Abstract
Among the alternative pre-mRNA splicing events that characterize protein 4.1R gene expression, one involving exon 2′ plays a critical role in regulating translation initiation and N-terminal protein structure. Exon 2′ encompasses translation initiation site AUG1 and is located between alternative splice acceptor sites at the 5′ end of exon 2; its inclusion or exclusion from mature 4.1R mRNA regulates expression of longer or shorter isoforms of 4.1R protein, respectively. The current study reports unexpected complexity in the 5′ region of the 4.1R gene that directly affects alternative splicing of exon 2′. Identified far upstream of exon 2 in both mouse and human genomes were 3 mutually exclusive alternative 5′ exons, designated 1A, 1B, and 1C; all 3 are associated with strong transcriptional promoters in the flanking genomic sequence. Importantly, exons 1A and 1B splice differentially with respect to exon 2′, generating transcripts with different 5′ ends and distinct N-terminal protein coding capacity. Exon 1A-type transcripts splice so as to exclude exon 2′ and therefore utilize the downstream AUG2 for translation of 80-kDa 4.1R protein, whereas exon 1B transcripts include exon 2′ and initiate at AUG1 to synthesize 135-kDa isoforms. RNA blot analyses revealed that 1A transcripts increase in abundance in late erythroblasts, consistent with the previously demonstrated up-regulation of 80-kDa 4.1R during terminal erythroid differentiation. Together, these results suggest that synthesis of structurally distinct 4.1R protein isoforms in various cell types is regulated by a novel mechanism requiring coordination between upstream transcription initiation events and downstream alternative splicing events.
Introduction
A family of protein 4.1R polypeptides is encoded by a single complex locus on human chromosome 1. Best characterized among the 4.1R isoforms is an 80-kDa polypeptide that functions as a key structural component of the membrane skeleton in mature red cells. Protein 4.1R's critical role in maintaining the specialized mechanical properties of the red cell plasma membrane is demonstrated by the abnormal morphology and increased membrane fragmentation of 4.1R-deficient red cells, occurring as a result of natural mutations in humans1,2 or by gene knock-out experiments in mice.3 A considerable body of work has defined a network of protein-protein interactions by which 4.1R participates in stabilization of the spectrin-based membrane skeleton and in connection of this skeleton to overlying plasma membrane. Moreover, recent crystal structure data have provided new insights into the structure and interactions of the membrane-binding domain.4
In contrast to the relatively simple expression pattern of 4.1R in mature red cells, a more complex array of 4.1R protein isoforms of varying sizes and proposed functions has been reported in erythroid progenitors and nonerythroid cells. Early erythroid progenitors, and many nonerythroid cells, express 4.1R isoforms differing in size and subcellular localization from the familiar erythroid form. Among the sites of function proposed for nonerythroid 4.1R are the nucleus, centrosomes, and spindle poles of dividing cells5-9; epithelial tight junctions10; and contractile sarcomeres in muscle.11 Characterization of 4.1R knock-outs in mice indicates a role in selected neurons that affect fine motor control.12 Finally, 4.1R interactions have been reported with a variety of other proteins, including translation and splicing factors,13-21 implying that 4.1R may have additional roles in the cell. The concept that 4.1R has a diverse set of cellular functions in addition to its role in red cells is thus well established. However, much remains to be learned about the structure and function of these 4.1R isoforms in nucleated cells.
A major challenge in the field is to understand how a single 4.1R gene can express such a variety of isoforms with diverse structures and functions. Molecular characterization of this complex gene has shown that it is approximately 240 kb in length (Huang et al22; Baklouti et al23; and our unpublished results, June 2002) and is subject to extensive regulation at the level of alternative pre-mRNA splicing (reviewed in Conboy24). At least 10 of the internal coding exons of the gene can be alternatively spliced, and several of these are tightly regulated in tissue- or developmental-specific patterns. As a direct consequence, different cell types can express different profiles of 4.1R mRNA isoforms,11,25 and thereby synthesize functionally different complements of 4.1R protein. Of particular relevance to erythroid differentiation and red cell function is alternative exon 16, which encodes an essential part of the spectrin-actin binding domain. Exon 16 is excluded in early erythroid progenitors but included in later progenitors, allowing the mature cells to produce 4.1R isoforms that can effectively stabilize the developing membrane skeleton.26 27
The current study focuses on novel aspects of protein 4.1R gene structure that play a critical role in regulating the balance in synthesis of 80-kDa versus 135-kDa 4.1R protein isoforms. Specifically, we report an unexpected complexity in 5′ exon/intron organization: 3 mutually exclusive 5′ exons map far upstream of the coding exons, and each of these alternative exons possesses its own transcriptional promoter, suggesting that they may represent alternative first exons of the 4.1R gene. Most important, exons 1A and 1B splice differentially to 2 acceptor sites downstream in exon 2′ in a manner that regulates expression of translation initiation site AUG1. Transcripts initiated at exon 1A splice to an internal acceptor site in exon 2, thereby skipping exon 2′ sequences and translation initiation site AUG1; the resulting spliced mRNAs encode 80-kDa isoforms of 4.1R protein by initiation at downstream AUG2 in exon 4. Conversely, transcripts initiated at exon 1B splice to include exon 2′, incorporating AUG1 into the mature mRNA and facilitating synthesis of 135-kDa protein 4.1R. These results suggest that there is a mechanism for functional coupling between upstream transcription and downstream alternative splicing events.
Materials and methods
Genetic database analyses
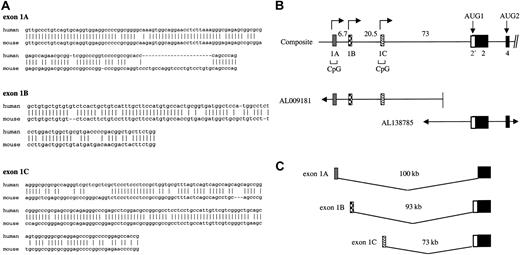
Identification of alternative 5′ exons in the 4.1R gene required analysis of both cDNA and high-throughput genomic sequence (htgs) information in Genbank. To identify candidate first exons, we used the human 4.1R exon 2 sequence as a probe for BLAST searches of Genbank. Among several human and mouse cDNA clones that contained sequence information upstream of exon 2, 3 unique sequences were found to splice properly to exon 2. These candidate exons were designated as exon 1A, exon 1B, and exon 1C. Exon 1A was identical to the 5′ end of the original human 4.1R cDNA (M61733), exon 1B was identical to mouse 4.1R cDNA (L00919), and exon 1C represented unique a sequence. In order to confirm that these were bona fide exons in the 4.1R gene, these sequences were used as probes in a BLAST search of the htgs database. All 3 sequences were found in bacterial artificial chromosome (BAC) clones upstream of known coding exons of the 4.1R gene. Of note, a human ortholog of mouse exon 1B was characterized by 79% identity over a region of 107 nucleotide (nt) (Figure1A). Although exon 1B has not been found in any human cDNA clones in the databases, its authenticity was confirmed by reverse transcriptase–polymerase chain reaction (RT-PCR) experiments demonstrating in many tissues the presence of 4.1R transcripts possessing exon 1B sequences properly spliced to exon 2 (Figure 3). In the current human genome assembly (June 2002), the coordinates for exon 1A are 29, 219, 768-29, 219, 837; for exon 1B: 29, 226, 261-29, 226, 464; for exon 1C: 29, 247, 204-29, 247, 395; and for exon 2: 29, 320, 058-29, 320, 532.
Alternative 5′ exons of the 4.1R gene.
(A) Sequence comparison of human versus mouse alternative 4.1R 5′ exons. The orthologous exons had the following sequence identities: 1A, 72% over a region of 142 nt; 1B, 79% over 107 nt; and 1C, 83% over 159 nt. Origins of sequences: for exon 1A, mouse sequence was from AA014918, and human from M61733 and BG943340. For exon 1B, mouse sequence was from L00919, while the human sequence was deduced by comparing the mouse 1B sequence to human genomic sequence downstream of exon 1A. That the predicted human 1B can splice to exon 2 was confirmed by RT-PCR analysis (Figure 3). For exon 1C, human sequence was from AL041809, while mouse sequence was derived from a 5′ RACE product from mouse spleen (results not shown). (B) Structure of the 5′ region of the 4.1R gene. The genomic map was constructed from overlapping BAC human genomic clones derived from high-throughput genomic sequences. Distances between exons (in kilobases) are indicated, and the location of CpG islands is shown. (C) Model showing independent splicing of alternative 5′ exons to downstream exon 2, to generate 3 4.1R transcript classes with different 5′ end sequences.
Alternative 5′ exons of the 4.1R gene.
(A) Sequence comparison of human versus mouse alternative 4.1R 5′ exons. The orthologous exons had the following sequence identities: 1A, 72% over a region of 142 nt; 1B, 79% over 107 nt; and 1C, 83% over 159 nt. Origins of sequences: for exon 1A, mouse sequence was from AA014918, and human from M61733 and BG943340. For exon 1B, mouse sequence was from L00919, while the human sequence was deduced by comparing the mouse 1B sequence to human genomic sequence downstream of exon 1A. That the predicted human 1B can splice to exon 2 was confirmed by RT-PCR analysis (Figure 3). For exon 1C, human sequence was from AL041809, while mouse sequence was derived from a 5′ RACE product from mouse spleen (results not shown). (B) Structure of the 5′ region of the 4.1R gene. The genomic map was constructed from overlapping BAC human genomic clones derived from high-throughput genomic sequences. Distances between exons (in kilobases) are indicated, and the location of CpG islands is shown. (C) Model showing independent splicing of alternative 5′ exons to downstream exon 2, to generate 3 4.1R transcript classes with different 5′ end sequences.
Amplification of 1A and 1B transcripts from human tissues
Splicing patterns of 1A- and 1B-type transcripts were characterized using RT-PCR techniques to amplify 4.1R mRNA from several different human tissue sources (Clontech, Palo Alto, CA). One μg total RNA was transcribed into cDNA using a specific antisense primer in exon 2 in a total volume of 10 μL. Then, 2 μL cDNA was amplified as described previously28 using the following primers: 1A-S, 5′-GCAAAGTGGCAGGAACCTCTTAAAG-3′ and 2-AS, 5′-CGAGGAGAATAGTCGTGAAAGTCC-3′; or 1B-S, 5′-GACTGGCTGCGTGACCCCGACGGCTG-3′ and 2-AS. Amplification (35 cycles) was performed under the following conditions: denaturation for 20 seconds at 94°C; annealing for 20 seconds at 60°C; and extension for 40 seconds at 72°C. DNA fragments were analyzed by 5% polyacrylamide gel electrophoresis. The identity of all major PCR products discussed in this paper was confirmed by DNA sequence analysis.
Construction of PCR standards
There were 2 potential 5′ cDNA structures, containing exon 1A spliced to the proximal 3′ splice site in exon 2 (“1A-proximal”) or exon 1B spliced to the distal 3′ splice site in exon 2 (“1B-distal”), that were never amplified from natural mRNA extracted from human tissues. However, it was essential to generate PCR size standards for these products in order to verify that such products would be detectable if they were present in a tissue. For the 1A-proximal product, we found that in vitro splicing of an artificial pre-mRNA construct (containing only exon 1A, a dramatically truncated intron, and exon 2) generated both 1A-proximal and 1A-distal products. A different approach using splice overlap techniques29 was required to generate the 1B-distal marker. Exon 1B was amplified using primer 1B-S and primer 2/1B-AS, 5′-CGGCCTCAGTCACTAAACTCTTCTCCAGAAGCAGCCGTCGGGGTCAC-3′. Exon 2 (excluding 2′) was amplified using primer 1B/2-S, 5′-GTGACCCCGACGGCTGCTTCTGGAGAAGAGTTTAGTGACTGAGGCCG-3′ and primer 2-end, 5′-GGCCTCTAGACTGTGTTTCTGCACTGCTTAAT-3′ (italicized region is an XbaI restriction site for cloning purposes). Underlined portions of the primers created a 24–base pair (bp) overlap between the fragments, which were mixed in equal proportions and amplified with the outside primers 1B-S and 2-AS to create the 1B-distal marker.
RNA blots
Northern blot analysis of RNA was performed as previously described, using 10 μg total RNA per lane. The full-length mouse 4.1R probe was prepared by amplification with the following primers: forward, 5′-ATGACAACAGAGAAGAGTTTAGTGGCTGAAGC-3′; reverse, 5′-TCACTC-CTCAGAGATCTCTGTCTCCTGGTGGACGACC-3′. The mouse exon 1A probe was also prepared by amplification with primers: forward, 5′-TCAGTGCAGGTGGAGGCCCCCGCGGG-3′; reverse, 5′-CTGGGCTGCACAGGACAGGGACCTGG-3′. The expression patterns of exons 1A, 1B, and 1C among human tissues were determined by hybridization to master RNA blots containing poly A+ RNA from a number of different tissues (Clontech). Probes used for RNA dot blots were prepared by PCR amplification of exon-specific DNA fragments using the following primers: exon 1A, forward, 5′-AAAGGGCGAGAGCGGCGCGGAG-3′ and reverse, 5′-GCTGGGTGCGGCGGGGA-3′; exon 1B, forward, 5′-TGCTGTCATTTGCTTCCATGTG-3′ and reverse, 5′-CGTCGGGGTCACGCAGCCAGTC-3′; and exon 1C, forward, 5′-CTCCCTCCGCTGGTGCGTTTAG-3′ and reverse, 5′-ACGAACAATGGCAGGAGGAG-3′.
Erythroblast cell procurement and culture
Erythroid cells obtained from the spleens of mice infected with the anemia-inducing strain of Friend erythroleukemia virus were isolated and cultured as previously described.30 31Cells at the 0 hour are mainly proerythroblasts, which then differentiate over approximately 48 hours into late-stage erythroblasts and enucleated reticulocytes. Total RNA was isolated from cell pellets using RNeasy columns according to the manufacturer's instructions (Qiagen, Valencia, CA).
Transcriptional promoter assays
A 1.5-kilobase (kb) region of genomic DNA upstream of exon 1A, and overlapping by approximately 50 nt the exon 1A sequence shown in Figure 1A, was cloned upstream of the firefly luciferase reporter in pGL2-basic. Analogous regions upstream of exons 1B and 1C were similarly cloned into pGL2-basic. For each construct, transcriptional activity of the putative promoter region was assessed following transfection into HEK293T (human embryonic kidney) cells by measurement of luciferase activity in comparison with the activity of a control promoterless construct, pGL2basic. As a positive control we used the pGL2-promoter vector containing an SV40 promoter (Promega, Madison, WI). Cells were cotransfected with a Renilla luciferase pRL-TK reporter to provide an internal control value to which the experimental firefly luciferase measurements can be normalized, to account for potential differences in transfection efficiency. Firefly and Renilla luciferase activities were measured using a dual luciferase assay system (Promega) according to the manufacturer's instructions.
Results
Evidence for multiple alternative 5′ exons in the protein 4.1R gene
The existence of multiple alternative 5′ exons was initially suggested by sequence comparisons between full-length cDNAs cloned from mouse (accession no. L00919) and human sources (accession no. M61733). These cDNAs exhibited high nucleotide sequence homology from exon 2 through the 3′ untranslated region (UTR), as expected for orthologous genes in human and mouse. However, the sequences upstream of exon 2 were quite distinct in these clones, consistent with the hypothesis that these sequences represent alternative first exons in the protein 4.1R gene. In order to explore this hypothesis further, we analyzed the 5′ end sequences of all available 4.1R cDNAs in the various genetic databases. The strategy was to identify all clones containing bona fide 4.1R exon 2 sequences, and then classify them according to the nature of the sequences upstream of exon 2. Only those strictly adhering to the following criteria were considered to represent bona fide 4.1R 5′ sequences: (1) at the mRNA level, a candidate alternative first exon must be joined, at proper exon junctions, to known downstream coding exon(s) of the 4.1R gene; and (2) at the genome level, such sequences must be located in the appropriate region of human chromosome 1 upstream of the coding exons. This approach was necessary to distinguish authentic new 4.1R transcripts from potential cloning or database artifacts.
Among the collection of 4.1R cDNAs in the databases, 3 distinct transcript classes possessed unique 5′ ends and satisfied the above criteria. The sequences comprising these 5′ ends were therefore designated as exons 1A, 1B, and 1C. The sequences of the corresponding exons in mouse versus human were derived from the database clones as well as from additional 5′ rapid amplification of cDNA ends (RACE) and RT-PCR experiments. As shown in Figure1A, the orthologous human and mouse exons are 72% to 83% identical at the nucleotide level. To date, there is no evidence that these exons are translated; however, differential utilization of these exons does affect protein structure via coupling to downstream alternative splicing (below).
Shown in Figure 1B is the 4.1R gene organization in the 5′ region, including the location of exons 1A, 1B, 1C, and 2. This map, derived by analysis of BAC clones from the htgs database, revealed that the 3 first exons are separated from one another by 6 to 20 kb and are located at least 70-kb upstream of the coding region. Physical linkage of these exons to the coding region was confirmed initially by the finding of 2 independent BAC clones containing both the exon 1 region and exon 2. More recently, these BAC sequences have been completed so that the linkage and precise distance among these various 5′ exons is now known. Notably, exons 1A and 1C are located in extensive CpG islands characteristic of many transcriptional promoter regions. Specifically, exon 1A resides in a region of 1.5 kb that is 68% C + G and has a CG/GC ratio of 0.73; exon 1C is located in a 1.2-kb region that is approximately 69% C + G and has a CG/GC ratio of 0.80 (data not shown). Exon 1B, in contrast, is located in a region with lower C + G content (55% over 0.5 kb) and exhibits a significantly lower CG/GC ratio (0.18). Nevertheless, transcription assays shown below demonstrate that exon 1B as well as 1A and 1C all possess active promoter regions.
Figure 1C summarizes the origins of the 3 different classes of 4.1R transcripts. According to this model, transcription can initiate at 3 distinct sites representing independent promoters for alternative 5′ exons 1A, 1B, and 1C, each of which can be spliced directly to exon 2. It is also important to note that exon 1A splices differently to exon 2 than do exons 1B and 1C. This latter point is explored in more detail below.
Evidence for transcriptional promoter activity associated with each alternative 5′ exon
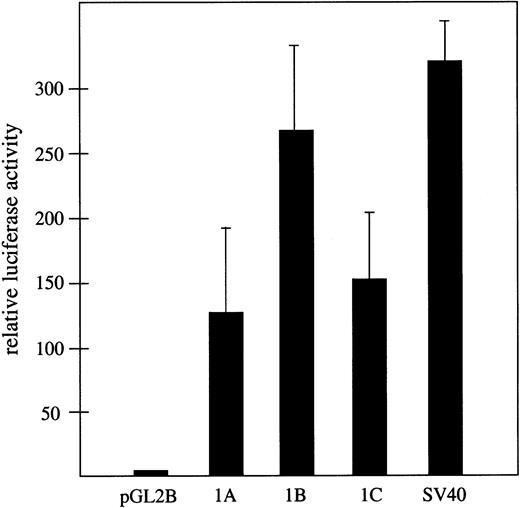
The hypothesis that the widely separated exons 1A, 1B, and 1C represent bona fide alternative first exons in the 4.1R gene predicts that each must be closely associated with a distinct transcriptional promoter activity. The finding that exons 1A and 1C are located in typical CpG islands is consistent with this model. Moreover, a computational approach using the PromoterInspector algorithm32 also predicted transcriptional promoters corresponding almost precisely with exons 1A and 1C (results not shown). However, the most direct experimental approach was to test for promoter activity using standard luciferase reporter vectors in transfected mammalian cells. For this purpose, 3 1.5-kb regions of genomic DNA upstream of exons 1A, 1B, and 1C respectively, were cloned into pGL2basic upstream of the luciferase gene (Figure2). The 3 candidate promoter regions exhibited strong transcriptional promoter activity relative to the promoterless luciferase reporter when tested in HEK293T cells (Figure2). All exhibited more than a 100-fold increase in luciferase expression compared with the promoterless control, and approximately 40% to 80% of the activity that was observed with an SV40-positive control promoter. These results demonstrate robust promoter activity associated with all 3 exons and strongly support the hypothesis that exons 1A, 1B, and 1C are authentic first exons in the 4.1R gene.
Transcriptional promoter activity associated with exons 1A, 1B, and 1C.
Shown is firefly luciferase activity assayed 48 hours after transfection of HEK293T cells, relative to the activity of cells transfected in parallel with the promoterless pGL2 basic control vector. Assays were performed in triplicate and transfection efficiency was normalized with respect to cotransfected Renilla luciferase activity in the same cultures. As a positive control, luciferase activity expressed from a standard SV40 promoter is shown.
Transcriptional promoter activity associated with exons 1A, 1B, and 1C.
Shown is firefly luciferase activity assayed 48 hours after transfection of HEK293T cells, relative to the activity of cells transfected in parallel with the promoterless pGL2 basic control vector. Assays were performed in triplicate and transfection efficiency was normalized with respect to cotransfected Renilla luciferase activity in the same cultures. As a positive control, luciferase activity expressed from a standard SV40 promoter is shown.
Evidence for coupling between alternative 5′ exon choice and alternative splicing at exon 2
Close examination of 4.1R cDNAs revealed a strong correlation between the identity of the 5′ exon, and the splice acceptor choice at exon 2. All reported 4.1R cDNAs that initiate at exon 1A exhibit splicing to exon 2 at the distal (downstream) acceptor site, thus excluding the 2′ region and AUG1. This conclusion was based on analysis of 20 cDNAs encompassing 7 cDNA sequences found in the databases, and 13 additional clones derived by 5′ RACE from mouse spleen (data not shown). The 1A transcripts would therefore be expected to encode exclusively 80-kDa isoforms of 4.1R protein. In contrast, all 1B transcripts identified to date, including a set of 96 products cloned from muscle by RT-PCR,11 are spliced to the proximal (upstream) acceptor site in exon 2. These transcripts include the 2′ region and encode 135-kDa isoforms of protein 4.1R initiated at AUG1. Finally, 3 cDNAs representing transcript 1C also spliced to the proximal site and should encode 135-kDa protein 4.1R.
Although this correlation between mutually exclusive 5′ exon use and splicing at exon 2 was intriguing, it was based on a relatively limited sample. A more systematic examination of 4.1R expression in a number of different cell types was therefore undertaken. For this approach we used RT-PCR techniques to amplify the exon 1A/2 and 1B/2 junctions, in order to distinguish between proximal and distal exon 2 splicing. Analysis of 1A transcripts was performed according to the scheme in Figure 3, which shows the predicted amplification products of 346 nt or 329 nt depending on whether proximal or distal splicing has occurred (upper panel). Importantly, amplification of RNA from 15 different human tissues yielded virtually identical results: in all cases only a single major PCR product was obtained, of a size consistent with splicing of exon 1A to the distal acceptor in exon 2. The identity of this product was confirmed by DNA sequence analysis. Control experiments showed that distal and proximal amplification products were resolvable under the experimental conditions used (Figure 3A, last lane). The major conclusion from this experiment is that 1A-type transcripts in all tissues examined are spliced to the distal 3′ splice site in a pattern that excludes exon 2′, and thus the resulting mature mRNA will encode the 80-kDa forms of 4.1R protein.
Coupling between alternative 5′ exons and alternative splicing at exon 2.
(Top) 1A transcripts were amplified from the indicated human tissues using primers in exons 1A and 2. Control shows migration of PCR products that include exon 2′ (346 nt; proximal splicing) or exclude exon 2′ (329 nt; distal splicing). (Bottom) 1B transcripts were amplified from the indicated tissues using primers in exons 1B and 2. Control shows migration of PCR products that include exon 2′ (295 nt; proximal splicing) or exclude exon 2′ (278 nt; distal splicing).
Coupling between alternative 5′ exons and alternative splicing at exon 2.
(Top) 1A transcripts were amplified from the indicated human tissues using primers in exons 1A and 2. Control shows migration of PCR products that include exon 2′ (346 nt; proximal splicing) or exclude exon 2′ (329 nt; distal splicing). (Bottom) 1B transcripts were amplified from the indicated tissues using primers in exons 1B and 2. Control shows migration of PCR products that include exon 2′ (295 nt; proximal splicing) or exclude exon 2′ (278 nt; distal splicing).
Similar analysis was performed on 1B transcripts (Figure 3B). In this case, amplification of RNA from numerous tissues yielded only a single product corresponding in size and nucleotide sequence to splicing at the proximal site. These results indicate that 4.1R exon 1B transcripts splice predominantly, if not exclusively, so as to include exon 2′. Translation of 1B mature mRNA transcripts will therefore generate selectively the 135-kDa isoforms of 4.1R protein. This result confirms and extends the previous studies of 4.1R mRNA structure showing that mouse skeletal muscle cDNAs, with 5′ ends corresponding to this newly defined exon 1B sequence, always include exon 2′.11
Differential expression of alternative 5′ exons in erythroid progenitor cells
Taken together, the data above support the idea that protein 4.1R transcripts can be expressed from one of several mutually exclusive alternative 5′ exons. To explore the relative expression levels of each transcript class during late erythropoiesis, we isolated RNA from differentiating erythroblasts obtained from the spleens of mice infected with the anemia-inducing strain of Friend virus.31 These cells differentiate to the point of enucleation over the course of 48 hours when cultured in the presence of erythropoietin, and have been shown to undergo expected changes in erythroid gene expression, including induction of globin synthesis,31 up-regulation of total protein 4.1R synthesis,33 and switching in protein 4.1R pre-mRNA splicing from exon 16 skipping to exon 16 inclusion.34
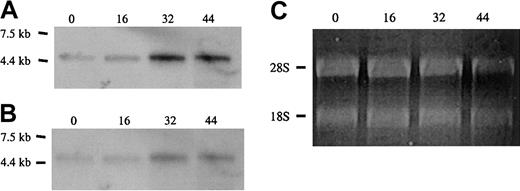
Approximately equal amounts of total RNA from cells at 0, 16, 30, and 44 hours of culture, normalized with respect to quantity of ribosomal RNA, were electrophoresed in agarose gels and subjected to Northern blot analysis with probes specific for protein 4.1R RNA (Figure4). Hybridization with a cDNA probe to the shared coding region, which should recognize all classes of 4.1R transcript, showed that total 4.1R mRNA levels progressively increased over the course of differentiation (Figure 4A). This result is consistent with previous observations of 4.1R protein accumulation at late stages of erythropoiesis.33 35 To determine the relative utilization of exons 1A, 1B, and 1C, Northern blotting was repeated with probes specific for each alternative 5′ exon. The 1A probe hybridized to a transcript identical in size and temporal pattern of expression to that detected with the full-length 4.1R probe; expression was relatively low at 0 hours and progressively increased with differentiation time (Figure 4B). Given the finding that 1A transcripts preferentially encode 80-kDa isoforms of 4.1R protein, this result is also consistent with previous observations that 80-kDa protein 4.1R is the predominant isoform in late erythroid cells. In contrast to the results with the 1A probe, hybridization of identical RNA blots with a probe specific for exon 1B did not detect any 4.1R mRNA corresponding to this isoform. Similarly, Northern blot analysis of mouse fetal liver total RNA revealed little or no 1B and 1C transcripts under conditions that permitted detection of abundant 1A-type 4.1R mRNA (results not shown). These results are in agreement with the low abundance of 135-kDa isoforms in late erythroid cells.
Expression of alternative 5′ exons in erythroid progenitor cells.
Shown are Northern blot analyses of total RNA from Friend virus anemia–induced erythroblasts following culture for the times indicated (in hours). (A) Full-length cDNA probe. (B) Exon 1A probe. Both probes detected a single 4.1R mRNA band that increased during the course of differentiation. No hybridization was detected with probes for exons 1B and 1C under identical conditions (not shown). (C) Ethidium-bromide staining of ribosomal RNA. Equal (0, 16, and 30 h) or slightly reduced (44 h) loads of RNA were applied for each time point, supporting the idea that a true increase in 4.1R RNA expression has occurred in these cells.
Expression of alternative 5′ exons in erythroid progenitor cells.
Shown are Northern blot analyses of total RNA from Friend virus anemia–induced erythroblasts following culture for the times indicated (in hours). (A) Full-length cDNA probe. (B) Exon 1A probe. Both probes detected a single 4.1R mRNA band that increased during the course of differentiation. No hybridization was detected with probes for exons 1B and 1C under identical conditions (not shown). (C) Ethidium-bromide staining of ribosomal RNA. Equal (0, 16, and 30 h) or slightly reduced (44 h) loads of RNA were applied for each time point, supporting the idea that a true increase in 4.1R RNA expression has occurred in these cells.
Differential expression of alternative 5′ exons in nonerythroid cells
The RT-PCR data in Figure 3 show that exons 1A and 1B are widely expressed among human tissues, consistent with previous reports that many nonerythroid tissues express a mixture of low- and high-molecular-weight isoforms of 4.1R protein. However, these PCR results do not give quantitative information regarding the relative abundance of these transcripts in different cell types. The abundance of each class of transcripts was therefore examined by RNA blot analysis using short exon-specific hybridization probes. As shown in Figure 5, exons 1A and 1C were expressed in many tissues. Exon 1B was generally expressed at a low level in many tissues, but appeared relatively more abundant in cells known to exhibit features of an early erythroid phenotype (K562 cells, coordinate C10) or in tissues with a significant component of early erythroid progenitors cells (fetal liver, bone marrow; coordinates D11 and G7, respectively).
Tissue-specific expression of alternative 5′ exons.
A master RNA blot (Clontech) containing poly A+ RNA from many human tissues and cell lines was blotted with probes for alternative exon 1A, exon 1B, or exon 1C. The 1A and 1C transcripts were particularly abundant in tissues with a high content of muscle cells (heart, A4; skeletal muscle, B7; and stomach, B5) as well as in pancreas (B9) and liver (A9). In contrast, exon 1B transcripts were most abundant in tissues enriched for early erythroid progenitors (fetal liver, D11; bone marrow, G7) and the erythroleukemia cell line K562 (C10). Lower panel shows the tissue map for all RNA sources tested.
Tissue-specific expression of alternative 5′ exons.
A master RNA blot (Clontech) containing poly A+ RNA from many human tissues and cell lines was blotted with probes for alternative exon 1A, exon 1B, or exon 1C. The 1A and 1C transcripts were particularly abundant in tissues with a high content of muscle cells (heart, A4; skeletal muscle, B7; and stomach, B5) as well as in pancreas (B9) and liver (A9). In contrast, exon 1B transcripts were most abundant in tissues enriched for early erythroid progenitors (fetal liver, D11; bone marrow, G7) and the erythroleukemia cell line K562 (C10). Lower panel shows the tissue map for all RNA sources tested.
Discussion
The results presented in this study indicate that the balance in synthesis of 135-kDa versus 80-kDa isoforms of 4.1R protein, which vary in N-terminal domain structure, is regulated by a novel mechanism involving coordination between transcriptional and alternative splicing events. Central to this model is the existence of mutually exclusive alternative 5′ exons that map upstream of, and can differentially splice to, alternative splice acceptor sites flanking exon 2′. Transcripts containing exon 1A splice to the distal 3′ splice site thus excluding exon 2′/AUG1 from the mature mRNA (Figure6A). This class of 4.1R mRNA initiates translation at the downstream AUG2 and encodes 80-kDa isoforms of 4.1R protein (Figure 6B). In contrast, transcripts possessing the 5′ exon 1B preferentially splice to the proximal 3′ splice site and include AUG1, thereby generating mRNAs that encode 135-kDa isoforms bearing extended N-terminal domains.
Model of protein 4.1R 5′ gene expression.
(A) Differential splicing of 5′ putative exons to alternative splice acceptor sites in exon 2. (B) The distinct protein isoforms resulting from translation of 1A- and 1B-type mRNAs. MBD indicates membrane binding domain; U2, unique region 2; SAB, spectrin-actin binding domain; CTD, C-terminal domain; and ss, splice site.
Model of protein 4.1R 5′ gene expression.
(A) Differential splicing of 5′ putative exons to alternative splice acceptor sites in exon 2. (B) The distinct protein isoforms resulting from translation of 1A- and 1B-type mRNAs. MBD indicates membrane binding domain; U2, unique region 2; SAB, spectrin-actin binding domain; CTD, C-terminal domain; and ss, splice site.
According to this model, cells of both erythroid and nonerythroid origin may regulate differentially the rate of gene expression at the alternative 5′ exons, as a primary mechanism for controlling their content of 4.1R protein that either includes (AUG1 isoforms) or excludes (AUG2 isoforms) the N-terminal headpiece domain. The demonstration that exons 1A, 1B, and 1C each possess an independent transcriptional promoter supports the model that these alternative 5′ sequences represent authentic first exons in the 4.1R gene, although direct mapping of 5′ cap sites will be required to confirm this hypothesis. Cells with high expression of 1A transcripts would be predicted to possess a higher content of 80-kDa 4.1R protein (or other isoforms initiated at AUG2); conversely, cells with high levels of 1B or 1C transcripts should express more 135-kDa protein. In accordance with these predictions, the observed patterns of 4.1R RNA expression during erythropoiesis are consistent with previously reported qualitative and quantitative changes in the cellular content of 4.1R protein.33,35 Early erythroid progenitors express low levels of total 4.1R protein and contain detectable amounts of 135-kDa isoforms.26 As expected, both exon 1A and exon 1B transcripts are detected in tissue sources enriched in early erythroid cells (bone marrow, fetal liver, and K562 cells; Figure 5). In contrast, late erythroblasts express much higher levels of 4.1R protein that is almost exclusively of the 80-kDa class. Northern blot analysis of differentiating mouse erythroid cells reveals a corresponding dramatic up-regulation in the expression of 1A transcripts in the absence of detectable 1B transcripts (Figure 4).
For nonerythroid cells, our results suggest that a complete characterization of 4.1R content in a given cell type will require probing at the RNA level for the presence of alternative 5′ sequences and at the protein level with antibodies that can detect both size classes of protein. Experiments performed prior to this recognition that 5′ exon choice strongly influences 4.1R protein synthesis, probably underestimated the complexity of 4.1R expression in a given cell. In one nonerythroid tissue in which this issue has been carefully analyzed, the kidney, there is a good correlation between the expression of exon 1A transcripts and the synthesis of 4.1R protein initiated at AUG2; these cells express neither exon 1B nor AUG1 isoforms of 4.1R protein (P. Gascard, personal oral communication, December 2001). The finding that most nonerythroid cells express both exon 1A and 1B transcripts of 4.1R is consistent with several reports demonstrating that such cells (including T lymphocytes,36 mammary epithelial cells,37 MDCK cells,38 and others39) do express both size classes of 4.1R protein.
In addition to the multiple promoters described here for the 4.1R gene, extensive precedence exists for multiple promoters/multiple first exons in a number of other erythroid genes. Examples include the genes for heme biosynthetic enzymes,40-44 erythroid skeletal proteins,45-47 cell-surface receptors,48 and transcription factors.49-51 In many of these genes, the alternative first exons are exclusively noncoding; transcriptional regulation therefore controls spatial and temporal patterns of expression without altering protein structure. In a few cases, including the genes for erythroid ankyrin and band 3, the alternative promoters/first exons are internally located, downstream of one or more coding exons, so that their respective transcripts will encode proteins with different N-termini.45-47 The situation for 4.1R is somewhat unique: although the alternative 5′ exons in the protein 4.1R gene are noncoding exons, their expression nevertheless dictates N-terminal protein structure via the coupling to downstream splicing.
Recent studies have evoked considerable interest in the coupling between transcription and several aspects of pre-mRNA processing including splicing and polyadenylation.52-55 The mechanism by which transcriptional events may be functionally coupled with downstream alternative splicing in the 4.1R gene is not known. Pre-mRNAs initiated at alternative promoters differ in several respects, and it is not yet clear which of these is most important in regulating the downstream splicing event. One critical factor may be the primary nucleotide sequence at the 5′ end of each 4.1R transcript class. These unique RNA sequences might contain binding sites for splicing regulatory proteins, or might adopt specific secondary structures, either of which could affect the accessibility of downstream acceptor sites in exon 2. Another intriguing possibility is that distinct transcriptional complexes formed at the alternative 5′ exons could directly affect downstream splicing events, perhaps by assembling and delivering specific splicing factors to the regulated splice sites in a cotranscriptional manner. Indeed, studies with model gene constructs have shown that the efficiency of alternative splicing can be modulated by changes in the transcriptional promoter, even in the absence of any alteration in the sequence of the transcribed pre-mRNA itself.56 57 Although few if any examples of coupling between transcription and alternative splicing have been reported previously in natural genes, it is likely that additional cases will be discovered as the genetic databases continue to grow. Our preliminary studies, in fact, indicate that a similar coupling of transcription and alternative splicing occurs in the protein 4.1B gene (data not shown).
Regardless of the precise mechanism, the regulated expression of 135- versus 80-kDa protein 4.1R isoforms is yet another example of complex expression in the 4.1R gene. An extensive array of tissue-specific alternative splicing events has been characterized in the gene, including the developmentally regulated splicing of exon 16 during erythroid differentiation, the regulated expression of exon 17B in epithelial cells,37 and the preferential expression of exon 17A in muscle cells.23,37 It seems reasonable to assume that precisely regulated expression of selected protein isoforms is indicative of an important function for these isoforms. This has already been shown in the case of exon 16, the inclusion of which results in synthesis of isoforms containing an intact spectrin-actin binding domain. While the function of the N-terminal extension in 4.1R protein isoforms is not well understood, it has been shown to influence subcellular localization,58 and may influence binding interactions of neighboring domains of the protein.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/blood-2002-06-1796.
Supported by National Institutes of Health (NIH) grants HL45182 and DK32094; by the Director, Office of Biological and Environmental Research, US Department of Energy under contract DE-AC03-76SF00098; and by a Merit Review Award from the Department of Veterans Affairs to M.J.K.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John G. Conboy, Lawrence Berkeley National Laboratory, Life Sciences Division, Mailstop 74-157, 1 Cyclotron Rd, Berkeley, CA 94720; e-mail: jgconboy@lbl.gov.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal