Abstract
In vivo distribution of myeloid transcription factors during granulopoiesis was investigated by Northern and Western blotting in 3 neutrophil precursor populations from human bone marrow: immature (myeloblasts [MBs] and promyelocytes [PMs]); intermediate mature (myelocytes [MCs] and metamyelocytes [MMs]); and mature neutrophil cells (band cells [BCs] and segmented neutrophil cells [SCs]). Nonneutrophil cells were removed with magnetic-bead–coupled antibodies against CD2, CD3, CD14, CD19, CD56, CD61, glycophorin-A, and CD49d (BCs/SCs) before RNA and protein extraction. Polymorphonuclear neutrophils (PMNs) from peripheral blood depleted with anti-CD49d antibodies were also included. Expression of acute myeloid leukemia 1b (AML-1b), c-myb, GATA-1, and CCAAT/enhancer binding protein γ (C/EBP-γ) was seen primarily in MBs/PMs, and little expression was found in more mature cells. The level of C/EBP-α was constant in the bone marrow–derived cells and decreased in PMNs. C/EBP-ϵ was found primarily in MCs/MMs and was almost absent in more mature cells. Expression of C/EBP-β, C/EBP-δ, and C/EBP-ζ was observed from the MC/MM stage onward, with peak levels in the most mature cells. The amount of PU.1 increased throughout maturation whereas the level of Elf-1 reached a nadir in MCs/MMs The PU.1 coactivator c-jun and c-jun's dimerization partner c-fos were both detectable in MCs/MMs and increased in amount with maturity. CCAAT displacement protein (CDP) was found at comparable levels at all stages of differentiation. This demonstrates a highly individualized expression of the transcription factors, which can form the basis for the heterogeneous expression of granule proteins during granulopoiesis and cell cycle arrest in metamyelocytes.
Introduction
Differentiation of granulocytes (granulopoiesis) takes place in the bone marrow over a period of 10 to 14 days. The first recognizable myeloid precursor is the myeloblast (MB), which differentiates to segmented neutrophils (SCs) through the morphologically distinct stages of promyelocytes (PMs), myelocytes (MCs), metamyelocytes (MMs), and band cells (BCs).1 Granulopoiesis is a complex process, in which a number of transcription factors play critical roles.2,3 This can be appreciated by recognizing that the differentiation stop associated with acute myeloid leukemia in most cases involves translocations or other mutations that disturb the function of a transcription factor.4,5 Further documentation comes from experiments involving ectopic expression of transcription factors in myeloid cell cultures6,7 and targeted disruption of genes encoding transcription factors in mice.8, 9, 10, 11 Experimental evidence has shown that some transcription factors, such as CCAAT/enhancer binding protein α (C/EBP-α) and acute myeloid leukemia 1 (AML-1),8,9 are important during early granulopoiesis whereas others, such as C/EBP-ϵ and CCAAT displacement protein (CDP),10,12 first exert their function in more mature neutrophil precursors. This demonstrates that timing of transcription factor expression and activation is also important for proper neutrophil differentiation.
To date, much of the information on the timed expression of transcription factors during neutrophil differentiation comes from investigation of in vitro–differentiated CD34+ bone marrow cells or myeloid cell lines.6,13, 14, 15, 16 These studies, however, have some inherent shortcomings. First, it is difficult to reproduce the endogenous milieu and stimuli encountered by the neutrophil precursors in the bone marrow, which is a prerequisite for proper granulocytic differentiation.6,14 Second, data have often been obtained from asynchronously differentiating cell cultures, which may obscure the disappearance of an early expressed transcription factor owing to a sustained presence of immature cells in the differentiated cell population.6,15,17 Third, many of the widely used neutrophil cell lines, such as HL60 and NB4 cells,18 lack the ability to express specific- and gelatinase-granule proteins. As the expression of these proteins is transcriptionally regulated,10,19, 20, 21 this strongly suggests that the transcriptional program is corrupted in these cells.
The physiologic role of transcription factors in granulopoiesis has also been analyzed by gene-targeting experiments.8, 9, 10, 11,22,23 Although the hematopoietic cells in these cases encounter the proper microenvironment, the nature of these experiments usually allows only the earliest effect of a knocked-out transcription factor to be observed. Since some transcription factors are required not only for hematopoiesis in general (eg, AML-1b and c-myb)9,11 or commitment of a hematopoietic precursor (eg, C/EBP-α and PU.1)8,22,23 but also for the development of the precursor along the neutrophil lineage, the specific expression pattern of the factor during granulopoiesis may not be observed owing to a lack of definitive hematopoiesis in these mice. Furthermore, the phenotype of the mutation may not always be a direct consequence of the targeted disruption but may instead be a secondary effect caused by the lack of a gene product induced by the transactivator as exemplified by the reduced C/EBP-δ expression in C/EBP-ϵ knockout mice.24 Finally, the resulting phenotype may depend on the design of the targeted disruption,25 as was observed for the 2 PU.1 knockout mouse models.22,23
To gain further insight into the in vivo expression of transcription factors during granulopoiesis, we have analyzed the mRNA and protein profiles of 14 transcription factors in neutrophil precursors from human bone marrow.
Materials and methods
Isolation of neutrophils and their precursors from bone marrow and peripheral blood
Bone marrow samples (15 mL) from healthy volunteers were used for isolation of neutrophil precursors by density centrifugation on a 2-layer Percoll (Amersham Biosciences, Little Chalfont, England) gradient, which resulted in the separation of bone marrow cells into 3 bands containing neutrophil precursors of different maturity: MBs/PMs, MCs/MMs, and BCs/SCs, as described previously.19,26 Contaminating erythrocytes in the BC/SC cell population were removed by 30 seconds of hypotonic lysis. Mature neutrophils from peripheral blood were isolated by centrifugation on Lymphoprep (Nygaard, Oslo, Norway) as described previously.26,27 Remaining erythrocytes were lysed by hypotonic lysis.
Removal of nonneutrophil hematopoietic cells by MACS depletion
Nonneutrophil bone marrow cells were depleted from the 3 populations of neutrophil precursors by incubation with mouse antibodies against human CD2, CD3, CD14, CD19, CD56, CD61, glycophorin-A, and CD49d (only BCs/SCs). Lymphoprep-purified polymorphonuclear neutrophils (PMNs) from peripheral blood were depleted for contaminating eosinophils by incubation with anti-CD49d antibodies. All antibodies were mouse immunoglobulin G (IgG) monoclonals (BD PharMingen, San Diego, CA). The antibodies were incubated with the cells according to the manufacturer's recommendations. Goat anti-mouse IgG microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) were added; following incubation and washing, the cells were layered on a depletion column of the appropriate size (AS or BS depletion columns; Miltenyi Biotech). The column was fixed in a magnet (VarioMACS; Miltenyi Biotech), and the flow-through collected. If isolation of immunopositive cells was required, the column was washed with a higher flow rate and removed from the magnet for elution of the cells. Then, 1 × 106 cells were saved for cytospin preparations and flow cytometric analysis. The remaining cells were used for RNA or protein purification.
Flow cytometric analysis
For flow cytometric analysis, the following phycoerythrin (PE)– and fluorescein isothiocyanate (FITC)–labeled mouse monoclonal antibodies and isotype negative controls were used: CD16-PE, CD19-PE, CD33-PE, CD34-PE, CD56-PE, IgG1-PE, and IgG1-FITC (Becton Dickinson, San Jose, CA); CD49d-PE (BD PharMingen); CD5-PE, CD10-PE, CD61-FITC, and glycophorin-A–PE (DAKO, Glostrup, Denmark); and CD14-PE and IgM-PE (Beckman Coulter, Miami, FL). Cells were incubated with antibody for 15 minutes at room temperature for labeling, washed twice in 0.5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) and fixed in 1% paraformaldehyde in PBS. Flow cytometric analysis was performed with the use of a FACScan (Becton Dickinson), for which settings and compensation were adjusted weekly by means of CaliBRITE Beads (Becton Dickinson). The data were analyzed by means of the CELLQuest and PAINT-A-GATE software (Becton Dickinson).
Staining of eosinophils
Cells resuspended in 1% BSA in PBS were cytocentrifuged onto a glass slide. Following methanol fixation, cells were stained 30 minutes in 0.2% Fast Green (Sigma-Aldrich, St Louis, MO), rinsed in water, and counter-stained in 1% Neutral Red (Sigma-Aldrich).
RNA isolation and Northern blotting
Total RNA was isolated with Trizol (Invitrogen, San Diego, CA). The RNA was ethanol-precipitated and resuspended in 0.1 mM EDTA (ethylenediami-netetraacetic acid). For Northern blotting, 5 μg RNA was run on a 1% agarose gel and transferred to a Hybond-N membrane (Amersham Biosciences) as described.19 Filters were prehybridized for 1 to 2 hours at 42°C in 6 mL ULTRAhyb (Ambion, Austin, TX) and hybridized overnight at 42°C after addition of a further 4 mL containing the 32P-labeled probe and sheared salmon sperm DNA (10 μg/mL). The membranes were washed as described19 and developed by a Fuji BAS2500 PhosphorImager (Tokyo, Japan). Membranes were stripped by boiling in 0.1% sodium dodecyl sulfate (SDS) before rehybridization. Probes used for hybridization were radiolabeled with [α-32P] deoxycytidine triphosphate (dCTP) by means of the Random Primers DNA Labeling System (Invitrogen). Sizes of the hybridizing bands were determined relative to 18S and 28S and found to be in accordance with previous reports (Table 1). For quantitative assessments, the intensities of the transcription factor signals were normalized to the hybridization intensity from a probe against 18S.
Synthesis of cDNA probes for Northern blot hybridization
Target . | GenBank accession no. . | Probe region . | Comment . | Primers . | Transcript size . |
|---|---|---|---|---|---|
| 18S | M10098 | 694-1301 | Structural | 5′-CATTGGAGGGCAAGTCTGG-3′ | 1.9 kb |
| 5′-TTCCTTTAAGTTTCAGCTTTGC-3′ | |||||
| AML-1 | D43968 | 6547-7259 | 3′ UTR | 5′-GGTTTGCTAAATACTGTAGGG-3′ | 6.8 + 7.2 kb |
| 5′-CTTGTCAAACTGTTTATTTGCC-3′ | |||||
| C/EBP-α | Y11525 | 1453-2132 | 3′ UTR | 700-bp EcoRI/HindIII fragment42 | 2.6 kb |
| C/EBP-β | X52560 | 1501-1864 | 3′ UTR | 5′-ATCTATATTTTGCCAACCAACC-3′ | 1.8 kb |
| 5′-AGATTCCCAAAATATACAGACG-3′ | |||||
| C/EBP-δ | S63168 | 953-1335 | 3′ UTR | 5′-CGGGAGAGACTCAGCAACG-3′ | 1.2 kb |
| 5′-GATTTCAAATGCTGCTTTATTC-3′ | |||||
| C/EBP-ϵ | U48865 | 3349-3592 | 3′ UTR + CR | 5′-CCTCCGCAACCTCTTCCG-3′ | 1.4 kb49 |
| 5′-ACAGTGCAACTTTATTCAGCC-3′ | |||||
| C/EBP-γ | U20240 | 569-996 | 3′ UTR + CR | 5′-AAAATCAAATTGCTGACCAAGG-3′ | 1 + 4 kb41 |
| 5′-GTCAGGATTTATTAAGAACATCG-3′ | |||||
| C/EBP-ξ | S40706 | 391-770 | 3′ UTR + CR | 5′-CAAGCACCTCCCAGAGCC-3′ | 0.9 kb |
| 5′-TCATTGGCACTAGTGAGAGG-3′ | |||||
| CASP | L12579 | 1093-1275 | CR | The same probe as for CDP | 3.4 kb59 |
| CDP | M74099 | 1084-1714 | CR | 5′-CCAGGCTGACTATGAAGAGG-3′ | 12 kb59 |
| 5′-GGCGATTTCTGCAGTGTCC-3′ | |||||
| c-fos | K00650 | 2907-3303 | CR | 5′-CTGCCTCTCCTCAATGACC-3′ | 2.6 kb |
| 5′-TGAGCGAGTCAGAGGAAGG-3′ | |||||
| c-jun | J04111 | 2844-3404 | 3′ UTR | 5′-TATTTCTTGTTTGTTTGTTTGGG-3′ | 3.3 kb |
| 5′-AATTCCTGCTTTGAGAATAAGC-3′ | |||||
| CLC | L01664 | 1-598 | CR | Provided by S. Ackerman, University of Illinois, Chicago | 0.9 kb |
| c-myb | M13665 | 2297-2840 | 3′ UTR | 5′-AAGGTACATTTATTGTACCAAACC-3′ | 2.5 + 3.8 kb76 |
| 5′-CAGTTGCAAACACAGGATCC-3′ | |||||
| Elf-1 | M82882 | 3013-3526 | 3′-end | 5′-GTTATGGGTCAAGATCTGCC-3′ | 3.6 kb |
| 5′-TACAAAAATGTTTAGAGGATTCC-3′ | |||||
| FMLP-R | M60626 | 31-551 | CR | 5′-CCAGGAGCAGACAAGATGG-3′ | 1.9 kb |
| 5′-CCAGGTACTGTAGTCACACG-3′ | |||||
| GATA-1 | X17254 | 175-677 | CR | 5′-TGCTCTGGTGTCCTCCACACC-3′ | 1.5 kb |
| 5′-TGGGAGAGGAATAGGCTGCT-3′ | |||||
| Glycophorin A | X08054 | 373-898 | 3′ UTR + CR | 5′-CGATCCTCTTAATTTCTTACGG-3′ | 0.7, 1.0, 1.7 + 2.6 kb77 |
| 5′-CTTCATCTAGGTATTTCAAGCC-3′ | |||||
| Lactoferrin | U07643 | 1864-2018 | CR | 5′-AGACTTTGCGCTGCTGTGCC-3′ | 2.4 kb |
| 5′-CAGTCAGATCCATTTCTCCC-3′ | |||||
| MPO | X04876 | 1683-1868 | CR | 5′-CTTCATGTTCCGCCTGGACA-3′ | 3.2 kb |
| 5′-CGGATCTCATCCACTGCAAT-3′ | |||||
| PU.1 | X52056 | 213-758 | CR | 5′-CCTGGGGAGACAGGCAGC-3′ | 1.4 kb |
| 5′-GAGTCCTGGAGGGAGGCG-3′ | |||||
| β-actin | X00351 | 1028-1719 | 3′ UTR + CR | 5′-CATTGCTCCTCCTGAGCGC-3′ | 1.8 kb |
| 5′-AGGTAAGCCCTGGCTGCC-3′ |
Target . | GenBank accession no. . | Probe region . | Comment . | Primers . | Transcript size . |
|---|---|---|---|---|---|
| 18S | M10098 | 694-1301 | Structural | 5′-CATTGGAGGGCAAGTCTGG-3′ | 1.9 kb |
| 5′-TTCCTTTAAGTTTCAGCTTTGC-3′ | |||||
| AML-1 | D43968 | 6547-7259 | 3′ UTR | 5′-GGTTTGCTAAATACTGTAGGG-3′ | 6.8 + 7.2 kb |
| 5′-CTTGTCAAACTGTTTATTTGCC-3′ | |||||
| C/EBP-α | Y11525 | 1453-2132 | 3′ UTR | 700-bp EcoRI/HindIII fragment42 | 2.6 kb |
| C/EBP-β | X52560 | 1501-1864 | 3′ UTR | 5′-ATCTATATTTTGCCAACCAACC-3′ | 1.8 kb |
| 5′-AGATTCCCAAAATATACAGACG-3′ | |||||
| C/EBP-δ | S63168 | 953-1335 | 3′ UTR | 5′-CGGGAGAGACTCAGCAACG-3′ | 1.2 kb |
| 5′-GATTTCAAATGCTGCTTTATTC-3′ | |||||
| C/EBP-ϵ | U48865 | 3349-3592 | 3′ UTR + CR | 5′-CCTCCGCAACCTCTTCCG-3′ | 1.4 kb49 |
| 5′-ACAGTGCAACTTTATTCAGCC-3′ | |||||
| C/EBP-γ | U20240 | 569-996 | 3′ UTR + CR | 5′-AAAATCAAATTGCTGACCAAGG-3′ | 1 + 4 kb41 |
| 5′-GTCAGGATTTATTAAGAACATCG-3′ | |||||
| C/EBP-ξ | S40706 | 391-770 | 3′ UTR + CR | 5′-CAAGCACCTCCCAGAGCC-3′ | 0.9 kb |
| 5′-TCATTGGCACTAGTGAGAGG-3′ | |||||
| CASP | L12579 | 1093-1275 | CR | The same probe as for CDP | 3.4 kb59 |
| CDP | M74099 | 1084-1714 | CR | 5′-CCAGGCTGACTATGAAGAGG-3′ | 12 kb59 |
| 5′-GGCGATTTCTGCAGTGTCC-3′ | |||||
| c-fos | K00650 | 2907-3303 | CR | 5′-CTGCCTCTCCTCAATGACC-3′ | 2.6 kb |
| 5′-TGAGCGAGTCAGAGGAAGG-3′ | |||||
| c-jun | J04111 | 2844-3404 | 3′ UTR | 5′-TATTTCTTGTTTGTTTGTTTGGG-3′ | 3.3 kb |
| 5′-AATTCCTGCTTTGAGAATAAGC-3′ | |||||
| CLC | L01664 | 1-598 | CR | Provided by S. Ackerman, University of Illinois, Chicago | 0.9 kb |
| c-myb | M13665 | 2297-2840 | 3′ UTR | 5′-AAGGTACATTTATTGTACCAAACC-3′ | 2.5 + 3.8 kb76 |
| 5′-CAGTTGCAAACACAGGATCC-3′ | |||||
| Elf-1 | M82882 | 3013-3526 | 3′-end | 5′-GTTATGGGTCAAGATCTGCC-3′ | 3.6 kb |
| 5′-TACAAAAATGTTTAGAGGATTCC-3′ | |||||
| FMLP-R | M60626 | 31-551 | CR | 5′-CCAGGAGCAGACAAGATGG-3′ | 1.9 kb |
| 5′-CCAGGTACTGTAGTCACACG-3′ | |||||
| GATA-1 | X17254 | 175-677 | CR | 5′-TGCTCTGGTGTCCTCCACACC-3′ | 1.5 kb |
| 5′-TGGGAGAGGAATAGGCTGCT-3′ | |||||
| Glycophorin A | X08054 | 373-898 | 3′ UTR + CR | 5′-CGATCCTCTTAATTTCTTACGG-3′ | 0.7, 1.0, 1.7 + 2.6 kb77 |
| 5′-CTTCATCTAGGTATTTCAAGCC-3′ | |||||
| Lactoferrin | U07643 | 1864-2018 | CR | 5′-AGACTTTGCGCTGCTGTGCC-3′ | 2.4 kb |
| 5′-CAGTCAGATCCATTTCTCCC-3′ | |||||
| MPO | X04876 | 1683-1868 | CR | 5′-CTTCATGTTCCGCCTGGACA-3′ | 3.2 kb |
| 5′-CGGATCTCATCCACTGCAAT-3′ | |||||
| PU.1 | X52056 | 213-758 | CR | 5′-CCTGGGGAGACAGGCAGC-3′ | 1.4 kb |
| 5′-GAGTCCTGGAGGGAGGCG-3′ | |||||
| β-actin | X00351 | 1028-1719 | 3′ UTR + CR | 5′-CATTGCTCCTCCTGAGCGC-3′ | 1.8 kb |
| 5′-AGGTAAGCCCTGGCTGCC-3′ |
Transcript sizes were found in the GenBank database or, where indicated, in publications.
CASP indicates cut alternatively spliced protein; CLC, Charcot-Leyden crystal; fMLP-R, formyl-Met-Leu-Phe receptor; MPO, myeloperoxidase; UTR, untranslated region; and CR, coding region.
Construction of probes for Northern blot
The probes were constructed by polymerase chain reaction (PCR) amplification with the use of a human bone marrow cDNA library (Clontech, Palo Alto, CA) or cDNA from HL60 cells as template (Table 1) and were cloned in pCRII (Invitrogen). Correctness of the inserts was confirmed by sequencing. Inserts were excised and gel-purified before use. The cDNA probes for AML-1 and β-actin were kindly donated by Dr Kim Theilgaard-Mönch, Rigshospitalet (University of Copenhagen, Denmark). The C/EBP-α probe was kindly provided by Dr Daniel Tenen, Harvard Medical School (Boston, MA), and the probe for CLC protein was kindly provided by Dr Steven Ackerman, University of Illinois, Chicago.
Protein isolation and Western blotting
The cells were pretreated with diisopropyl fluorophosphate (DFP) and subsequently isolated by a guadinium-hydrochloride–based method to rapidly inactivate cellular proteases according to the manufacturer's recommendations (Trizol; Invitrogen). Equal amount of protein lysate was applied on 7% to 14% SDS gels and transferred to nitrocellulose membranes (Amersham Biosciences). The source of the antibodies and the reaction conditions are shown in Table 2. The immune complexes were visualized by the enhanced chemiluminiscence (ECL) reaction (Amersham Biosciences). Equal loading was assessed by probing with an antibody against α-tubulin.
Source and reaction conditions of antibodies for Western blot analysis
Target . | Antibody source (antibody) . | Dilution of primary antibody; blocking . | Size of protein . |
|---|---|---|---|
| AML-1 | OR (Ab-2) | 1:40; 5% milk | 53-55 kDa78 |
| c-myb | SC (H-141) | 1:1000; 3% BSA | 75 kDa |
| C/EBP-α | SC (14-AA) | 1:1000; 3% BSA | 42 and 30 kDa |
| C/EBP-β | SC (C-19) | 1:1000; 3% BSA | 43 and 20 kDa48 |
| C/EBP-δ | SC (C-22) | 1:1000; 3% BSA | 33 kDa79 |
| C/EBP-ϵ | SC (C-22) Koeffler and colleagues80 * | 1:1000; 3% BSA | 32, 27, and 14 kDa43,80 |
| C/EBP-γ | Koeffler and colleagues83 † | 1:1000; 5% milk | 18-20 kDa66 |
| C/EBP-ζ | SC (B-3) | 1:1000; 3% BSA | 29 kDa81 |
| CDP | Koeffler and colleagues58 ‡ (α-861) | 1:1000; 2.5% BSA, 5% milk | 160-180 kDa82 |
| c-fos | BD (G54-9.9) | 1:1000; 5% BSA | 55, 57, 60, and 62 kDa |
| c-jun | BD (clone 3) | 1:1000; 3% BSA | 39 kDa |
| PU.1 | SC (T-21) | 1:1000; 3% BSA | 40 kDa |
| Elf-1 | SC (C-20) | 1:1000; 2.5% milk | ≈95 kDa, as a double band |
| α-tubulin | SA (DM 1A) | 1:4500; 3% BSA | 55 kDa |
Target . | Antibody source (antibody) . | Dilution of primary antibody; blocking . | Size of protein . |
|---|---|---|---|
| AML-1 | OR (Ab-2) | 1:40; 5% milk | 53-55 kDa78 |
| c-myb | SC (H-141) | 1:1000; 3% BSA | 75 kDa |
| C/EBP-α | SC (14-AA) | 1:1000; 3% BSA | 42 and 30 kDa |
| C/EBP-β | SC (C-19) | 1:1000; 3% BSA | 43 and 20 kDa48 |
| C/EBP-δ | SC (C-22) | 1:1000; 3% BSA | 33 kDa79 |
| C/EBP-ϵ | SC (C-22) Koeffler and colleagues80 * | 1:1000; 3% BSA | 32, 27, and 14 kDa43,80 |
| C/EBP-γ | Koeffler and colleagues83 † | 1:1000; 5% milk | 18-20 kDa66 |
| C/EBP-ζ | SC (B-3) | 1:1000; 3% BSA | 29 kDa81 |
| CDP | Koeffler and colleagues58 ‡ (α-861) | 1:1000; 2.5% BSA, 5% milk | 160-180 kDa82 |
| c-fos | BD (G54-9.9) | 1:1000; 5% BSA | 55, 57, 60, and 62 kDa |
| c-jun | BD (clone 3) | 1:1000; 3% BSA | 39 kDa |
| PU.1 | SC (T-21) | 1:1000; 3% BSA | 40 kDa |
| Elf-1 | SC (C-20) | 1:1000; 2.5% milk | ≈95 kDa, as a double band |
| α-tubulin | SA (DM 1A) | 1:4500; 3% BSA | 55 kDa |
The membranes were blocked in PBS with 0.5% Tween with the indicated amount of BSA and/or skimmed milk prior to addition of primary antibody. Protein sizes are as reported on the data sheets of the antibodies or, where indicated, in publications.
OR indicates Oncogene Research (Boston, MA); SC, Santa Cruz Biotechnology (Santa Cruz, CA), BD, Becton Dickinson; and SA, Sigma-Aldrich.
University of California—Los Angeles (UCLA) School of Medicine.
National Cancer Institute (NCI)—Frederick, MD.
McGill University, Montreal, QC, Canada.
Results
Isolation of neutrophil precursor populations
Previously, our laboratory developed a method for purification of neutrophil precursors from human bone marrow by Percoll density centrifugation to study the expression of neutrophil-specific granule proteins.19,26 By this method, the neutrophil precursors were separated into 3 cell populations enriched in MBs/PMs, MCs/MMs, or BCs/SCs.19,26 For the investigation of neutrophil transcription factors it was, however, important that contamination with nonneutrophil cells be minimized since some transcription factors are shared among different hematopoietic lineages.9,11,28 An immuno-magnetic isolation step was therefore applied to the cells purifed on Percoll density gradients. Since no neutrophil-specific membrane marker that is expressed at all stages of granulopoiesis exists, we decided to use a depletion protocol with antibodies directed against plasma membrane proteins present on the erythroid (glycophorin-A), B-lymphoid (CD19), T-lymphoid (CD2 and CD3), monocytic (CD14), megakaryocytic (CD61), and natural killer (NK) cell (CD56) lineages. The membrane marker CD49d is expressed on mature eosinophils and on neutrophil precursors up to the metamyelocyte stage.29 Antibodies against CD49d were therefore included when purifying the BC/SC population to remove contaminating eosinophils. Neutrophil granulocytes from peripheral blood (PMNs) were also included in the study as they represent the most mature form of this cell. The PMNs were isolated by Lymphoprep density centrifugation and further purified by removal of contaminating eosinophils on a magnetically activated cell sorter (MACS) column with the use of anti-CD49d antibodies.
By inclusion of the immunobead purification step, the mean percentage of contaminating cells was reduced from 68%, 22%, 16%, and 4.0% to 7.7%, 2.0%, 2.2%, and 0.7% for the MBs/PMs, MCs/MMs, BCs/SCs, and PMNs, respectively, as determined by flow cytometric analysis (Figures 1, 2A; Table 3).
Flow cytometric analysis of the 4 neutrophil cell populations before and after MACS purification. (A) MBs/PMs. (B) MCs/MMs. (C) BCs/SCs. (D) PMNs. Separations are from Percoll-separated human bone marrow cells before (Ai,Bi,Ci,Di) and after (Aii,Bii,Cii,Dii) depletion of nonneutrophil cells.
Flow cytometric analysis of the 4 neutrophil cell populations before and after MACS purification. (A) MBs/PMs. (B) MCs/MMs. (C) BCs/SCs. (D) PMNs. Separations are from Percoll-separated human bone marrow cells before (Ai,Bi,Ci,Di) and after (Aii,Bii,Cii,Dii) depletion of nonneutrophil cells.
Cytospin and Northern blot analysis of the neutrophil cell populations before and after MACS purification. (A) Cytospin preparations of cells from the 3 Percoll-separated bone marrow populations (MBs/PMs, MCs/MMs, and BCs/SCs) and peripheral blood granulocytes (PMNs) before and after MACS depletion of nonneutrophil cells. Cells were stained with May-Grünwald-Giemsa. (B) Cytospins of granulocytic populations from peripheral blood stained with May-Grünwald-Giemsa (MGG) and Fast Green/Nuclear Red. (Top) Total granulocytes isolated from peripheral blood by means of Lymphoprep. Neutrophils are dominant, but eosinophils are visible (characterized by their bilobed nucleus, intense red granules in MGG staining, and green granules in Fast Green staining,84 respectively). (Middle) Total granulocytes depleted for CD49d-expressing cells. (Bottom) The CD49d+ fraction from total granulocytes highly enriched in eosinophils. (C) Northern blot of RNA from the 3 granulocytic populations. The blot was hybridized to C/EBP-ϵ, CLC, and 18S.
Cytospin and Northern blot analysis of the neutrophil cell populations before and after MACS purification. (A) Cytospin preparations of cells from the 3 Percoll-separated bone marrow populations (MBs/PMs, MCs/MMs, and BCs/SCs) and peripheral blood granulocytes (PMNs) before and after MACS depletion of nonneutrophil cells. Cells were stained with May-Grünwald-Giemsa. (B) Cytospins of granulocytic populations from peripheral blood stained with May-Grünwald-Giemsa (MGG) and Fast Green/Nuclear Red. (Top) Total granulocytes isolated from peripheral blood by means of Lymphoprep. Neutrophils are dominant, but eosinophils are visible (characterized by their bilobed nucleus, intense red granules in MGG staining, and green granules in Fast Green staining,84 respectively). (Middle) Total granulocytes depleted for CD49d-expressing cells. (Bottom) The CD49d+ fraction from total granulocytes highly enriched in eosinophils. (C) Northern blot of RNA from the 3 granulocytic populations. The blot was hybridized to C/EBP-ϵ, CLC, and 18S.
Flow cytometric determination of contaminating cells in the neutrophil cell preparations
Cell type* . | MBs/PMs, % . | MBs/PMs depleted, % . | MCs/MMs, % . | MCs/MMs depleted, % . | BCs/SCs, % . | BCs/SCs depleted, % . | PMNs, % . | PMNs depleted, % . |
|---|---|---|---|---|---|---|---|---|
| B cells | 6.5 (3.8-8.1) | 0.53 (0-1.2) | 1.3 (0-3.4) | 0.11 (0-0.4) | 0.17 (0-0.5) | 0.19 (0-1.1) | ND | ND |
| T cells | 20 (13-28) | 1.3 (0-3.3) | 5.6 (0-10) | 0.09 (0-0.5) | 0.46 (0-1.0) | 0.02 (0-0.1) | ND | ND |
| NK cells | 6.3 (1.6-10) | 3.8 (0-10) | 0.43 (0-1.6) | 0.23 (0-1.1) | 0.01 (0-0.3) | 0.14 (0-0.9) | ND | ND |
| Erythroid cells | 12 (6.8-21) | 0.01 (0-0.06) | 12 (8.4-13) | 0.28 (0-1.1) | 5.9 (0-17) | 0.34 (0-0.8) | ND | ND |
| Monocytes | 14 (8.6-19) | 1.3 (0.6-2.2) | 1.6 (0.1-2.5) | 0.91 (0.5-2.3) | 1.3 (0.5-1.9) | 0.40 (0-1.4) | ND | ND |
| Megakaryocytes | 9.3 (4.5-21) | 0.74 (0-2.4) | 1.4 (0.6-2.7) | 0.39 (0-0.8) | 2.1 (0-3.9) | 0.52 (0-1.3) | ND | ND |
| Eosinophils | ND | ND | ND | ND | 5.7 (3.1-8.7) | 0.57 (0.1-2.0) | 4.0 (2.6-6.7) | 0.69 (0-1.5) |
| Total contamination | 68 (55-75) | 7.7 (3.2-13) | 22 (11-30) | 2.0 (1.0-3.4) | 16 (9.5-24) | 2.2 (0.4-6.6) | 4.0 (2.6-6.7) | 0.7 (0-1.5) |
Cell type* . | MBs/PMs, % . | MBs/PMs depleted, % . | MCs/MMs, % . | MCs/MMs depleted, % . | BCs/SCs, % . | BCs/SCs depleted, % . | PMNs, % . | PMNs depleted, % . |
|---|---|---|---|---|---|---|---|---|
| B cells | 6.5 (3.8-8.1) | 0.53 (0-1.2) | 1.3 (0-3.4) | 0.11 (0-0.4) | 0.17 (0-0.5) | 0.19 (0-1.1) | ND | ND |
| T cells | 20 (13-28) | 1.3 (0-3.3) | 5.6 (0-10) | 0.09 (0-0.5) | 0.46 (0-1.0) | 0.02 (0-0.1) | ND | ND |
| NK cells | 6.3 (1.6-10) | 3.8 (0-10) | 0.43 (0-1.6) | 0.23 (0-1.1) | 0.01 (0-0.3) | 0.14 (0-0.9) | ND | ND |
| Erythroid cells | 12 (6.8-21) | 0.01 (0-0.06) | 12 (8.4-13) | 0.28 (0-1.1) | 5.9 (0-17) | 0.34 (0-0.8) | ND | ND |
| Monocytes | 14 (8.6-19) | 1.3 (0.6-2.2) | 1.6 (0.1-2.5) | 0.91 (0.5-2.3) | 1.3 (0.5-1.9) | 0.40 (0-1.4) | ND | ND |
| Megakaryocytes | 9.3 (4.5-21) | 0.74 (0-2.4) | 1.4 (0.6-2.7) | 0.39 (0-0.8) | 2.1 (0-3.9) | 0.52 (0-1.3) | ND | ND |
| Eosinophils | ND | ND | ND | ND | 5.7 (3.1-8.7) | 0.57 (0.1-2.0) | 4.0 (2.6-6.7) | 0.69 (0-1.5) |
| Total contamination | 68 (55-75) | 7.7 (3.2-13) | 22 (11-30) | 2.0 (1.0-3.4) | 16 (9.5-24) | 2.2 (0.4-6.6) | 4.0 (2.6-6.7) | 0.7 (0-1.5) |
Data are presented as mean (variation).
ND indicates not determined.
Cell types include precursors.
A differential count of the 3 neutrophil precursor cell populations showed a distribution profile similar to that reported previously,19,26 only demonstrating a slightly higher percentage of band cells in the MC/MM fraction in this study (Table 4). To further ensure that the distribution profile of the neutrophil precursors was not biased by the MACS purification step, cells from the 3 bone marrow populations and PMNs were analyzed for the presence of granule protein mRNAs known to be found specifically in MBs/PMs (myeloperoxidase); MCs/MMs (lactoferrin); and BCs/SCs and PMNs (the fMLP-receptor).19,26,30 As shown in Figure 3A-B, the distribution profile of the granule markers was as expected and in accordance with previously published distribution profiles.19,26,30
Distribution of neutrophil precursors in the 3 Percoll-separated and MACS-depleted bone marrow populations
Population . | Myeloblasts, % . | Promyelocytes, % . | Myelocytes, % . | Metamyelocytes, % . | Bands, % . | Segmented neutrophils, % . |
|---|---|---|---|---|---|---|
| MBs/PMs | 3.0 | 90.4 | 1.5 | 0.5 | 2.0 | 2.6 |
| MCs/MMs | 0 | 1.0 | 26.7 | 39.2 | 28.1 | 5.0 |
| BCs/SCs | 0 | 0 | 0 | 1.5 | 23.5 | 75.0 |
Population . | Myeloblasts, % . | Promyelocytes, % . | Myelocytes, % . | Metamyelocytes, % . | Bands, % . | Segmented neutrophils, % . |
|---|---|---|---|---|---|---|
| MBs/PMs | 3.0 | 90.4 | 1.5 | 0.5 | 2.0 | 2.6 |
| MCs/MMs | 0 | 1.0 | 26.7 | 39.2 | 28.1 | 5.0 |
| BCs/SCs | 0 | 0 | 0 | 1.5 | 23.5 | 75.0 |
Northern blot analysis of the 4 neutrophil cell populations after MACS purification. (A) Northern blot of total RNA from the 4 MACS-depleted cell populations. The blot was hybridized with probes against the granule proteins MPO, lactoferrin, and fMLP-R, which are expressed specifically in MBs/PMs, MCs/MMs, and BCs/SCs and PMNs, respectively. (B) Schematic representation of the hybridization intensities from panel A. For each probe, the cell population showing maximal expression is given the value 1; the expression levels in the other cell populations are shown relative to this. All expression levels are normalized to the 18S level. (C) Relative hybridization intensities of 18S and β-actin in the 4 cell populations. Data are based on 12 (18S) or 17 (β-actin) filters blotted with 5 μg RNA from each cell population. For each hybridization, the cell population with the strongest signal was given the value 1, and the other signal intensities were related to this.
Northern blot analysis of the 4 neutrophil cell populations after MACS purification. (A) Northern blot of total RNA from the 4 MACS-depleted cell populations. The blot was hybridized with probes against the granule proteins MPO, lactoferrin, and fMLP-R, which are expressed specifically in MBs/PMs, MCs/MMs, and BCs/SCs and PMNs, respectively. (B) Schematic representation of the hybridization intensities from panel A. For each probe, the cell population showing maximal expression is given the value 1; the expression levels in the other cell populations are shown relative to this. All expression levels are normalized to the 18S level. (C) Relative hybridization intensities of 18S and β-actin in the 4 cell populations. Data are based on 12 (18S) or 17 (β-actin) filters blotted with 5 μg RNA from each cell population. For each hybridization, the cell population with the strongest signal was given the value 1, and the other signal intensities were related to this.
To normalize transcript levels in Northern blots, hybridization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, or 18S transcripts is usually performed. Previously, we found that the level of GAPDH mRNA decreases with neutrophil maturation, and we therefore examined β-actin and 18S RNA levels to determine whether they were a better choice. Relative hybridization intensities of 12 (18S) and 17 (β-actin) Northern filters, each containing 5 μg total RNA from the 4 neutrophil cell populations, are shown in Figure 3C. Levels of β-actin mRNA were approximately 45% lower in MBs/PMs and PMNs than in the 2 other cell populations, whereas 18S appeared to be more evenly expressed, demonstrating, at most, 25% difference between any 2 cell populations. For this reason, we chose 18S for normalization of our transcripts. All data presented are the combined result of 3 hybridizations, each normalized to 18S. The distribution of transcription factors was also examined by Western blotting. In this case, equal loading was assured by probing with an antibody against α-tubulin.
When analyzing transcript levels, one should bear in mind that only the relative hybridization intensities of one probe with the RNA on a single filter can be compared. Comparison of hybridization levels of one transcript with another (eg, PU.1 with C/EBP-α) is not possible, as a stronger signal for one of the transcripts does not necessarily reflect a higher level of that particular mRNA population, but may instead reflect differences in the size and labeling efficiency of the probe used. The same considerations apply for immunoblots where the immune complexes are visualized by ECL.
Transcript profiles for c-myb, AML-1, CDP, and GATA-1
Studies have demonstrated that c-myb, AML-1, CDP, and GATA-1 are involved in neutrophil gene regulation prior to the promyelocyte-myelocyte transition. AML-1 and c-myb are required for hematopoietic development9,11 and expression of early neutrophil markers such as the azurophil granule proteins MPO and elastase.31, 32, 33 GATA-1 is important for erythroid and eosinophil development but can also be found in early myeloid precursors.34,35 CDP is a transcriptional repressor that blocks expression of the genes encoding specific granule proteins and glycoprotein 91phox (gp91phox) in MBs and PMs.12
A similar distribution profile was found for the AML1b (approximately 7 kilobase [kb]) and AML1c (approximately 6.5 kb) transcripts, which arise from the AML1 gene by use of 2 different promoters,36 with highest mRNA levels in MBs/PMs and lower levels in more mature cells (Figure 4A-B). Also for c-myb, transcripts of 2 different sizes were observed (Figure 4A). The 3.8-kb mRNA encodes a transcriptional activator, and the 2.5-kb mRNA an inhibitor.37,38 Both splice products were abundant in the 2 most immature cell populations and almost undetectable from the band cell stage onward. The amount of the largest c-myb transcript (3.8 kb) was 3- to 4-fold larger in the MBs/PMs relative to the MCs/MMs, whereas the opposite was the case for the 2.5-kb c-myb transcript, which increased 3-fold in the MCs/MMs compared with the MB/PM population. In contrast, a strong hybridization signal was measured for CDP mRNA in all 4 cell populations and demonstrated no significant change in the transcript level with maturity (Figure 4A-B). The approximately 3.4-kb alternatively spliced transcript encoding the protein CASP, which lacks the DNA-binding domains of CDP,39 is most prominent in the MBs/PMs and decreases with maturity of the neutrophil granulocyte. Transcripts for GATA-1 were detected in MBs/PMs, but not in more mature granulocytic cells. The signal was not due to contaminating erythroid cells, which strongly express GATA-1,34 as no hybridization to the erythroid-specific transcript glycophorin-A was observed in the granulocytic precursors, whereas RNA from bone marrow cells depleted of neutrophil cells (but still containing erythrocytic precursors) hybridized strongly (Figure 4A-B). Contamination of MBs/PMs by eosinophils, which also express GATA-1 throughout differentiation,35 cannot be ruled out. The nondetection of GATA-1 in the other cell populations, however, demonstrates that the more mature neutrophilic cells are uncontaminated by eosinophils.
Northern and Western blot of AML-1, c-myb, CDP, CASP, GATA-1, and glycophorin-A. (A) Northern blot of total RNA from mature neutrophils and the 3 MACS-depleted populations of neutrophil precursors. The blot was hybridized with probes against AML-1, c-myb, CDP, GATA-1, and glycophorin-A. CASP is a splice variant of CDP. RNA from erythrocytes (ERY) was included as a positive control for glycophorin-A and GATA-1. (B) Schematic representation of the hybridization intensities from panel A. For each probe, the cell population showing maximal expression is given the value 1; the expression levels in the other cell populations are shown relative to this. All expression levels are mean values of 18S-normalized transcript levels from 3 different subjects. Standard deviations (SDs) are shown as error bars. (C) Western blot of AML-1, c-myb, and CDP. Equal loading was assessed by probing with an antibody against α-tubulin. The arrows indicate the bands of the target proteins.
Northern and Western blot of AML-1, c-myb, CDP, CASP, GATA-1, and glycophorin-A. (A) Northern blot of total RNA from mature neutrophils and the 3 MACS-depleted populations of neutrophil precursors. The blot was hybridized with probes against AML-1, c-myb, CDP, GATA-1, and glycophorin-A. CASP is a splice variant of CDP. RNA from erythrocytes (ERY) was included as a positive control for glycophorin-A and GATA-1. (B) Schematic representation of the hybridization intensities from panel A. For each probe, the cell population showing maximal expression is given the value 1; the expression levels in the other cell populations are shown relative to this. All expression levels are mean values of 18S-normalized transcript levels from 3 different subjects. Standard deviations (SDs) are shown as error bars. (C) Western blot of AML-1, c-myb, and CDP. Equal loading was assessed by probing with an antibody against α-tubulin. The arrows indicate the bands of the target proteins.
Transcript profiles for the family of C/EBP transcription factors
Members of the C/EBP transcription factor family are required for proper neutrophil differentiation. Since homodimerization and heterodimerization of different C/EBP family members as well as functional substitution of one C/EBP dimer with another can occur,40 we decided to examine the transcript profile of the entire C/EBP family (ie, C/EBP-α, -β, -δ, -ϵ, -γ, and -ζ).40 Owing to high homology in the coding region, C/EBP probes were designed toward the 3′-region of the mRNA. The specificity of the probes was confirmed by a BLAST search (http://www.ncbi.nlm.nih.gov/blast/), which demonstrated that cross-reaction with other genes should not be possible. The amount of C/EBP-α transcript was highest in MBs/PMs and then declined to a 50% lower level in the more mature cells (Figure 5A-B). The 2 transcripts for C/EBP-γ, which probably encode the same protein,41 had identical distribution profiles with strong expression in MBs/PMs and barely detectable levels from myelocytes onward (Figure 5A-B). The peak level of the C/EBP-ϵ mRNA was found in MCs/MMs, with a considerably lower level in the other cell populations (Figure 5A-B). Transcripts for C/EBP-β, C/EBP-δ, and C/EBP-ζ were only abundant in the most mature neutrophil precursors of the bone marrow and in peripheral blood granulocytes (Figure 5A-B). A 5000-nucleotide (nt) band was observed following prolonged exposure of the C/EBP-β blot. The distribution profile of this band was similar to that of C/EBP-α (data not shown).
Northern and Western blot of C/EBP-α,-β,-δ,-ϵ,-γ, and -ζ. (A) Northern blot of total RNA from mature neutrophils and the 3 MACS-depleted populations of neutrophil precursors. The blot was hybridized with probes against C/EBP-α, -β, -δ, -ϵ,-γ, and -ζ. (B) Schematic representation of the hybridization intensities from panel A as in Figure 4. (C) Western blot of C/EBP-α,-β,-δ,-ϵ,-γ, and -ζ. Equal loading was assessed by probing with an antibody against α-tubulin (not shown). The arrows indicate the bands of the target proteins. The asterisk denotes an unspecific band in the C/EBP-γ blot.
Northern and Western blot of C/EBP-α,-β,-δ,-ϵ,-γ, and -ζ. (A) Northern blot of total RNA from mature neutrophils and the 3 MACS-depleted populations of neutrophil precursors. The blot was hybridized with probes against C/EBP-α, -β, -δ, -ϵ,-γ, and -ζ. (B) Schematic representation of the hybridization intensities from panel A as in Figure 4. (C) Western blot of C/EBP-α,-β,-δ,-ϵ,-γ, and -ζ. Equal loading was assessed by probing with an antibody against α-tubulin (not shown). The arrows indicate the bands of the target proteins. The asterisk denotes an unspecific band in the C/EBP-γ blot.
The 2 C/EBPs examined most extensively with regard to their role in granulocytic differentiation are C/EBP-α and C/EBP-ϵ. The data presented here for C/EBP-α fit those of earlier reports,17,42 whereas the expression profile of C/EBP-ϵ contrasts with previous findings in which an expression of C/EBP-ϵ in peripheral blood granulocytes was demonstrated.15,43 Since the only major cellular contaminant of standard Lymphoprep-isolated granulocytes is eosinophils, we examined whether the previously reported C/EBP-ϵ expression in the PMN fraction could result from these cells. As shown in Figure 2B-C, this was indeed the case: RNA from CD49+ cells (ie, eosinophils) hybridized strongly to the C/EBP-ϵ probe, whereas no C/EBP-ϵ transcript was detected in the CD49– cells (ie, neutrophils). The presence of eosinophils only in the CD49+ and PMN cell populations, and not in the CD49– cells, was confirmed by the strong expression of the eosinophil-specific transcript CLC only in the former cell populations.44
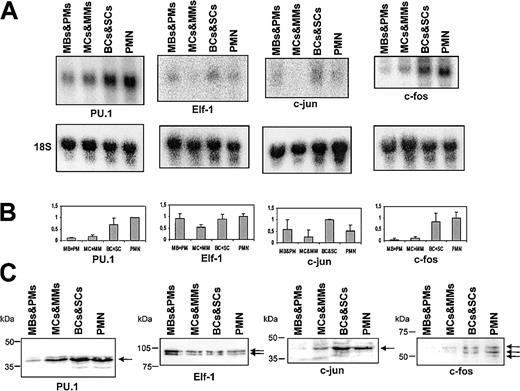
Transcript profiles for PU.1, Elf-1, c-jun, and c-fos
The ets factors PU.1 and Elf-1 are both found in neutrophils, and a requirement for PU.1 in early granulopoiesis has been demonstrated.20,22,23 We therefore chose to examine the expression profile of these 2 transcription factors. As c-jun acts as a coactivator of PU.1 in monocytic differentiation,45 we decided to include this factor, as well as c-fos, a well-documented dimerization partner of c-jun.46 PU.1 transcript was found at all stages of granulopoiesis and increased gradually with maturity, reaching the highest level in peripheral blood PMNs (Figure 6A-B). The transcript profile of Elf-1 was more complex, with a lower mRNA level in MCs/MMs compared with the other cell populations. A similar pattern was observed for the c-jun mRNA, where the transcript level also diminishes at the MC/MM stage. The c-fos transcript, on the other hand, was prominent only in BCs/SCs and peripheral blood granulocytes (Figure 6A-B).
Northern and Western blot of PU.1, Elf-1, c-jun, and c-fos. (A) Northern blot of total RNA from mature neutrophils and the 3 MACS-depleted populations of neutrophil precursors. The blot was hybridized with probes against PU.1, Elf-1, c-jun, and c-fos (B) Schematic representation of the hybridization intensities from panel A as in Figure 4. (C) Western blot of PU.1, Elf-1, c-jun, and c-fos. Equal loading was assessed by probing with an antibody against α-tubulin (not shown). The arrows indicate the bands of the target proteins.
Northern and Western blot of PU.1, Elf-1, c-jun, and c-fos. (A) Northern blot of total RNA from mature neutrophils and the 3 MACS-depleted populations of neutrophil precursors. The blot was hybridized with probes against PU.1, Elf-1, c-jun, and c-fos (B) Schematic representation of the hybridization intensities from panel A as in Figure 4. (C) Western blot of PU.1, Elf-1, c-jun, and c-fos. Equal loading was assessed by probing with an antibody against α-tubulin (not shown). The arrows indicate the bands of the target proteins.
Protein profiles of transcription factors
To examine whether the protein profiles matched the mRNA profiles for the 14 transcription factors, we also performed Western blot analysis. The protein profiles of AML-1, PU.1, Elf-1, c-jun, c-fos, CDP, C/EBP-γ, and C/EBP-δ were very similar to their mRNA profiles (Figures 4, 5, 6). For c-myb, only the large (75-kDa) activator form37 was observed, indicating either that the 2.5-kb mRNA also encodes the 75-kDa activator or that the concentration of the inhibitory form is below the detection limit of our antibody (Figure 4C). Although some mRNA for c-myb is found in MCs/MMs, we were able to detect a strong c-myb signal only by Western blotting in MBs/PMs. We were unable to detect CASP with our CDP antibody as the antibody was directed against a part of CDP that is not contained in CASP.
Both the 42- and 30-kDa isoforms of C/EBP-α, representing the transactivation-competent full-length and the N-truncated transdominant repressor forms of the protein, respectively,47 were detected (Figure 5C). The larger isoform was most prominent in all cell populations, indicating that the transactivating competence of C/EBP-α was maintained thoughout granulopoiesis. Two forms of C/EBP-β have been reported in liver cells (a 43-kDa activator and a 20-kDa form that attenuates transcriptional activation of the 43-kDa isoform).48 Our antibody should detect both proteins, but only the 43-kDa type was seen, indicating that only the transactiving type of C/EBP-β is present in the neutrophil precursors. Three isoforms of C/EBP-ϵ, of 32, 27, and 14 kDa, were seen in the neutrophil precursors. As for the mRNA, the vast majority of C/EBP-ϵ protein was found in MCs/MMs (Figure 5C). The 32-kDa transactivation-competent form was the most prominent of the 3 C/EBP-ϵ isoforms, and the 27- and 14-kDa forms, which lack transactivating potential, constituted only a very minor portion of the C/EBP-ϵ proteins.43,49 The 14-kDa form was seen only following prolonged exposure. The same expression pattern of C/EBP-ϵ was found with another C/EBP-ϵ antibody (Table 2).
Discussion
It is well established that a finely tuned timing of transcription factor expression is pivotal for correct neutrophil differentiation. Expression of the receptors for granulocyte colony-stimulating factor (G-CSF) and interleukin 6 (IL-6),50 the azurophil and specific granule proteins,10,20,31, 32, 33 and transcriptional regulators such as PU.1, C/EBP-δ, and C/EBP-ϵ7,24,51 is governed by the emergence and/or disappearance of key transcription factors. Current knowledge about the expression pattern of transcription factors during granulopoiesis has been obtained primarily from work with neutrophil cell lines, bone marrow–derived CD34+ cells, and murine knockout models. Although much information can be gained from such experiments, the data may not always be a true representation of the in vivo situation owing to the previously mentioned limitations of these model systems. For this reason, we decided to examine the in vivo profile of transcription factors during granulopoiesis in 4 distinct cell populations of different maturity from human bone marrow and peripheral blood.
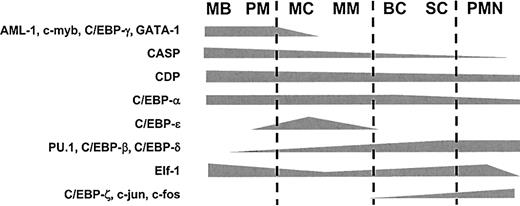
It is possible to obtain a finely tuned transcription factor profile in relation to neutrophil maturation on the basis of the mRNA and protein levels in the 4 neutrophil cell populations if the following assumptions are fulfilled: First, it is assumed that the relative level of 18S mRNA is constant in the cells in such a way that it can be used to normalize the levels of mRNA for the transcription factors (Figure 3C). Second, if the level of a specific mRNA is low in one cell population and increases in the next cell population, which contains more mature cells, then it is assumed that the low mRNA level in the more immature cells is not due to a uniformly low level in all the cells, but instead to a high level in the most mature cells of this cell population, resulting in a gradual increase in the mRNA level. The alternative interpretation would be that uniform transcript levels exist in all cells of the population, despite the fact that these cover a span of maturation artificially sampled into the same population by the limitations set by the density of the separating Percoll medium,52 and that steep changes in the mRNA level occur at the transition between the different cell populations. Third, although the presence of a particular transcript does not necessarily imply the synthesis of the concurrent protein and, conversely, the disappearance of a given mRNA does not always mean that the protein it encodes also disappears, coexpression of an mRNA and its cognate protein is usually observed for cytosolic proteins.53 Our protein expression data demonstrate that this indeed is the case for the transcription factors investigated here. This is in contrast to granular proteins such as MPO and lactoferrin, which are stored in granules (and thus protected from degradation) until exocytosed.30 In this case, the protein can be detected long after the transcript has disappeared.19,26 If the assumption is made that gradual changes of mRNA and protein levels occur, then the hypothetical distribution profile depicted in Figure 7 can be made. Although alternative splicing and translation, as well as posttranslational modifications such as phosphorylation, acetylation, and proteolytic processing, may also influence the activity of a transcriptional regulator and thus have to be taken into consideration, we believe that the in vivo expression pattern of the 14 transcription factors presented here can to a large extent explain the temporal regulation of stage-specifically expressed genes during granulopoiesis.
Hypothetical distribution scheme of neutrophil transcription factors during granulopoiesis. Theoretical distribution scheme of the transcription factors examined in this work based on the data presented in Figures 4, 5, and 6.
In accordance with previous reports,31,54 AML-1 and c-myb were found to be strongly expressed at the early stages of neutrophil differentiation, which fits with the requirement of both these transcription factors for the expression of azurophil granule proteins such as MPO and elastase.31, 32, 33 Down-regulation of c-myb after the myelocyte stage was anticipated since c-myb blocks differentiation and induces proliferation and was therefore expected to disappear at the stage of differentiation where cell cycle arrest and initiation of terminal neutrophil differentiation takes place.1 The same consideration is likely to apply to AML-1, as it also stimulates proliferation (in part by repressing transcription of the cell cycle inhibitor p21Cip1).55
The level of CDP remained almost unaltered during neutrophil differentiation, which was surprising since CDP activity was expected to disappear at the myelocyte stage where expression of specific granule proteins is initiated. The repressive effect of CDP in granulopoiesis has been demonstrated in HL60 cells and murine 32Dcl3 cells where transcription of the genes encoding gp91phox56 and specific granule proteins was inhibited by a continued expression of CDP—both by direct inhibition of the genes12,57 and indirectly by inhibiting the synthesis of C/EBP-ϵ,51 which is essential for specific granule protein gene expression.10,21 Recently, it has been shown that CDP protein is synthesized in HL60 cells during the entire process of granulocytic differentiation. However, the binding capability of CDP disappears in the more mature cells owing to phosphorylation of the protein.58 A similar inactivation of CDP may occur in myelocytes in vivo. This suggests that the observed repression of specific granule gene activity in cell cultures that overexpress CDP is due to an overload of the phosphorylating capacity of these cells rather than to a continued CDP synthesis. No function has been ascribed to CASP, but it has been speculated that it may regulate CDP activity since CDP and CASP are able to heterodimerize.39,59
C/EBP-α is required for granulopoiesis, as demonstrated by the selective block in neutrophil differentiation in mice with a targeted disruption of the C/EBP-α gene8 and the induction of terminal granulocytic differentiation in 32Dcl3 cells in response to induced expression of C/EBP-α.7 C/EBP-α is also needed for gene expression of both the early neutrophil markers MPO and elastase33,60 and of the receptors for IL-6 and G-CSF, which are also expressed at later stages of neutrophil development.7,50 In agreement with these requirements, C/EBP-α is found throughout neutrophil differentiation. A similar expression pattern for C/EBP-α has been observed for HL60 cells and NB4 cells induced to granulocytic differentiation by retinoic acid.42 This is in contrast to the transcript levels found during differentiation of adipocytes and hepatocytes, where a steep increase in the C/EBP-α mRNA level is observed when proliferation ceases and terminal differentiation is initiated.61 The proposed antiproliferative effect of C/EBP-α seen in the latter cell types62 is therefore not likely to be relevant in neutrophils.
The levels of both C/EBP-β, C/EBP-δ, and C/EBP-ζ increased dramatically at the stage where proliferation ceases in neutrophils (ie, metamyelocytes1 ). Since in other cell systems these proteins are associated with termination of proliferation,63,64 a role for these C/EBPs (rather than C/EBP-α) in neutrophil cell cycle arrest can be envisaged. It is, however, also possible that C/EBP-β, C/EBP-δ, and/or C/EBP-ζ are required for transcriptional initiation of late neutrophil markers such as gelatinase, fMLP-receptor, CD35, and Toll-like receptor 4 (TLR4).19,26,65 C/EBP-γ lacks a transactivating domain and can inhibit the transactivating potential of other C/EBPs.40,66 A high expression of C/EBP-γ has been associated with proliferation although the mechanism by which it governs this is unknown.66,67 The strong expression of C/EBP-γ in only the highly proliferative immature neutrophil precursors supports such a function also in granulopoiesis.
Previous publications have demonstrated peak levels of C/EBP-ϵ mRNA in promyelocyte and late myeloblast-like cell lines and a sustained expression in PMNs from peripheral blood.15,43 This is in contrast to the findings in this report in which the peak C/EBP-ϵ level is found in MCs/MMs. Our data, however, fit the phenotype of C/EBP-ϵ knockout mice10 and patients with specific granule deficiency,21 where the earliest steps of neutrophil differentiation are unaffected whereas expression of specific granules at the myelocytic stage is completely abolished owing to the lack of C/EBP-ϵ. The peak expression of C/EBP-ϵ in MCs/MMs and its disappearance in more mature cells could thus explain the confined expression of the specific granule genes in myelocytes and metamyelocytes.19 The disparity between the C/EBP-ϵ mRNA profiles in bone marrow–derived neutrophil precursors and in cell cultures may be due to an erroneous induction of C/EBP-ϵ transcription by retinoic acid14 or G-CSF6 as both these substances are commonly added to the growth media to induce neutrophil differentiation of cell cultures. Our data furthermore indicate that the previously reported presence of C/EBP-ϵ transcript in peripheral blood neutrophils is caused by contaminating eosinophils since a complete lack of measurable C/EBP-ϵ signal by Northern blot analysis was demonstrated following removal of the eosinophils from this cell population (Figure 2B-C). We only detected a C/EBP-ϵ mRNA of approximately 1.4 kb by Northern blotting and were not able to identify the earlier reported C/EBP-ϵ splice variant at 2.4 kb, even following prolonged exposure of the Northern blot.43 As our probe was designed to recognize the 3′-end of the C/EBP-ϵ mRNA, which is included in all splice variants, our data indicate either that the 2.4 kb-splice product is not made in vivo or that the amount produced is so low that a more sensitive detection method such as reverse transcriptase–PCR (RT-PCR) is required for its identification.
Targeted disruption of PU.1 abolishes or severely impairs the production of neutrophils.22,23 The few neutrophils formed in the PU.1 knockout mice are able to differentiate morphologically to PMNs but fail to express specific granule genes and gp91phox.20 Although mRNAs for azurophil granule proteins were detected in the PU.1 null mice, demonstrating that PU.1 was not essential for their transcription, other experiments have shown that PU.1 is required for optimal gene expression of early neutrophil markers such as MPO, proteinase-3, and elastase33,60,68,69 as well as of very late markers such as TLR4.65,70 Together, these data fit the transcript pattern observed here where PU.1 is found at all stages of neutrophil differentiation. The expression profile of the second ets factor included in this study, Elf-1, was quite different from that of PU.1, demonstrating that ets factors are also expressed in an individual manner. Elf-1 is unable to functionally substitute for PU.1 and has been found to cooperate with PU.1 in the expression of the nicotinamide adenine dinucleotide phosphate (NADPH)–oxidase component gp91phox, indicating an individual regulatory specificity of this transcription factor.71,72
Transcriptional competence of PU.1 during monocytic differentiation is dependent upon c-jun as a non–DNA-binding coactivator.73 The same may apply to neutrophil differentiation, and the expression pattern demonstrated here indicates that c-jun would be available for such a function during late granulopoiesis. Expression of c-fos, which is often found heterodimerized with c-jun in the transcriptional transactivator complex AP-1,46 is also primarily found at the late stages of development and in PMNs from peripheral blood. It is possible that c-fos, through heterodimerization with c-jun, can modulate the transcriptional capacity of c-jun—either by reducing the transactivating potential of PU.1 or by forming a transcriptional active AP-1 complex or by both.73 Since the “active concentration of PU.1” in the cell is critical for its transactivating potential,28 a careful titration of the c-jun/PU.1 level may be required for proper transcriptional regulation by PU.1.
On the basis of the data presented here and in the literature, the following course of events during granulopoiesis can be envisaged: In myeloblasts, low concentrations of PU.1 and high amounts of C/EBP-α favor granulocytic differentiation over monocytic differentiation.42,73 AML-1 and c-myb direct the expression of azurophil granule protein genes such as MPO and NE. Following onset of granulopoiesis, C/EBP-α may stimulate additional PU.1 transcription, which, when initiated, increases further, owing to a positive-feedback mechanism.74 In myelocytes, the decrease in AML-1 and c-myb levels halts expression of azurophil granule protein genes. Furthermore, reduction of the active CDP level causes its repression of the genes for C/EBP-ϵ, gp91phox, and the specific granule proteins to be relieved. C/EBP-ϵ then induces transcription of the specific granule protein genes and of C/EBP-δ.24 In band cells, down-regulation of C/EBP-ϵ causes the cessation of specific granule protein gene transcription. Cell cycle arrest and initiation of terminal neutrophil differentiation in metamyelocytes may be a result of the combined disappearance of the pro-proliferative transcription factors c-myb, AML-1, C/EBP-γ, and CDP (which inhibits p21cip1)75 and the emergence of potential antiproliferative factors such as C/EBP-δ and C/EBP-ζ. At present, it is unknown which transcription factors regulate the timed onset of very late neutrophil markers such as the fMLP-receptor, TLR2, TLR4, and CD35, which are all preferentially expressed in band cells, segmented cells, and PMNs. The data presented in this work indicate that C/EBP-β, C/EBP-δ, and C/EBP-ζ may play a role. In conclusion, the transcription factor profiles described here may to a large extent explain the timed gene expression of granule protein genes and cell cycle arrest during granulocytic differentiation.
Prepublished online as Blood First Edition Paper, January 30, 2003; DOI 10.1182/blood-2002-03-0835.
Supported by grants from The Danish Medical Research Council, The Carlsberg Foundation, The Danish Cancer Society, Copenhagen University Hospital, The Danish Medical Association Research Fund, and The Danish Foundation for Cancer Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The expert technical assistance of Inge Kobbernagel and Charlotte Horn is greatly appreciated. Bo Porse, Kim Theilgaard-Mönch, Ole Sorensen, and Mikkel Faurschou are thanked for helpful comments to the manuscript. Dr Steven Ackerman, University of Illinois, is thanked for advice regarding staining of eosinophils.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal