by fluorescence-activating cell sorter (FACS) analysis. The cells were stained with biotinylated F(ab′)2 fragments of OKT8 followed by FITC-conjugated streptavidin and analyzed by FACS. Dotted lines indicate fluorescence of unstained cells. (B,C) The RAW264.7 transfectants were stimulated with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin. Whole-cell lysates (B) or SHIP immunoprecipitates (C) were separated by SDS-PAGE and blotted with antiphosphotyrosine (anti-pTyr). The filter in C was reprobed with anti–SHIP antibody (lower panel). (D) The expression of CD8 chimeras in stable THP-1 transfectants was examined by FACS analysis using biotinylated F(ab′)2 fragments of OKT8 followed by FITC-conjugated streptavidin. (E,F) THP-1 transfectants were stimulated with biotinylated F(ab′)2fragments of OKT8 followed by streptavidin. Whole-cell lysates (E) or SHIP immunoprecipitates (F) were blotted with anti-pTyr. In panel F, untransfected cells were also stimulated with Fab fragments of IV.3 antibody followed by F(ab′)2 fragments of goat anti–mouse antibody (2 lanes on the farthest right). The membrane was reprobed with anti–SHIP antibody (lower panel).
The human-restricted FcγRIIa is unique among immunoreceptors and consists of a single polypeptide chain containing ITAM in its cytoplasmic tail. We therefore asked whether the ITAM of the human-restricted FcγRIIa was also sufficient to induce SHIP phosphorylation, like the murine γ-chain ITAM. To test this possibility, we expressed in human THP-1 monocytes a chimera of the ITAM of human-restricted FcγRIIa fused to the extracellular region of CD8 (CD8/IIa). Flow cytometry analysis of CD8 expression shows that the cells express CD8 or the CD8/IIa chimera (Figure 4D). Stimulation of the transfected CD8+ THP-1 cells with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin as above revealed tyrosine phosphorylation appearing in whole-cell lysates (Figure 4E) in the cells expressing CD8IIa, but not in the untransfected population or the cells expressing CD8 alone. SHIP tyrosine phosphorylation was similarly increased in cells expressing the CD8/IIa chimera (Figure 4F). As an additional control, we stimulated endogenous FcγRIIa in THP-1 cells with specific monoclonal antibody IV.3 (Looney et al37; Figure 4F; untransfected). Earlier studies showed the IV.3 mAb recognizes FcγRIIa and not FcγRIIb in hematopoietic cells.38 These data clearly demonstrate that the ITAM of the γ-chain associated with activating FcγRs or the ITAM of FcγRIIa in macrophages is capable of efficiently promoting SHIP phosphorylation.
SHIP directly binds to the ITAM-containing receptor, FcγRIIa, with an affinity comparable to the ITIM-containing receptor, FcγRII(b)
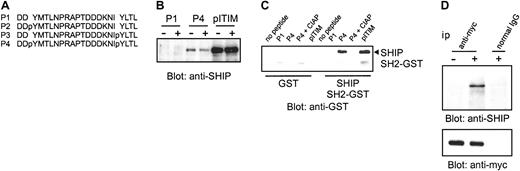
Although SHIP was phosphorylated upon clustering of γ-chain or FcγRIIa in the absence of FcγRII(b), the mechanism by which SHIP is recruited to the ITAM-containing phagocytic receptors is unclear. To begin to address this issue, we tested the in vitro binding of SHIP to doubly phosphorylated peptide derived from ITAM of FcγRIIa (P4; Figure 5A) in the cell lysates of THP-1 cells with or without stimulation of IV.3 antibody. SHIP bound to P4 as well as to phosphopeptides of FcγRII(b) ITIM (pITIM), but not unphosphorylated peptide of FcγRIIa ITAM (P1) in vitro (Figure 5B). Because the binding of SHIP to P4 or pITIM did not require prior cell stimulation, binding in this case indicated that SHIP is capable of direct association to the ITAM of FcγRIIa and did not involve a phosphorylated adapter molecule(s). We examined the binding between FcγRIIa peptides and purified, recombinant GST-SH2-SHIP fusion protein by in vitro peptide binding assay. The GST-SH2-SHIP fusion protein bound to P4 as well as pITIM, but not to P1 or to P4 after dephosphorylation by alkaline phosphatase (Figure 5C). To explore whether SHIP could associate with FcγRIIa in cells, we examined coimmunoprecipitation of endogenous SHIP with the ITAM of FcγRIIa in THP-1 cells expressing CD8/IIa. We found (Figure 5D) that SHIP coimmunoprecipitated with CD8/IIa in an activation-dependent manner. Equal amounts of the immunoprecipitated CD8/IIa was verified by immunoblot with anti–myc antibody. These data demonstrate that SHIP is capable of binding to ITAM-containing FcγRIIa in cells and in vitro.
SHIP binds directly to the ITAM of FcγRIIa in vitro and in vivo.
(A) Sequences of peptides used are shown and described previously.29 (B) THP-1 cells were stimulated with Fab fragments of IV.3 antibody followed by F(ab′)2 fragments of goat anti–mouse antibody. The lysates were incubated with biotinylated peptides indicated and purified by Neutravidin beads. The precipitates were resolved by SDS-PAGE, and blotted with anti–SHIP antibody. (C) The recombinant GST-SH2-SHIP fusion protein (right 5 lanes) or GST protein (left 5 lanes) was incubated with biotinylated peptides indicated, precipitated with Neutravidin beads, resolved by SDS-PAGE, and blotted with anti–GST antibody. The phosphorylated peptides were pretreated with calf intestinal alkaline phosphatase (CIAP) before the incubation with recombinant proteins as indicated. The position of GST-SH2-SHIP was indicated at right. (D) THP-1 transfectants were stimulated with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin and lysates were immunoprecipitated with anti-myc. The immunoprecipitates (ip) were separated by SDS-PAGE and probed with antibody to SHIP (upper panel) and reprobed with anti-myc (lower panel).
SHIP binds directly to the ITAM of FcγRIIa in vitro and in vivo.
(A) Sequences of peptides used are shown and described previously.29 (B) THP-1 cells were stimulated with Fab fragments of IV.3 antibody followed by F(ab′)2 fragments of goat anti–mouse antibody. The lysates were incubated with biotinylated peptides indicated and purified by Neutravidin beads. The precipitates were resolved by SDS-PAGE, and blotted with anti–SHIP antibody. (C) The recombinant GST-SH2-SHIP fusion protein (right 5 lanes) or GST protein (left 5 lanes) was incubated with biotinylated peptides indicated, precipitated with Neutravidin beads, resolved by SDS-PAGE, and blotted with anti–GST antibody. The phosphorylated peptides were pretreated with calf intestinal alkaline phosphatase (CIAP) before the incubation with recombinant proteins as indicated. The position of GST-SH2-SHIP was indicated at right. (D) THP-1 transfectants were stimulated with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin and lysates were immunoprecipitated with anti-myc. The immunoprecipitates (ip) were separated by SDS-PAGE and probed with antibody to SHIP (upper panel) and reprobed with anti-myc (lower panel).
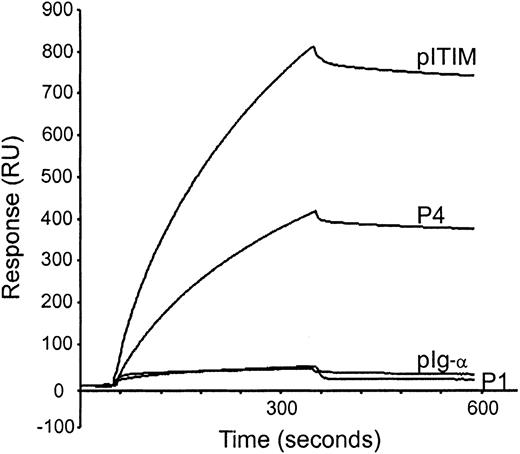
To measure the affinity between SHIP and FcγRIIa ITAM, surface plasmon resonance measurement was performed using purified GST-SH2-SHIP protein and phosphopeptides immobilized on sensor chips, listed in Figure 5A. GST-SH2-SHIP bound to P4 with a slow association rate and a comparable dissociation rate compared with pITIM (Figure6). We found no significant binding of GST-SH2-SHIP to P1 or pIg-α, derived from ITAM of the Ig-α chain of BCR. Kinetic parameters for the interaction of GST-SH2-SHIP with singly phosphorylated peptides (P2 and P3), P4, pITIM, and pIg-α were determined by sensor measurements using at least 5 different concentrations of GST-SH2-SHIP (Table 1). Although GST-SH2-SHIP associated with P4 at slower rate (association constant [kon], 2616 M−1s−1) than pITIM (kon, 3370 M−1s−1), it dissociated from P4 at a rate (dissociation constant [koff], 0.188 ms−1) similar to that of pITIM (koff, 0.154 ms−1). These binding kinetics translate into comparable affinities of P4 (affinity constant [KD], 71.0 nM) compared with pITIM (KD, 47.2 nM). Singly phosphorylated peptides P2 and P3 also showed moderate affinities to GST-SH2-SHIP, 149 nM and 75.1 nM of KD, respectively. The pIg-α ITAM peptide showed an approximately 10-fold lower affinity (KD, 402 nM) than P4 or pITIM, which may account for earlier findings that SHIP is phosphorylated only when BCR is coclustered with FcγRII(b).4
Measurements of affinities between GST-SH2-SHIP fusion protein and phosphopeptides by surface plasmon resonance.
Biotinylated phosphopeptides indicated were captured on a streptavidin-coated sensor chip, and GST-SH2-SHIP was injected for 5 minutes at a flow rate of 30 μL/min at 25°C. The chip was washed with binding buffer for a further 5 minutes to examine dissociation rates.
Measurements of affinities between GST-SH2-SHIP fusion protein and phosphopeptides by surface plasmon resonance.
Biotinylated phosphopeptides indicated were captured on a streptavidin-coated sensor chip, and GST-SH2-SHIP was injected for 5 minutes at a flow rate of 30 μL/min at 25°C. The chip was washed with binding buffer for a further 5 minutes to examine dissociation rates.
Kinetic parameters for the interaction of GST-SH2-SHIP with synthetic peptides
| Peptides . | kon (M−1s−1) . | koff (ms−1) . | KD (nM) . |
|---|---|---|---|
| P1 | NM | NM | NM |
| P2 | 1295 | 0.181 | 149.0 |
| P3 | 1709 | 0.125 | 75.1 |
| P4 | 2616 | 0.188 | 71.0 |
| pITIM | 3370 | 0.154 | 47.2 |
| pIg-α | 1285 | 0.520 | 402.0 |
| Peptides . | kon (M−1s−1) . | koff (ms−1) . | KD (nM) . |
|---|---|---|---|
| P1 | NM | NM | NM |
| P2 | 1295 | 0.181 | 149.0 |
| P3 | 1709 | 0.125 | 75.1 |
| P4 | 2616 | 0.188 | 71.0 |
| pITIM | 3370 | 0.154 | 47.2 |
| pIg-α | 1285 | 0.520 | 402.0 |
The measurements were performed at 30 μL/min of flow rate at 25°C. The chip was regenerated by washing with phosphate buffered saline containing 0.05% sodium dodecyl sulfate. The data were calculated from at least 5 different concentrations by BIAevaluation software. NM indicates not measurable.
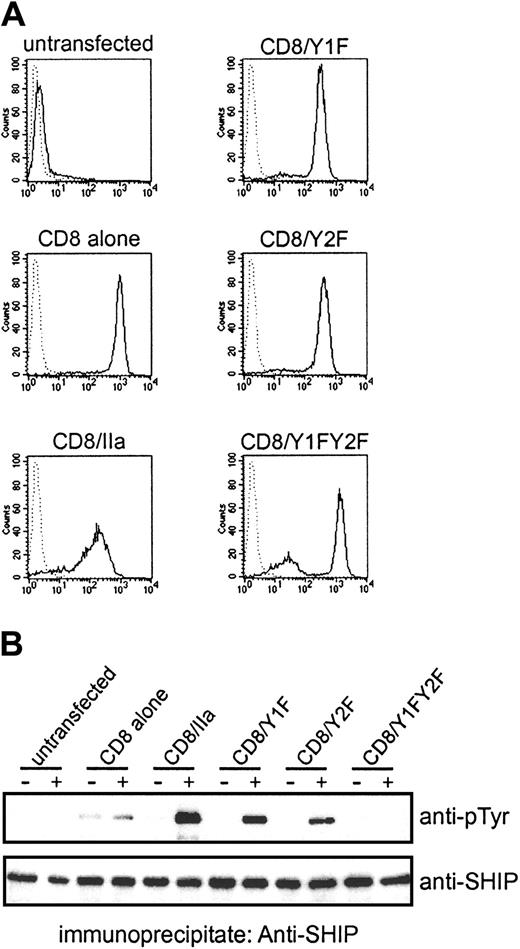
Because SHIP has been found to bind to both P2 and P3 in vitro with moderate affinities, we also examined SHIP phosphorylation upon stimulation of OKT8 in THP-1 cells expressing CD8/IIa. In these experiments, we used a mutant construct in which the ITAM tyrosine residues positioned at amino acid 288 (CD8/Y1F), or at amino acid 304 (CD8/Y2F), or at both (CD8/Y1FY2F) were substituted with phenylalanines. The mutant receptor chimeras were used to test which tyrosine residues within the ITAM of FcγRIIa are responsible for SHIP phosphorylation in vivo. FACS analysis showed comparable expressions of CD8 chimeras in THP-1 cells (Figure 7A). SHIP phosphorylation was induced upon stimulation of OKT8 in THP-1 expressing CD8/Y1F or CD8/Y2F, but the level was less than that induced by the wild-type chimera. However, the cells expressing CD8/Y1FY2F were incapable of phosphorylating SHIP (Figure 7B). These results indicate that either single tyrosine residue within ITAM of FcγRIIa are capable of recruiting SHIP. This finding is consistent with the in vitro data showing that GST-SH2-SHIP is able to bind P2 and P3 (Table1).
Both tyrosine residues in FcγRIIa ITAM are responsible for SHIP phosphorylation in vivo.
(A) The expression of CD8 chimeras with substitutions of tyrosine residues with phenylalanine was examined by FACS analysis using biotinylated F(ab′)2 fragments of OKT8 followed by FITC-conjugated streptavidin. Dotted lines indicate fluorescence of unstained cells. (B) The THP-1 transfectants were stimulated with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin. The lysates from the cells were immunoprecipitated with anti–SHIP antibody, separated on SDS-PAGE gels, and blotted with antiphosphotyrosine (anti-pTyr) antibody. The filter was reprobed with anti–SHIP antibody.
Both tyrosine residues in FcγRIIa ITAM are responsible for SHIP phosphorylation in vivo.
(A) The expression of CD8 chimeras with substitutions of tyrosine residues with phenylalanine was examined by FACS analysis using biotinylated F(ab′)2 fragments of OKT8 followed by FITC-conjugated streptavidin. Dotted lines indicate fluorescence of unstained cells. (B) The THP-1 transfectants were stimulated with biotinylated F(ab′)2 fragments of OKT8 followed by streptavidin. The lysates from the cells were immunoprecipitated with anti–SHIP antibody, separated on SDS-PAGE gels, and blotted with antiphosphotyrosine (anti-pTyr) antibody. The filter was reprobed with anti–SHIP antibody.
SHIP negatively regulates FcγR-mediated phagocytosis independently of ITIM-containing FcγRII(b)
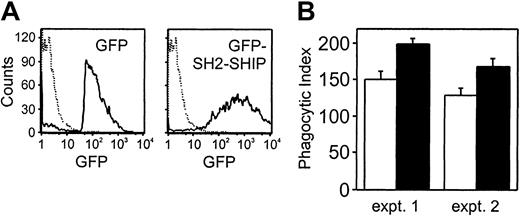
The findings shown above demonstrate that SHIP inhibits macrophage function through ITAM tyrosines in activating FcγRs. To address the contribution of SHIP to inhibition of FcγRIIa-triggered macrophage function, we transiently introduced GFP-SH2-SHIP into THP-1 cells. The SH2 domain has been shown to function as a dominant-negative form by inhibiting endogenous SHIP in B cells.39 Thus, cells expressing the SH2 domain should show enhanced function in FcγRIIa-stimulated macrophages. The cells expressing GFP or GFP-SH2-SHIP were isolated by cell sorting and used for phagocytosis assay. After sorting, the population of THP-1 cells expressing GFP-SH2-SHIP or GFP alone was 95% or 98%, respectively (Figure8A). For the phagocytosis assay, we used RBCs coated with Fab fragment of IV.3 which binds to only FcγRIIa on THP-1, and not to FcγRII(b).38 This assay system allows us to direct phagocytosis to FcγRIIa and thereby exclude a contribution by FcγRII(b). The average of duplicate samples of 2 separate experiments is shown in Figure 8B. We found that the phagocytic ability of THP-1 cells expressing GFP-SH2-SHIP was significantly enhanced compared with control transfectants. However, the extent of increase was less than that seen in either SHIP−/− or FcγRII−/−. These data indicate that SHIP is able to function as a negative regulator directly through ITAM-containing phagocytic receptors and independently of FcγRII(b). Additionally, SHIP might have functions in macrophages that are induced independently of its SH2 domain.
The introduction of SH2-SHIP enhances the phagocytic abilities in the absence of FcγRII(b).
(A) THP-1 cells were transiently transfected with GFP or GFP-SH2-SHIP. The GFP-positive cells were sorted and analyzed by FACS analysis. Dashed lines indicate untransfected THP-1 cells. (B) The sorted GFP-positive cells were incubated with RBCs coated with Fab fragments of IV.3 antibody for 20 minutes at 37°C. The results were expressed as the number of the internalized RBCs per 100 cells which phagocytosed at least one RBC (phagocytic index). Open and closed bars represent phagocytic indexes for GFP-expressing cells and GFP-SH2-SHIP–expressing cells, respectively. Results are shown as the averages of duplication and are representative of 2 independent experiments. Bars represent standard errors of duplicate samples.
The introduction of SH2-SHIP enhances the phagocytic abilities in the absence of FcγRII(b).
(A) THP-1 cells were transiently transfected with GFP or GFP-SH2-SHIP. The GFP-positive cells were sorted and analyzed by FACS analysis. Dashed lines indicate untransfected THP-1 cells. (B) The sorted GFP-positive cells were incubated with RBCs coated with Fab fragments of IV.3 antibody for 20 minutes at 37°C. The results were expressed as the number of the internalized RBCs per 100 cells which phagocytosed at least one RBC (phagocytic index). Open and closed bars represent phagocytic indexes for GFP-expressing cells and GFP-SH2-SHIP–expressing cells, respectively. Results are shown as the averages of duplication and are representative of 2 independent experiments. Bars represent standard errors of duplicate samples.
Discussion
Recent studies indicate that FcR-mediated phagocytosis is negatively regulated by FcγRII(b)21 and SHIP.22 However, the molecular mechanism of the negative regulation for phagocytosis has not been elucidated. In this report, we directly compared phagocytic rates and signal transduction events of macrophages from FcγRII(b)−/− and SHIP−/−mice, as well as wild-type mice. We found that macrophages of both FcγRII(b)−/− mice and SHIP−/− mice displayed a similar elevated phenotype regarding phagocytic potential, suggesting that FcγRII(b) contributes to SHIP function in macrophages as it does in B cells. However, in contrast to this possibility, macrophages of SHIP−/− but not FcγRII(b)−/− showed elevated intracellular Ca++ influx. Additionally, macrophages of FcγRII(b)−/− mice displayed efficient tyrosine phosphorylation of SHIP. These findings suggest that FcγRII(b) and SHIP function independently of each other, but both inhibit phagocytosis. In support of the notion that SHIP functions independently of FcγRII(b), we found that clustering a receptor chimera containing the ITAM tyrosine residues of the FcγR γ-chain or human-restricted FcγRIIa and without coengagement of FcγRII(b) is sufficient for SHIP phosphorylation in a macrophage cell line.
In vitro studies using phosphopeptides indicated that the SH2 domain of SHIP is able to interact with phosphorylated ITAM peptides, such as γ-, δ-, ε-, and ζ-chains of T-cell receptor (TCR)/CD3 complex.40 It is unclear whether phosphopeptide binding reflects an event occurring in stimulated cells. Previous experiments in T cells showed minimal SHIP phosphorylation under stimulation conditions promoting efficient phosphorylation of ITAM tyrosines in TCR/CD3,41 suggesting that SHIP has low affinity for these ITAMs in live cells. We demonstrated efficient SHIP phosphorylation caused by clustering ITAM-bearing molecules, and that SHIP directly binds to the ITAM of FcγRIIa with a comparable affinity to that of FcγRII(b). Furthermore, we show using dominant-negative mutants, that SHIP negatively regulates FcγRIIa-mediated phagocytosis independently of FcγRII(b). Our findings are consistent with the possibility that the direct binding of SHIP to the phosphorylated ITAM-containing receptor, followed by SHIP phosphorylation, leads to the negative regulation of FcγR-mediated phagocytosis.
Although direct binding of SHIP to phosphorylated FcγRIIa ITAM tyrosines represents the most simple explanation for our findings, we do not rule out the possibility of indirect binding through an adapter molecule. SHIP is known to interact with phosphorylated Shc42 and with Grb2.30 31 Either of these proteins might play an important role in SHIP recruitment to phosphorylated FcγRIIa. The phosphopeptide pull-down experiment shown in Figure 5 indicated that SHIP was able to bind FcγRIIa phospho-ITAM without cell stimulation. Although this observation argues against the need for an activation-induced adapter protein, the finding is based on synthetic peptides, which might disclose nonphysiologic interactions. Nevertheless, whether direct or indirect, our findings show that SHIP in macrophages engages activating receptors. This situation is in contrast to that in other hematopoietic cells where inhibitory phosphatases require pairing of ITIM- and ITAM-bearing receptors.
Macrophages from SHIP- and FcγRII(b)-deficient mice show similar enhanced phagocytic abilities. Our data show that SHIP is a mediator of ITAM-stimulated signal transduction and biology. It is unclear what enzyme is the mediator of FcγRII(b)-induced inhibition. Analysis of synthetic phosphopeptides corresponding to the phosphorylated ITIM of FcγRII(b) revealed binding to the protein phosphatases SHP-1 and SHP-2.43 It is possible that the negative regulation of phagocytosis by FcγRII(b) is mediated by SHP-1. Indeed, FcγR-triggered functions are enhanced in macrophages frommotheaten viable mice, which are deficient in expression of SHP-1.44 However, FcγRII(b) is expressed on monocytes/macrophages, and SHIP binds to the phosphorylated ITIM of FcγRII(b) with 2-fold higher affinity than that of FcγRIIa. Therefore, it is still possible that SHIP contributes to negative regulation for FcγR-mediated phagocytosis by FcγRII(b).
Our observations indicate that negative signaling involving SHIP is concomitant with the positive, phagocytic signaling; that is, both positive and negative effects are induced through the identical ITAM-containing receptors. The situation is therefore distinct from other paired inhibitory receptors that function only under conditions of receptor coclustering. Based on these and previous findings, we propose that hematopoietic cells employ the 2 suppressor phosphatases SHP-1 and SHIP in very different ways, and that they are induced under very different conditions. One manner of employment is a general one which regulates but does not abort signal transduction and biology. The other manner of employment is a specific one that critically involves coclustering of an ITIM-bearing receptor, and completely blocks cell activation. Regulator phosphatases are induced by directly or indirectly engaging the ITAM of the activating receptor. Regulator phosphatases act to control the biochemical and biologic output, but cannot completely abort the response. These features are consistent with SHIP behavior in myeloid cells where it appears to be directly recruited to the receptor, does not require the ITIM-containing FcγRII(b), and controls but does not block biologic function. Our earlier findings in B cells suggest that SHP-1 behaves similarly: SHP-1 is indirectly recruited to the activating receptor, its recruitment does not require the ITIM-containing FcγRII(b), and SHP-1 controls but does not block biologic function.45
Lymphoid or myeloid cells can also employ phosphatases as specific inhibitors. Phosphatases working in this way act only under conditions of paired ITAM/ITIM coclustering, as in the cases of BCR/FcγRII,4 BCR/PIR-B,12FcεR/gp49,13 or any of several activating NK cell receptors coclustered with KIR (reviewed in Taylor et al11). Thus, the specific inhibitory phosphatases require an ITIM-containing receptor for their induction. The mechanism for such exclusivity is phosphorylation of the ITIM tyrosine residue, and the selectivity of the SH2 domain of SHIP for phosphorylated tyrosines in an ITIM rather than the B-cell ITAM configuration, as shown in the affinity measurements summarized in Table 1. However, phosphatases working in this way are more potent and can completely block cell activation. These features are consistent with SHIP behavior in B lymphocytes. Experiments of signal transduction by granulocyte–colony-stimulating factor reveal a situation more like macrophage FcγRs where SHIP behaves as a regulator phosphatase. PtdIns 3-kinase and SHIP are recruited by distinct positive and negative growth regulatory domains in the cytokine receptor and coordinately regulate growth signaling.46 SHIP appears to function as a general regulator phosphatase in other situations of cytokine-induced biology47,48 (reviewed in Liu et al49). Thus, SHIP appears to play a regulatory role to prevent hyperinflammation. This notion is consistent with the SHIP−/− animal, which displays a pronounced hyperinflammatory phenotype,47,48 but a mild B-cell phenotype.50
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2002-03-0787.
Supported by National Institutes of Health grants CA64268 and AI41447. K.M.C. is a scholar of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
K. M. Coggeshall, The Oklahoma Medical Research Foundation, Program in Immunobiology and Cancer, 825 NE 13th St, Oklahoma City, OK 73104; e-mail: mark-coggeshall@omrf.ouhsc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal