Chronic lymphocytic leukemia (CLL) has a variable clinical course. CD38 expression and IgVH gene mutational status are independent predictors of prognosis, but their relationships and the CD38 cutoff level are unknown. Using cytofluorography, we analyzed CD38 in 148 patients, in 108 of whom we were able to evaluate IgVH mutations, make correlations with disease history, and assess cumulative survival. Three different patient groups were identified by the CD38 expression pattern: a group homogeneously CD38−, a group homogeneously CD38+, and a group characterized by a bimodal profile, because of the concomitant presence of variable proportions of 2 distinct populations, one CD38+ and one CD38−. In CD38 bimodal expression patients the CD38+ subset was significantly more represented in the bone marrow than in the peripheral blood. For IgVH mutations, 11.4% of CD38−, 84.6% of CD38+, and 68.0% of CD38 bimodal expression patients had no mutation. CD38 expression, IgVH mutational status, and traditional prognostic factors were concordant. The progression rate was 12.9% for CD38−, 75.0% for CD38+, and 63.3% for CD38 bimodal expression patients. Only 25.8% of the CD38−patients but 63.3% of the bimodal and 75.0% of CD38+patients were treated. The presence of a CD38+ population, albeit small, correlated with the development of autoimmune manifestations. The CD38− group has not yet reached the median survival, which is 183 months in the CD38+ group and 156 months in the CD38 bimodal expression group, regardless of the size of the CD38+ population. The presence of a distinct CD38+ population within the leukemic clone, rather than a numerical cutoff definition, correlates with IgVH gene mutational status and, irrespective of its size, identifies CLL patients who will have progressive disease.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most frequent leukemia in the Western world. It is characterized by a variable clinical course1 with some patients having an aggressive malignancy and others a slow, nonprogressive disease and a virtually normal life expectancy. The possibility of establishing at diagnosis the most likely outcome of individual patients would provide the basis for a differential management policy. Several biologic risk factors, ranging from serum markers2 to cytogenetics,3 have been used to assess a patient's prognosis. Two parameters have emerged as potent predictors. One is the mutational status of the immunoglobulin variable region (IgVH) genes. Retrospective studies have established that patients whose cells carry somatic mutations with less than 98% sequence homology with the nearest germ line gene have a prognosis significantly better than those presenting with germ line IgVH genes.4-6 Such a difference may be accounted for by the observation that patients with germ line IgVH genes tend to harbor high-risk genomic aberrations and p53 dysfunctions.7-9 The other parameter is the cell's expression of CD38, which has been shown to be an independent marker that correlates with a shorter median survival and more need of treatment.4 10-13
Despite a general agreement on the role of both IgVH gene mutational status and CD38 expression in independently predicting the prognosis, 2 major issues remain controversial. First is the CD38 cutoff level used for risk stratification. For some groups the threshold is 30% positive cells,11,12 whereas for others it is 20%.10,13 This is not a trivial point because numerous cases are in the “gray” zone and are not allowed a precise prognostic categorization. Also a few patients appear to modify their percentage of CD38+ cell expression over time,10,12 thereby crossing the cutoff border. Second, it has been initially suggested,4 but later challenged,12 14 that the expression of CD38 might correlate with IgVH mutational status, meaning that patients with 30% or more CD38+ cells are also the ones who carry germ line IgVH genes, whereas those with less than 30% CD38+ cells have somatically mutated IgVH genes. Thus, it is not established which, if any, relationship exists between the presence or absence of IgVHmutations and the expression of CD38 nor how the 2 markers can be used to define prognosis in individual patients. Finally, it cannot be overlooked that the cytofluorometric evaluation of CD38 expression is an easy and rapid technique used in most laboratories, whereas the detection of IgVH gene mutations can be reliably used only in a minority of sophisticated centers.
We have analyzed the expression of CD38 and the occurrence of IgVH somatic mutations in an unselected cohort of patients and we have correlated these parameters with the patients' disease history. According to the pattern of CD38 expression we have identified 3 groups of patients. CLL cells can be homogeneously negative for CD38 (CD38− group), or they can homogeneously express CD38 at high levels (CD38+ group). A third distinct subset of patients shows a bimodal expression profile of CD38, with the concomitant presence of one population expressing high levels of CD38 and a second population completely negative (CD38 bimodal expression group). Irrespective of the percentage of positive cells the bimodal subset resembles the CD38+ group in terms of risk of disease progression and median survival time. Our data suggest that the pattern of CD38 expression, that is, the presence within the leukemic clone of an albeit small CD38+ cell population, rather than a numerically defined percentage of positive cells, correlates with IgVH genes mutational status and can identify patients who will have a progressive disease. Of interest, the CD38+subset in CD38 bimodal expression patients is significantly more represented in the bone marrow (BM) than in the peripheral blood (PB), suggesting that the cells that are associated with an adverse outcome tend to congregate in the BM environment, a well-known privileged site of disease relapse.
Patients, materials, and methods
Patient population
A group of 148 consecutive, unselected patients with CLL who met the diagnostic criteria of the National Cancer Institute-Working Group (NCI-WG),15 93 men and 55 women with a mean age of 70 years (range, 33-92 years), were studied for the expression of CD38.
Within this cohort, 67 men and 41 women (mean age, 70 years) were selected on the basis of a disease history lasting more than 1 year (median follow-up time, 90 months; range, 16-306 months) to allow the evaluation of clinical progression (Table1). These patients were studied in detail by analyzing the following parameters measured at diagnosis or during the follow-up: lymphocyte and platelet counts; lymphocyte doubling time (LDT); hemoglobin (Hb), immunoglobulin and β2-microglobulin serum levels; liver and spleen enlargement; size of lymph nodes involved; extranodal localizations; autoimmune manifestations; disease stage according to Rai modified criteria16; history of treatment; and progressive or stable disease as defined by the NCI-WG.15
Characteristics of CLL patients according to CD38 expression
| . | All patients . | CD38− . | CD38 bimodal expression . | CD38+ . | P . |
|---|---|---|---|---|---|
| No. of cases (%) | 108 | 62 (57.4) | 30 (27.8) | 16 (14.8) | NA |
| Mean age, y | 70 | 68 | 73 | 70 | NA |
| Sex, M/F | 67/41 | 36/25 | 21/9 | 9/7 | NA |
| Mean duration of follow-up, mo | 90 | 90 | 88 | 95 | NA |
| Risk status | <.0011-153 | ||||
| Low risk*(%) | 78/108 (72.2) | 53/62 (85.5) | 18/30 (60.0) | 7/16 (43.8) | |
| Intermediate risk*(%) | 22/108 (20.4) | 8/62 (12.9) | 10/30 (33.3) | 4/16 (25.0) | |
| High risk*(%) | 8/108 (7.4) | 1/62 (1.6) | 2/30 (6.7) | 5/16 (31.3) | |
| Disease status | <.0011-153 | ||||
| Stable disease (%) | 69/108 (63.9) | 54/62 (87.1) | 11/30 (36.7) | 4/16 (25.0) | |
| Progressive disease (%) | 39/108 (36.1) | 8/62 (12.9) | 19/30 (63.3) | 12/16 (75.0) | |
| Mutation | <.0011-153 | ||||
| Somatically Ig mutated (%) | 49/82 (59.8) | 39/44 (88.6) | 8/25 (32.0) | 2/13 (15.4) | |
| Somatically Ig unmutated (%) | 33/82 (40.2) | 5/44 (11.4) | 17/25 (68.0) | 11/13 (84.6) | |
| Autoimmune manifestations (%) | 20/108 (18.5) | 6/20 (30) | 9/20 (45) | 5/20 (25) | .0291-153 |
| Treatment | <.0011-153 | ||||
| Not requiring treatment (%) | 61/108 (56.5) | 46/62 (74.2) | 11/30 (36.7) | 4/16 (25.0) | |
| Requiring treatment (%) | 47/108 (43.5) | 16/62 (25.8) | 19/30 (63.3) | 12/16 (75.0) | |
| Blood parameters | |||||
| Mean β2-microglobulin count, mg/L† | 2.85 | 2.1 | 3.9 | 4.1 | <.0011-155 |
| Mean Hb count, g/dL | 12.9 | 13.6 | 12.1 | 11.5 | <.0011-155 |
| Mean platelet count, per μL | 167 000 | 188 500 | 140 100 | 134 500 | .0021-155 |
| Mean lymphocyte count, per μL | 36 400 | 25 700 | 46 300 | 58 900 | .0051-155 |
| Mean LDT, %‡ | 94.6 | 33.0 | 123.0 | 282.0 | .0031-155 |
| . | All patients . | CD38− . | CD38 bimodal expression . | CD38+ . | P . |
|---|---|---|---|---|---|
| No. of cases (%) | 108 | 62 (57.4) | 30 (27.8) | 16 (14.8) | NA |
| Mean age, y | 70 | 68 | 73 | 70 | NA |
| Sex, M/F | 67/41 | 36/25 | 21/9 | 9/7 | NA |
| Mean duration of follow-up, mo | 90 | 90 | 88 | 95 | NA |
| Risk status | <.0011-153 | ||||
| Low risk*(%) | 78/108 (72.2) | 53/62 (85.5) | 18/30 (60.0) | 7/16 (43.8) | |
| Intermediate risk*(%) | 22/108 (20.4) | 8/62 (12.9) | 10/30 (33.3) | 4/16 (25.0) | |
| High risk*(%) | 8/108 (7.4) | 1/62 (1.6) | 2/30 (6.7) | 5/16 (31.3) | |
| Disease status | <.0011-153 | ||||
| Stable disease (%) | 69/108 (63.9) | 54/62 (87.1) | 11/30 (36.7) | 4/16 (25.0) | |
| Progressive disease (%) | 39/108 (36.1) | 8/62 (12.9) | 19/30 (63.3) | 12/16 (75.0) | |
| Mutation | <.0011-153 | ||||
| Somatically Ig mutated (%) | 49/82 (59.8) | 39/44 (88.6) | 8/25 (32.0) | 2/13 (15.4) | |
| Somatically Ig unmutated (%) | 33/82 (40.2) | 5/44 (11.4) | 17/25 (68.0) | 11/13 (84.6) | |
| Autoimmune manifestations (%) | 20/108 (18.5) | 6/20 (30) | 9/20 (45) | 5/20 (25) | .0291-153 |
| Treatment | <.0011-153 | ||||
| Not requiring treatment (%) | 61/108 (56.5) | 46/62 (74.2) | 11/30 (36.7) | 4/16 (25.0) | |
| Requiring treatment (%) | 47/108 (43.5) | 16/62 (25.8) | 19/30 (63.3) | 12/16 (75.0) | |
| Blood parameters | |||||
| Mean β2-microglobulin count, mg/L† | 2.85 | 2.1 | 3.9 | 4.1 | <.0011-155 |
| Mean Hb count, g/dL | 12.9 | 13.6 | 12.1 | 11.5 | <.0011-155 |
| Mean platelet count, per μL | 167 000 | 188 500 | 140 100 | 134 500 | .0021-155 |
| Mean lymphocyte count, per μL | 36 400 | 25 700 | 46 300 | 58 900 | .0051-155 |
| Mean LDT, %‡ | 94.6 | 33.0 | 123.0 | 282.0 | .0031-155 |
Rai modified stage at diagnosis.
β2-microglobulin serum count was obtained in 83 patients.
Lymphocyte doubling time (as increasing percent in 6 months).
P for the comparison among CD38−, CD38 bimodal, and CD38+ subgroups was calculated using the χ2 test.
P refers to 1-way ANOVA test.
In 82 of 108 cases the analysis of IgVH genes mutational status was performed on cDNA obtained from the same cell samples analyzed for CD38 expression. In selected patients further in vitro studies aimed at investigating the biologic basis of clinical differences were performed.
Immunophenotypic analysis
Immunophenotypic analysis was performed on fresh blood samples or on cells cryopreserved at diagnosis after having assessed that fresh and cryopreserved cells did not yield different results. In 63 cases the analysis was repeated over time either before or after any treatment. In individual cases the analysis was concomitantly performed on both PB and BM.
The following antibodies were used: allophycocyanin (APC)–conjugated anti-CD19 and phycoerythrin (PE)–labeled anti-CD23 from Caltag Laboratories (San Francisco, CA); fluorescein isothiocyanate (FITC)–labeled anti-CD20, PE-conjugated anti-CD5, and PE-conjugated anti-CD38 from Becton Dickinson (San Jose, CA); FITC-labeled anti-CD79b from Dako (Glostrup, Denmark); and FITC-conjugated FMC-7 (Serotec, Oxford, United Kingdom). FITC-labeled anti-IgM, anti-IgG, anti-IgA heavy chain were obtained from Southern Biotechnology Associates (Birmingham, AL); FITC-conjugated F(ab)2–anti-κ and PE–anti-λ light chain were purchased from Dako.
To determine the percentage of CD38+ events, CLL cells were incubated with FITC-conjugated anti-CD5 (1:20 final dilution), PE-conjugated anti-CD38 (1:50 final dilution), and APC-conjugated anti-CD19 (1:20 final dilution). CD19+CD5+lymphocytes were identified by properly gating on both fluorescent and light scatter parameters. Staining and acquisition conditions were standardized allowing reproducibility of the results, regardless of the actual operator.
For each sample at least 10 000 events were acquired on a FACSCalibur equipped with 488 argon ion laser and 635 red diode laser (Becton Dickinson) and analyzed with the CellQuest software system (Becton Dickinson).
In selected experiments CD38+ and CD38− CLL cells were further stained with a number of different monoclonal antibodies (mAbs) aimed at defining CLL cell differentiation status, aggressive behavior, and homing and migratory capacity. The mAbs against the following markers were used: CD10, CD21, CD23, CD24, CD27, CD31, CD44, CD69, CD71, CD95 (Caltag Laboratories); CD9, CD11c, CD20, CD22, CD70, CD72, CD74, CD75, CD78, CD80, CD81, CD86, CXCR3 (BD-Pharmingen, San Jose, CA); CD25, CD79b, CD103, CD43, CD6, CD11a, CD18 (Dako); CD40, CD49d, CD54, CD100, FMC-7 (Immunotech, Marseille, France); CCR6 (R & D Systems, Minneapolis, MN); CD52 (Serotec); and CD138 (IQ Products, Groningen, Holland).
Determination of mutational status of IgVHgenes
Total RNA was extracted using guanidinium thiocyanate method (TRIzol; Invitrogen Life Technologies, Paisley, United Kingdom). RNA was reverse-transcribed into cDNA using 200 U Superscript II, Rnase H− Reverse Transcriptase (Invitrogen-Life Technologies Italia, San Giuliano Milanese, Italy). After determining the IgVH gene family used by the leukemic cells, the VH gene sequences were determined by amplifying 2.5 μL cDNA by polymerase chain reaction (PCR) using the appropriate sense VH leader primer in combination with the appropriate antisense CH primer, as described previously.17
PCR products were sequenced directly after purification with PCR preps (Promega, Madison, WI) using an automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequences were compared with those present both in the V BASE sequence directory (http://www.mrc-cpe.cam.ac.uk/imt-doc/vbase-home-page.htlm)18and in the IMGT/V-QUEST database (http://imgt.cines.fr).19The sequences with a germ line homology 98% or higher were considered unmutated, and those with a homology less than 98% mutated.4 5
CD40 stimulation of CLL cells
Mononuclear cells from patient PB were isolated by density centrifugation over Lymphoprep (Nycomed Pharma, Oslo, Norway). Cells were washed with phosphate-buffered saline (PBS) solution and resuspended in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS; Life Technologies-Italia), 2 mMl-glutamine, and 15 μg/mL gentamicin. Cells cultured for up to 3 days at a concentration of 1 × 106 cells/mL with or without 0.1 μg/mL soluble recombinant human CD40L plus 1 μg enhancer, according to manufacturer's instructions (Alexis, San Diego, CA), were collected and phenotypically analyzed.
Statistical analysis
CD38 expression on CLL cells has been compared to other parametric series (lymphocyte count, Hb, LDT, β2-microglobulin) using the Spearman correlation test. The Student t test and one-way ANOVA test were applied to determine the statistical significance of mean value difference among series.
Patients were grouped into 3 subsets depending on CD38 expression (negative, bimodal, positive) and in 2 subsets depending on the IgVH gene mutation status (unmutated, mutated). The Fisher exact test (2-tailed) and χ2 test were performed to determine the difference between the observed proportions of events among patients' subsets (nonparametric analysis). The events examined were the stage of disease, presence or absence of disease progression, history of treatment, and history of hematologic autoimmune disease.
Cumulative survival analyses were obtained according to Kaplan- Meier method.
Results
The pattern of expression of CD38 identifies 3 groups of CLL patients
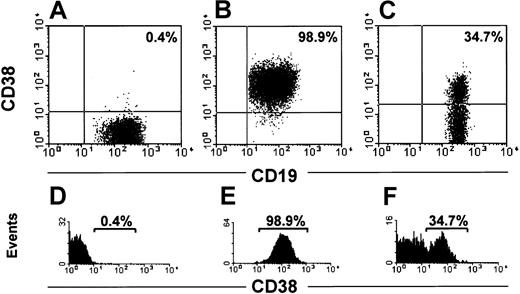
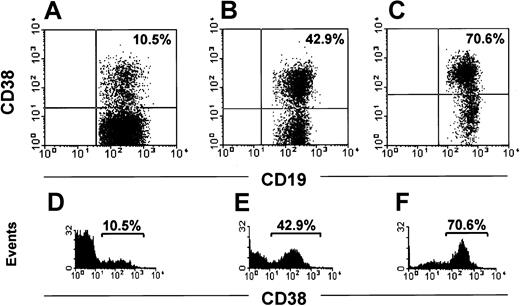
The range of CD38+ CLL cells in 148 patients was 0.1% to 99%. In 82 cases (Figure 1A,D) the cells were homogeneously negative with exceedingly rare and scattered positive elements (negative group; < 2%). In 26 patients most cells clustered in a homogeneously positive population (Figure 1B,E; positive group). In 40 patients a clear-cut bimodal expression of CD38 was evident (Figure 1C,F) with 2 distinct cell populations, one expressing high levels of CD38 and the other completely negative (CD38 bimodal expression group). The proportion of each population varied in individual patients (Figure 2A-E). In our cohort of CD38 bimodal patients the CD38+ subset ranged between a minimum of 8% to a maximum of 78%. It remains to be determined whether a distinct bimodal profile can also be observed in patients presenting a lower percentage of CD38+ cells (eg, < 5%).
Pattern of CD38 expression displayed by CLL cells.
CLL cells were analyzed for surface CD38 expression after incubation with anti-CD19, anti-CD38, and anti-CD5 conjugated with different fluorochromes. Dot plots (A-C) were obtained by gating lymphocytes (identified on side and forward scatter profiles) expressing CD19 and CD5. The intensities of CD19 and CD38 are shown on the x-axis and y-axis, respectively. The histograms in panels D-F show the profile of CD38 expression. The intensity of CD38 and the number of events are shown on the x-axis and y-axis, respectively. In panels A and D, a representative patient negative for CD38 is depicted, whereas panels B and E illustrate a representative patient homogeneously positive for CD38. A patient with a bimodal expression of CD38 is shown in panel C, with 2 distinct cell populations: one positive and the other negative (F).
Pattern of CD38 expression displayed by CLL cells.
CLL cells were analyzed for surface CD38 expression after incubation with anti-CD19, anti-CD38, and anti-CD5 conjugated with different fluorochromes. Dot plots (A-C) were obtained by gating lymphocytes (identified on side and forward scatter profiles) expressing CD19 and CD5. The intensities of CD19 and CD38 are shown on the x-axis and y-axis, respectively. The histograms in panels D-F show the profile of CD38 expression. The intensity of CD38 and the number of events are shown on the x-axis and y-axis, respectively. In panels A and D, a representative patient negative for CD38 is depicted, whereas panels B and E illustrate a representative patient homogeneously positive for CD38. A patient with a bimodal expression of CD38 is shown in panel C, with 2 distinct cell populations: one positive and the other negative (F).
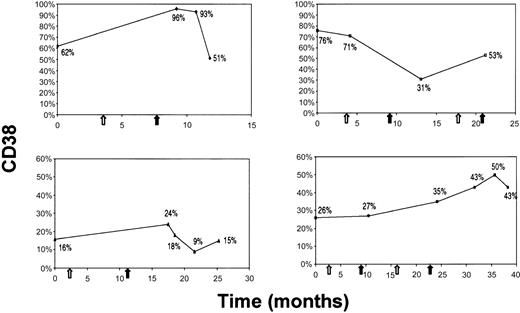
In 63 cases, blood samples were sequentially analyzed over time (12-86 months). No significant modifications of CD38 expression were observed in 59 of 63 cases either in the absence of treatment (42 cases) or after therapy (17 cases). In the remaining 4 cases the CD38 expression was modified after chemotherapy. All 4 patients who had modified CD38 expression belonged to the CD38 bimodal expression group; the pattern of modification was not clear-cut (Figure 3A-D); no one became CD38−.
The pattern of expression of CD38 correlates with IgVHgene mutational status
The IgVH mutational status was analyzed in 82 patients, 76 surface IgM+ (sIgM+) and 6 surface IgG+ (sIgG+), and confirmed that: (1) the most frequently encountered VH genes areV3-23 (9 of 82, 11.0%), V1-69 (7 of 82, 8.5%),V3-07 (7 of 82, 8.5%), 3.30 (6 of 82, 7.3%), and V4-34 (5 of 82, 6.1%) genes5,17; (2) there is a preferential use of D3 (23 of 82, 28.0%),D2 (15 of 82, 18.3%), and D6 (13 of 82, 15.8%) genes17; and (3) the most frequently used JHsegments are JH4 (45 of 82, 54.9%), JH6 (17 of 82, 20.7%) and JH3 (12 of 82, 14.6%).17
Thirty-three patients (40.2%) showed no IgVHmutation.17 20 The remaining 49 (59.8%) patients showed evidence of somatic hypermutation: 41 of 49 (83.7%) cases differed 5% or more from germ line genes. All IgG cases were mutated.
We evaluated the IgVH mutational status in the 3 groups of patients defined according to the pattern of CD38 expression. Most patients (39 of 44, 88.6%) of the CD38− group carried mutated VH genes. Within the CD38+ group 11 of 13 were unmutated (84.6%). Interestingly, 17 of 25 (68.0%) patients belonging to the CD38 bimodal expression group carried no mutation (Table 1).
We then analyzed the expression of CD38 in patients categorized as mutated or unmutated observing that 39 of 49 mutated cases (79.6%) were CD38−, 2 were CD38+ (4.1%), and 8 were bimodal (16.3%). In the unmutated group, 28 of 33 cases were either CD38+ (11 of 33, 33.3%) or bimodal (17 of 33, 51.5%); only 5 were CD38− (5 of 33, 15.2%).
These data indicate a statistically significant correlation between the expression of CD38 and the mutational status of IgVH genes (P < .001). Strong positive predictive values (PPVs) predicting either the presence or the absence of somatic mutations are found in the CD38− group (PPV = 88.6%) and in the CD38+ group (PPV = 84.6%). In contrast, a bimodal expression does not predict the status of the IgVHgene status.
The 2 subsets of CD38 bimodal expression patients are differentially represented in the BM
We asked which (if any) of the 2 subpopulations (CD38+and CD38−) detected in the PB of the CD38 bimodal expression group might be prevalent in infiltrated tissues. To this end, we compared the expression of CD38 in CLL cell samples concomitantly obtained from both PB and BM in 14 CD38 bimodal expression patients. Interestingly, in 10 of 14 CD38 bimodal expression patients a significantly higher amount of CD38+cells could be seen in the BM with an increase ranging between 28% and 97% (Table2).
Increased difference in CD38 expression between PB and BM in 14 CD38 bimodal expression patients
| Patient no. . | % CD38+ PB . | % CD38+ BM . | Increase, % . |
|---|---|---|---|
| 1 | 20.9 | 41.2 | 97 |
| 2 | 39.6 | 68.6 | 73 |
| 3 | 47.9 | 70.2 | 46 |
| 4 | 11.0 | 16.0 | 45 |
| 5 | 27.9 | 40.1 | 44 |
| 6 | 41.6 | 58.8 | 41 |
| 7 | 26.5 | 37.5 | 41 |
| 8 | 24.3 | 33.0 | 36 |
| 9 | 62.4 | 81.4 | 30 |
| 10 | 69.0 | 88.5 | 28 |
| 11 | 11.2 | 11.8 | — |
| 12 | 12.8 | 14.0 | — |
| 13 | 16.3 | 16.5 | — |
| 14 | 35.0 | 33.7 | — |
| Patient no. . | % CD38+ PB . | % CD38+ BM . | Increase, % . |
|---|---|---|---|
| 1 | 20.9 | 41.2 | 97 |
| 2 | 39.6 | 68.6 | 73 |
| 3 | 47.9 | 70.2 | 46 |
| 4 | 11.0 | 16.0 | 45 |
| 5 | 27.9 | 40.1 | 44 |
| 6 | 41.6 | 58.8 | 41 |
| 7 | 26.5 | 37.5 | 41 |
| 8 | 24.3 | 33.0 | 36 |
| 9 | 62.4 | 81.4 | 30 |
| 10 | 69.0 | 88.5 | 28 |
| 11 | 11.2 | 11.8 | — |
| 12 | 12.8 | 14.0 | — |
| 13 | 16.3 | 16.5 | — |
| 14 | 35.0 | 33.7 | — |
Phenotypic characterization of leukemic cells from CD38 bimodal expression patients
To evaluate whether the 2 subpopulations (CD38+ and CD38−) detected in the PB of the CD38 bimodal expression group might also differ for the expression of other markers we further characterized by cytofluorography the surface phenotype of the 2 subpopulations in 10 bimodal expression patients. We have analyzed the expression of: (1) CD10, CD20, CD21, CD22, CD24, CD25, CD27, CD40, CD69, CD70, CD72, CD74, CD75, CD78, CD79b, CD80, CD81, CD86, CD95, CD103, CD138, based on the hypothesis that these 2 subsets could differ for their differentiation status; (2) CD23, CD43, CD52, CD6, CD9, CD71, FMC-7 based on the hypothesis that CD38 expression might correlate with a more aggressive phenotype; and (3) CD11a, CD11c, CD18, CD31, CD44, CD49d, CD54, CD100, CXCR3, CCR6 based on the hypothesis that the 2 subsets might have a different homing and migratory capacity.
Most markers were homogeneously either positive or negative on the leukemic clones as a whole; that is, no differential expression could be detected in the 2 cell subsets. Few markers (eg, CD11c, CD49d, CD69, CD78, CCR6, CXCR3) had a more dispersed expression with a mixture of positive and negative cells but the pattern of expression was superimposable in both CD38+ and CD38−subpopulations.
Functional characterization of leukemic cells from CD38 bimodal expression patients
Next we investigated whether the presence of 2 subsets in the PB of CD38 bimodal expression patients might be associated with a different response to microenvironmental stimuli. We have previously described that cells obtained from PB of unselected CLL patients stimulated through CD40 can have a different outcome.21Leukemic cells from the majority of the patients (66%) respond to such stimulation by prolonging their survival in vitro and up-regulating CD80 and CD95 on the surface (CD40L responders), whereas the remaining (33%) are not modified by this stimulus (CD40L nonresponders). Therefore, we cultured CLL cells from PB of 46 CLL patients (31 CD38−, 5 CD38+, and 10 CD38 bimodal expression) in the presence or the absence of soluble human CD40 ligand (sCD40L) and analyzed cell survival and CD80/CD95 expression after up to 3 days of culture. Twenty-three of the 31 CD38− as well as all 5 of the CD38+ patients were CD40L responders. Among the 10 CD38 bimodal expression patients, 3 were CD40L nonresponders and the remaining 7 had a homogeneous up-regulation of both CD80 and CD95 on the surface. In these cases, after up to 3 days of culture, both CD38+ and CD38− compartments prolonged their survival, and the relative ratios of the 2 subsets were unmodified.
The phenotypic and functional homogeneity shown by both subsets of CLL cells in CD38 bimodal expression patients is also matched by the molecular evidence that in 25 CD38 bimodal expression patients specifically analyzed the cells belonging to the 2 CD38 subsets carry the same IgVH rearrangement. Repetitive reverse transcription-PCR amplification produced only one discrete band from each sample and the direct sequencing of this band, performed in different occasions from both 5′ and 3′ ends, always produced the same nucleotide sequence.
Concordance of CD38 pattern of expression, IgVHmutation status, and traditional prognostic factors
The increasing value of CD38 significantly correlated with low platelet counts (r = −0.278; P = .004), low Hb levels (r = −0.360; P < .001), and high β2-microglobulin serum levels (r = 0.372;P = .001).
CD38+ and CD38 bimodal expression patients had significantly higher β2-microglobulin mean levels than CD38− cases (respectively 4.1 mg/L and 3.9 mg/L versus 2.6 mg/L). A significant difference was also observed in lymphocyte counts, platelet counts, Hb, and LDT percent mean values comparing CD38−, CD38+, and CD38 bimodal expression patients (Table 1). No statistically significant correlation was observed with the other parameters analyzed.
Likewise, patients belonging to the unmutated group had significantly higher β2-microglobulin levels, LDT percent values, lymphocyte counts, and significantly lower Hb levels than patients belonging to the mutated group (Table3). These data support the concordance among the pattern of expression of CD38, IgVHmutation status, and traditional CLL prognostic factors.
Characteristics of CLL patients according to the IgVH mutational status
| . | All patients . | Somatically mutated . | Somatically unmutated . | P . |
|---|---|---|---|---|
| No. of cases (%) | 82 | 49 (59.8) | 33 (40.2) | NA |
| Mean age, y | 69 | 68 | 71 | NA |
| Sex, M/F | 49/33 | 30/19 | 19/14 | NA |
| Mean duration of follow-up, mo | 92.1 | 98.5 | 82.4 | NA |
| Risk status | .1773-153 | |||
| Low risk3-150(%) | 59/82 (72.0) | 39/49 (79.6) | 20/33 (60.6) | |
| Intermediate risk3-150(%) | 18/82 (22.0) | 8/49 (16.3) | 10/33 (30.3) | |
| High risk3-150 (%) | 5/82 (6.1) | 2/49 (4.1) | 3/33 (9.1) | |
| Disease status | <.0013-153 | |||
| Stable disease (%) | 48/82 (58.5) | 38/49 (77.6) | 10/33 (30.3) | |
| Progressive disease (%) | 34/82 (41.5) | 11/49 (22.4) | 23/33 (69.7) | |
| Autoimmune manifestations (%) | 15/82 (18.3) | 6/15 (40.0) | 9/15 (60.0) | .0773-153 |
| Treatment | ||||
| Not requiring treatment (%) | 39/82 (47.6) | 30/49 (61.2) | 9/33 (27.3) | .0023-153 |
| Requiring treatment (%) | 43/82 (52.4) | 19/49 (38.8) | 24/33 (72.7) | |
| Blood parameters | ||||
| Mean β2-microglobulin count, mg/L3-151 | 3.0 | 2.3 | 3.9 | .0023-155 |
| Mean Hb count, g/dL | 12.8 | 13.4 | 11.9 | <.0013-155 |
| Mean lymphocyte count, per μL | 43 700 | 29 800 | 64 300 | <.0013-155 |
| LDT, %3-152 | 121.5 | 58.1 | 215.6 | .0143-155 |
| . | All patients . | Somatically mutated . | Somatically unmutated . | P . |
|---|---|---|---|---|
| No. of cases (%) | 82 | 49 (59.8) | 33 (40.2) | NA |
| Mean age, y | 69 | 68 | 71 | NA |
| Sex, M/F | 49/33 | 30/19 | 19/14 | NA |
| Mean duration of follow-up, mo | 92.1 | 98.5 | 82.4 | NA |
| Risk status | .1773-153 | |||
| Low risk3-150(%) | 59/82 (72.0) | 39/49 (79.6) | 20/33 (60.6) | |
| Intermediate risk3-150(%) | 18/82 (22.0) | 8/49 (16.3) | 10/33 (30.3) | |
| High risk3-150 (%) | 5/82 (6.1) | 2/49 (4.1) | 3/33 (9.1) | |
| Disease status | <.0013-153 | |||
| Stable disease (%) | 48/82 (58.5) | 38/49 (77.6) | 10/33 (30.3) | |
| Progressive disease (%) | 34/82 (41.5) | 11/49 (22.4) | 23/33 (69.7) | |
| Autoimmune manifestations (%) | 15/82 (18.3) | 6/15 (40.0) | 9/15 (60.0) | .0773-153 |
| Treatment | ||||
| Not requiring treatment (%) | 39/82 (47.6) | 30/49 (61.2) | 9/33 (27.3) | .0023-153 |
| Requiring treatment (%) | 43/82 (52.4) | 19/49 (38.8) | 24/33 (72.7) | |
| Blood parameters | ||||
| Mean β2-microglobulin count, mg/L3-151 | 3.0 | 2.3 | 3.9 | .0023-155 |
| Mean Hb count, g/dL | 12.8 | 13.4 | 11.9 | <.0013-155 |
| Mean lymphocyte count, per μL | 43 700 | 29 800 | 64 300 | <.0013-155 |
| LDT, %3-152 | 121.5 | 58.1 | 215.6 | .0143-155 |
Rai modified stage at diagnosis.
β2-microglobulin serum count was obtained in 63 patients.
Lymphocyte doubling time (as increasing percent in 6 months).
P for the comparison among mutated and unmutated subgroups was calculated using the χ2 test.
P refers to 1-way ANOVA test.
The pattern of CD38 expression predicts CLL patients at risk of disease progression
At diagnosis the majority of patients (78 of 108, 72.2%) were in the low-risk group16: specifically, 53 of 62 (85.5%) of CD38− patients, 18 of 30 (60.0%) of CD38 bimodal expression group, and 7 of 16 (43.8%) of the CD38+group (Table 1).
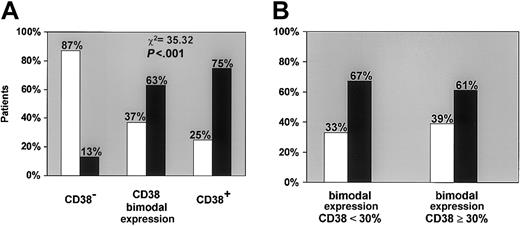
When disease progression criteria were correlated with CD38 expression (Figure 4A) the vast majority of CD38− patients (54 of 62, 87.1%) had stable disease. In 5 of 8 CD38− patients in progression, the only criterion was LDT. In contrast, most patients of the CD38+ group (12 of 16, 75.0%) had progression during the follow-up period. A high progression rate was also evident in the CD38 bimodal expression group (19 of 30, 63.3%), regardless of the percentage of CD38+ cells. As shown in Figure 4B, the high progression rate was evident even when CD38 bimodal expression patients were analyzed according to the 30% positivity cutoff, indicating that this correlation is independent of the percentage of CD38+cells. Because the CD38 bimodal expression group patients with stable disease (11 of 30, 36.7%) had a follow-up shorter than patients who progressed (65 versus 109 months), the possibility of a later progression cannot be ruled out. The degree of association between CD38 expression (irrespective of the proportion of the positive population) and disease progression is strongly significant (P < .001; Figure 4A).
The CD38+ population size differs in individual patients of the bimodal expression group.
Three representative patients with a bimodal surface expression of CD38 are depicted (A-C). The intensity of CD19 and CD38 are shown on the x-axis and y-axis, respectively. The histograms in panels D-F show the profile of CD38 expression. The intensity of CD38 and the number of events are shown on the x-axis and y-axis, respectively.
The CD38+ population size differs in individual patients of the bimodal expression group.
Three representative patients with a bimodal surface expression of CD38 are depicted (A-C). The intensity of CD19 and CD38 are shown on the x-axis and y-axis, respectively. The histograms in panels D-F show the profile of CD38 expression. The intensity of CD38 and the number of events are shown on the x-axis and y-axis, respectively.
The CD38+ population size may change on treatment in individual patients of the bimodal expression group.
CD38 expression (y-axis) at different time points of the disease course (x-axis) is depicted in relation to treatment. White and black arrows indicate, respectively, the beginning and the end of therapy.
The CD38+ population size may change on treatment in individual patients of the bimodal expression group.
CD38 expression (y-axis) at different time points of the disease course (x-axis) is depicted in relation to treatment. White and black arrows indicate, respectively, the beginning and the end of therapy.
The expression of CD38 correlates with disease progression.
(A) When disease progression criteria were correlated with CD38 expression, the vast majority of CD38− patients showed a strong tendency to maintain a stable disease (■). In contrast, most of CD38+ patients had a progressive disease (▪) as did a large proportion of patients belonging to the bimodal expression group. The differences among the 3 groups are highly significant (P < .001). (B) The strong tendency to develop progressive disease was not related to the percentage of CD38+ cells as shown by the analysis of CD38+bimodal expression patients divided according to the 30% positivity cutoff (< 30% and ≥ 30%).
The expression of CD38 correlates with disease progression.
(A) When disease progression criteria were correlated with CD38 expression, the vast majority of CD38− patients showed a strong tendency to maintain a stable disease (■). In contrast, most of CD38+ patients had a progressive disease (▪) as did a large proportion of patients belonging to the bimodal expression group. The differences among the 3 groups are highly significant (P < .001). (B) The strong tendency to develop progressive disease was not related to the percentage of CD38+ cells as shown by the analysis of CD38+bimodal expression patients divided according to the 30% positivity cutoff (< 30% and ≥ 30%).
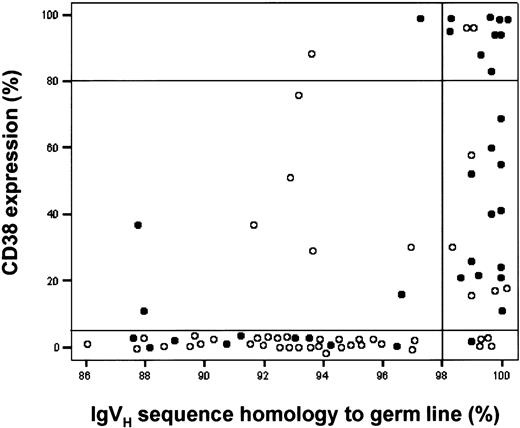
The relationship of CLL cell IgVH mutation status with disease progression was statistically significant (P < .001), though at a lower degree than CD38 expression alone (Figure 5; Table 2; PPV = 77.6% for mutated and stable disease; PPV = 69.7% for unmutated and progressive disease). The concomitant analysis of both somatic mutations and CD38 expression improved the capacity in predicting disease progression only in the presence of the association of unmutated Ig and CD38+ or bimodal expression cells (PPV = 90.0% and 81.3%, respectively).
IgVH mutation status in relation to CD38 expression and progressive or stable disease.
Data on 82 patients are plotted according to IgVH sequence homology to germ line (x-axis), CD38 expression on CLL cells (y-axis), and status of the disease (○ indicates a patient with stable disease; ●, one with progressive disease). For clarity, 2 horizontal lines encompass the CLL patients with a CD38 bimodal expression profile and a vertical line divides mutated (< 98%) from unmutated (≥ 98%) CLL patients.
IgVH mutation status in relation to CD38 expression and progressive or stable disease.
Data on 82 patients are plotted according to IgVH sequence homology to germ line (x-axis), CD38 expression on CLL cells (y-axis), and status of the disease (○ indicates a patient with stable disease; ●, one with progressive disease). For clarity, 2 horizontal lines encompass the CLL patients with a CD38 bimodal expression profile and a vertical line divides mutated (< 98%) from unmutated (≥ 98%) CLL patients.
Only 16 of 62 (25.8%) CD38− patients were treated during their disease history and all reached a complete or partial response, whereas in 19 of 30 (63.3%) patients of the bimodal expression group and in 12 of 16 (75.0%) of the CD38+ group at least one course of treatment was administered (P < .001), strengthening the relationship between disease progression and CD38 expression. In addition, 24 of 33 unmutated cases (72.7%) required therapy, confirming that germ line configuration has a tendency toward progressive disease. The majority (30 of 49, 61.2%) of mutated cases were untreated.
Among the 20 patients who presented autoimmune hemolytic anemia7 or thrombocytopenia,13 5 were CD38+ and 9 were CD38 bimodal expression, indicating that patients presenting with even a small population of CD38+cells are more prone to develop autoimmune manifestations (P = .03; Table 1).
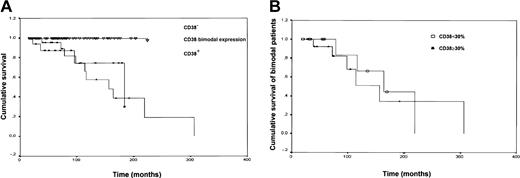
The analysis of cumulative survival curves (Figure6A) indicates that the median survival for the bimodal expression group, regardless of the proportion of CD38+ cells, is 156 months and overlaps the median survival of the CD38+ group (183 months). As shown in Figure 6B, the median survival of CD38 bimodal expression patients did not vary significantly when the patients were analyzed according to the 30% positivity cutoff. The mean survival of the CD38−population has not yet been reached during the follow-up. The difference among the 3 curves is statistically significant (P = .002). Similarly patients carrying IgVHsomatic mutations have not reached the median survival time, which is 183 months in the unmutated group (P < .001). Such a prognostic difference does not correlate with the stage at diagnosis as shown by the fact that considering only low-risk (stage 0) patients, CD38+ and bimodal expression patients have a median survival of 183 and 163 months, respectively, whereas CD38− patients do not reach median survival (P = .001). The same holds true for the survival of intermediate-risk and high-risk patients classified according to the expression of CD38. Median survival of CD38+ and bimodal expression intermediate- and high-risk patients is 174 and 156 months, respectively; again it has not yet been reached by CD38− patients.
The survival curve of CD38 bimodal CLL patients overlaps that of CD38+ patients.
(A) Kaplan-Meier curves show cumulative survival of CD38−, CD38 bimodal expression, and CD38+ patients. CD38− patients have a significantly longer survival (median survival not reached) than CD38 bimodal expression (median survival, 156 months; P < .001) and CD38+(median survival, 183 months; P = .001) subsets. The differences among the 3 curves are statistically significant (P = .002). (B) No significant differences were observed when the survival curves of CD38 bimodal expression patients were analyzed according to the 30% positivity cutoff (< 30% and ≥ 30%).
The survival curve of CD38 bimodal CLL patients overlaps that of CD38+ patients.
(A) Kaplan-Meier curves show cumulative survival of CD38−, CD38 bimodal expression, and CD38+ patients. CD38− patients have a significantly longer survival (median survival not reached) than CD38 bimodal expression (median survival, 156 months; P < .001) and CD38+(median survival, 183 months; P = .001) subsets. The differences among the 3 curves are statistically significant (P = .002). (B) No significant differences were observed when the survival curves of CD38 bimodal expression patients were analyzed according to the 30% positivity cutoff (< 30% and ≥ 30%).
Discussion
CLL is a frequently diagnosed blood malignancy whose management still defies clear-cut practice guidelines. One major reason for this failure is the clinical heterogeneity of the disease. Predicting prognosis would allow patients optimal counseling and more adequate treatment planning. Starting from the classical staging systems,22 23 many efforts have been devoted to the identification of prognostic parameters that might help dissect the clinical heterogeneity and could indicate at diagnosis which natural history can be reasonably expected in individual patients.
The rationale of focusing on CLL cell expression of CD38 and IgVH mutational status is that the clinical relevance of these 2 markers is matched by their biologic significance. In vitro studies have shown that the CLL cell expression of CD38 defines 2 groups of patients strikingly differently in their in vitro response to anti-Ig antibodies.24,25 In clinical studies CD38 has proved to be an independent and reliable prognostic marker.10-13 The IgVH mutational status is used to classify the cellular origin of blood malignancies26 because IgVH somatic mutations appear to occur only in B cells that have been somehow exposed to antigenic stimulation. In CLL it has been shown4,5 and confirmed6 that the presence of IgVH somatic mutations carries a favorable prognosis, whereas its absence is an unfavorable marker. The use of these parameters in clinical practice would be extremely useful provided that 2 major discrepancies in the literature are clarified. The first concerns the relationship between immunoglobulin somatic mutations and CD38 expression that some authors have observed,4 but others12,27 have questioned. The second even more important problem is the numerical definition of which cutoff value of CD38+ cells should be used to have the most useful clinical information. Different authors have used 30%, 20%, or even 7% of CD38+ cells as the cutoff point that provides the best separation for prognostic subgroups.4,7 10-13 A nonsecondary corollary of the whole issue is that cytofluorographic determination of CD38 is an easily manageable test, whereas the definition of IgVH somatic mutations requires a definitely less common expertise.
We have shifted the perspective under which the cytofluorometric analysis of CD38 expression in CLL has to be assessed. In this work we demonstrate that the important clinical discriminant is not a CD38 numerical value, but rather it is its pattern of expression that correlates with IgVH gene mutational status and allows us to predict which patients are at risk for disease progression. We started from the observation that CLL patients can be divided into 3 different subsets: a CD38− subset where CLL cells are homogeneously CD38− (Figure 1A,D); a CD38+subset where CLL cells are homogeneously CD38+ (Figure1B,E); and a third subset (Figure 1C,F) characterized by the concomitant presence of 2 populations, one expressing high levels of CD38 and the second completely CD38−. The bimodal expression profile of CD38 in these cases led us to call them CD38 bimodal expression group. The bimodal expression group is the likely equivalent of the mixed CD38 expression that has been recently recognized.7 We next observed that within the bimodal expression group the proportions of the 2 populations (CD38+ and CD38−) may be highly variable in individual patients (Figure 2A-E) because the percentage of CD38+ cells ranged between 8% and 76%. It is presently impossible from our series to determine whether even smaller percentages of CD38+ cells (eg, < 5%) could exist with a distinct bimodal phenotype. Very low percentages of CD38+cells may be accounted for by normal activated CD5+ B cells, so that technical refinements like 4- or 5-color combinations may be needed and prospective studies of the clinical outcome of patients presenting with tiny populations of CD38+ cells are necessary. Even if the existence of very unusual cases cannot be denied a priori and the influence of chemotherapy on CD38 expression has to be explored in detail, the expression of CD38 appears to be essentially stable over time. Because it has been reported that the expression of CD38 may vary during the course of the disease,12 we studied 63 cases over time. No transition from a CD38− to a CD38+ status was observed during the period of observation nor was a chemotherapy-induced shift from a CD38+ to a CD38− pattern of expression found. A shift was observed in a minority of the cases examined (4 of 63, 6.3%), all belonging to the bimodal expression group, on treatment (Figure 3A-D). These observations led us to evaluate the clinical outcome of our patients avoiding cutoff threshold numbers but relying on the biologic observation of the presence of a well-defined population of CD38+ cells irrespective of its size.
Using this approach it has become evident that the CD38−group is characterized by CLL cells that usually (88.4%) carry mutated VH genes. CD38− patients have not reached the median survival after a follow-up of 89 months and most of them (74.2%) have not required treatment during this time. The CD38+ group is characterized by CLL cells that most frequently (84.6%) carry unmutated VH genes. CD38+ patients have a median survival of 183 months and most of them (75.0%) have required treatment during a follow-up of 98 months. The CD38 bimodal expression group is characterized by cells that tend to have somatically unmutated VH genes (68.0%). In clinical terms, this group of patients resembles the CD38+ group in terms of risk of disease progression (63.3%), required treatment during a follow-up period of 87 months, and especially of median survival time (156 months). These differences cannot simply be accounted for by the prevalence of stage 0 low-risk patients in our series, but rather they imply that the presence of 2 distinct populations is an adverse prognostic factor irrespective of the stage at presentation. Further, the presence of a CD38+population is more likely associated with the development of autoimmune complications (Table 1).
The relevance of our observation is 2-fold. First, it indicates that a reproducible, well-disseminated technique can be used to identify patients who, irrespective of the clinical stage at presentation, will have progressive disease. This approach may be used to design new trials aimed at differentially managing patients whose outcome can be reasonably foreseen. It is irrelevant whether the CD38+population is present in a high or in a low proportion (Figures 4B and6B). What really matters is the presence of a CD38+population whose existence proves to be a simple and effective prognostic parameter that can be also used to reconcile the literature discrepancies. The second point concerns the biologic background of these clinical observations. It becomes obvious to ask whether the 2 populations detected in the PB of the bimodal expression group may have distinctive features. We here show that the 2 populations carry the same IgVH rearrangement and do not differ in terms of expression of differentiation status, aggressive phenotype, homing, and migratory capacity markers. Likewise no differences have been observed in terms of response to microenvironmental stimuli represented by the in vitro response to CD40L. Of interest, the CD38+ subset of bimodal patients appears to be increased in infiltrated BM as compared to PB, suggesting that the BM environment, which is known to be a privileged site of relapse in CLL patients, more easily hosts cells that are associated with an adverse outcome. The quest for the biologic bases of the observed clinical differences will need painstaking experiments of cell culture and molecular investigation of extensively purified populations.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-06-1801.
Supported by AIRC (Associazione Italiana per la Ricerca sul Cancro), Milano, by Ministero dell'Università e Ricerca Scientifica (MIUR), Roma, and by MDACC Laboratory Study Agreement LS01-039 (Houston, TX). G.G. and C.S. are recipients of a fellowship from Comitato Gigi Ghirotti (Torino).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Federico Caligaris-Cappio, Ospedale Mauriziano Umberto I, Largo Turati 62, 10128 Torino, Italy; e-mail:fcaligaris@mauriziano.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal