We determined cytotoxic T lymphocyte (CTL) epitopes through screening with a computer-assisted algorithm and an enzyme-linked immunospot (ELISPOT) assay using in vitro–reactivated polyclonal Epstein-Barr virus (EBV)–specific CD8+ T cells as responders. In addition, to confirm that the epitopes were generated after endogenous processing and presentation of the EBV proteins, a novel T-cell receptor (TCR) down-regulation assay was introduced, in which a fluorescent tetrameric major histocompatibility complex (MHC)/peptide complex was employed for detecting TCR down-regulation after stimulation with the epitope presented on antigen-presenting cells. Through such screening, 3 HLA A*2402–restricted epitopes were identified: IYVLVMLVL, TYPVLEEMF, and DYNFVKQLF, derived from LMP2, BRLF1, and BMLF1 proteins, respectively. TCR down-regulation assays disclosed that, in contrast to the other 2 epitopes, IYVLVMLVL was not presented on HLA A24–positive fibroblast cells infected with recombinant vaccinia viruses expressing LMP2. Furthermore, ELISPOT assays with an epitope-specific CTL clone demonstrated that the presentation was partially restored by pretreatment of the fibroblast cells with interferon-γ. The epitope was presented on transporters associated with antigen processing (TAP)–negative T2 cells transfected with plasmids encoding HLA A*2402 and the minimal epitope, indicating that the presentation is TAP independent. In conclusion, the 3 epitopes thus defined could be useful for studying EBV-specific CD8+ T-cell responses among populations positive for HLA A*2402.
Introduction
Epstein-Barr virus (EBV) is associated with malignancies, such as a subset of Hodgkin disease, Burkitt lymphomas, immunoblastic lymphomas seen in immunocompromised hosts, nasopharyngeal carcinomas (NPCs), and some gastric carcinomas.1 Primary infection is usually asymptomatic, although some individuals may suffer from infectious mononucleosis, followed by a strong HLA class I–restricted, virus-specific CD8+ cytotoxic T lymphocyte (CTL) response.1,2 The latter is believed to play an important role in controlling the virus both during primary infection and in the long-term carrier state, whereby EBV persists for life in a subset of B cells.3 Both lytic- and latent-cycle EBV proteins are known to be recognized by EBV-specific CTLs targeting short peptides presented on cell surfaces by HLA class I molecules.2
The importance of EBV-specific T cells for control of latently infected B cells has been re-emphasized by observations in patients with immunoblastic lymphomas or lymphoproliferative disease (LPD) after bone marrow transplantation.4,5 Adoptive transfer of EBV-specific CTLs or unfractionated donor lymphocytes can result in remission of the LPD.6,7 Withdrawal of immunosuppressive drugs may cause regression of EBV-associated LPD in some patients after bone marrow transplantation.8-12
In contrast to immunoblastic lymphomas seen in immunocompromised hosts, whereby the full panel of the EBV latent proteins is expressed, virus protein expression in other EBV-associated malignancies is more restricted. In Hodgkin disease, NPC, and EBV-positive gastric carcinomas, the only EBV proteins expressed are EBNA1, latent membrane protein 1 (LMP1), and LMP2.1,13,14 Of these, LMP1 and LMP2 have drawn attention as potential target antigens to establish CTL-based immunotherapy.15-23 Indeed, T cells isolated from biopsy samples of Hodgkin disease demonstrate MHC-restricted cytotoxicity to target cells expressing LMP2.17 Dendritic cells, infected with recombinant adenovirus expressing LMP2,18,19 or transfected with mRNA encoding the LMP2 gene,20 have been studied for effective and selective stimulation of LMP2-specific CTLs.
Some viral proteins synthesized in infected cells are degraded, primarily by the proteasome system, and the resultant peptides are translocated into the endoplasmic reticulum by transporters associated with antigen processing (TAPs). Stable major histocompatibility complex (MHC) class I molecules then move to the cell surface for presentation of peptides to CD8+ T cells.24 A number of antigenic peptides have been identified within the amino acid sequences of LMP2.15,16,21 These peptides can be pulsed on dendritic cells to elicit a peptide-specific polyclonal CTL response.22 In addition, such peptides have the potential to elicit EBV-specific CTL responses in vivo when used as vaccines before EBV challenge or as immunotherapeutic reagents for treatment of EBV-associated malignancies.1 Another attribute of the antigenic peptide is to produce fluorescent-labeled tetrameric MHC-peptide complexes to enumerate and/or to sort peptide-specific CTLs for large-scale expansion.25-28 One limitation of using such epitope peptides is that their presentation to T cells is HLA class I allele specific. In many ethnic groups, HLA A24 is one of the most common alleles in Japan,29,30 being the most frequently encountered HLA class I allele with a genotype almost exclusively A*2402.31 An LMP2 epitope presented by HLA A*2402 has been reported.16 To establish epitope-based immunotherapy, however, multiple epitopes are preferred because of the emergence of escaping mutant cells after single epitope vaccination,32 and the nature of processing of such epitopes needs to be extensively studied to obtain a maximal clinical response.
We have adopted an approach for determination of CTL epitopes through screening with a computer-assisted algorithm and an enzyme-linked immunospot (ELISPOT) assay by which a major human cytomegalovirus epitope presented by HLA A*2402 molecules has been successfully identified.33 In addition, we introduce here a novel assay using relevant tetramers for detecting down-regulation of T-cell receptor (TCR) expression after stimulation with HLA A24–positive fibroblast cells expressing each of the EBV genes, in order to confirm endogenous processing and presentation. In this paper, the identification through such screening of 2 HLA A*2402–restricted epitopes derived from EBV lytic-cycle proteins, and an unusually presented LMP2 epitope, together with characterization of the epitope processing is described in detail.
Materials and methods
Cell lines
HLA A*2402–transfected, TAP-negative T2-A24 cells33 were cultured in RPMI 1640 (GIBCO, Grand Island, NY) supplemented with 10% fetal calf serum (FCS), 2 mMl-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, and 5 × 10−5M β-mercaptoethanol (referred to as culture medium) in the presence of 800 μg/mL G418 (GIBCO). HEK293T cells (American Type Culture Collection, Manassas, VA) and HLA A24–positive dermal fibroblast cell lines cultured in Dulbecco modified Eagle medium (GIBCO) supplemented with 10% FCS, 2 mMl-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin.
CD40-activated B cells (CD40-Bs) were generated from the peripheral blood mononuclear cells (PBMCs) of HLA A24–positive donors as previously described.34 Briefly, PBMCs were cultured with irradiated human CD40L-transfected NIH3T3 cells (kindly provided by Dr Gordon Freeman, Dana-Farber Cancer Institute, Boston, MA), recombinant interleukin 4 (IL-4) (Genzyme, Cambridge, MA), and cyclosporine A (Sandoz, Basel, Switzerland) in the culture medium. Expanding B cells were stimulated twice a week.
EBV-carrying lymphoblastoid cell lines (LCLs) were prepared by transforming PBMCs with B95-8 cell culture supernatant as previously described11 and propagated in culture medium. Small populations, between 0.3% and 4%, of the LCLs used for establishment of CTLs, expressed the BZLF1 protein determined by indirect immunofluorescent analysis.
EBV-specific polyclonal CTLs were generated as described earlier.7,11 To establish EBV-specific T-cell clones, CD8+ T cells were plated in wells of 96-well round-bottom plates at 0.3 cell/well and 1 cell/well with γ-irradiated allogeneic PBMCs, LCLs, anti-CD3 monoclonal antibody (mAb; Ortho Diagnostics, Raritan, NJ), and IL-2.33
Construction and transfection of plasmids
For construction of vectors expressing full-length LMP2 and EBNA3A genes, cDNAs were generated using the THERMOScript RT-PCR System (GIBCO, Gaithersburg, MD) from cells infected 16 hours earlier with recombinant vaccinia viruses coding each of the EBV genes (kindly donated by Dr A. B. Rickinson, University of Birmingham, United Kingdom). LMP2 cDNAs were amplified by polymerase chain reaction (PCR) using a sense primer, 5′-TTAGATCTGCCACCATGGGGTCCCTAGAAATGG-3′, and an antisense primer, 5′-TTGAATTCTTATACAGTGTTGCGATATGGGGTCG-3′. PCR products were cloned and inserted into pcDNA3 (Invitrogen, Carlsbad, CA) using itsBamHI and EcoRI sites. EBNA3A cDNAs were amplified using a sense primer, 5′-TTAAGCTTGCCACCATGGACAAGGACAGGCCG-3′, and an antisense primer, 5′-ACCTCTAGATTAGGCCTCATCTGGAGG-3′, and inserted into pcDNA3 using its HindIII and XbaI sites. To express minimal epitopes, the following oligonucleotides were inserted into the BamHI/EcoRI cut pcDNA3: IYVLVMLVL sense 5′-GATCTGCCACCATGATTTACGTTCTGGTGATGCTTGTGCTCTAAG-3′ and antisense 5′-AATTCTTAGAGCACAAGCATCACCAGAACGTAAATCATGGTGGCA-3′; RYSIFFDYM sense 5′-GATCTGCCACCATGCGCTACAGTATTTTCTTTGACTATATGTAAG-3′ and antisense 5′-AATTCTTACATATAGTCAAAGAAAATACTGTAGCGCATGGTGGCA-3′. Cloned genes were sequenced to verify their identity.
Each plasmid and one expressing HLA A*240233 were transfected using LIPOFECTAMINE PLUS (GIBCO) into HEK293T cells. T2-A24 cells were electroporated in the presence of each plasmid using a GENE PULSER II (Bio-Rad Laboratories, Hercules, CA). Transfected cells were incubated in medium with 10% FCS for 48 hours and applied as antigen-presenting cells (APCs) in the ELISPOT assay.
Peptides
To identify the potential HLA A24–binding peptides within B95-8 strain EBV proteins (accession no. V01555),35 a computer-based program was applied with access through the website of BioInformatics & Molecular Analysis Section (BIMAS) HLA Peptide Binding Predictions.36,37 Most peptides were synthesized with a Cleaved PepSet from Mimotope (Melbourne, Australia). There were 4 HLA A24–restricted CTL epitope peptides derived from EBNA3A,38 EBNA3B,2 LMP2,16 and HIV envelope protein39 that were synthesized by Toray Research Center (Kamakura, Japan). The peptides used in this study are listed in Table 1.
Details for synthetic oligopeptides and results with the MHC stabilization assay
| Peptide no. . | Amino acid sequence . | Protein name . | Start position . | Amino acid length . | Score* . | Percentage MFI increase† . |
|---|---|---|---|---|---|---|
| 1 | FYNIPPMPL | EBNA2 | 282 | 9 | 300 | 390 |
| 2 | GYDVGHGPL | EBNA2 | 119 | 9 | 200 | 44 |
| 3 | DYMAIHRSL | EBNA3A | 271 | 9 | 420 | 142 |
| 4 | VYDSMLQSDL | EBNA3B | 114 | 10 | 240 | 66 |
| 5 | RYEDPDAPL | EBNA3C | 721 | 9 | 720 | 41 |
| 6 | GYQEPPAHGL | EBNA3C | 775 | 10 | 360 | 36 |
| 7 | DYSQGAFTPL | EBNA3C | 923 | 10 | 240 | 72 |
| 8 | DYDASTESEL | EBNA3C | 982 | 10 | 220 | 40 |
| 9 | RYAREAEVRF | EBNA3C | 239 | 10 | 200 | 80 |
| 10 | GYRTATLRTL | EBNA3C | 183 | 10 | 200 | 40 |
| 11 | IYVLVMLVL | LMP2 | 222 | 9 | 420 | 86 |
| 12 | SYAAAQRKL | LMP2 | 170 | 9 | 220 | 54 |
| 13 | LYALALLLL | LMP2 | 357 | 9 | 200 | 213 |
| 14 | AYRRRWRRL | LMP2 | 234 | 9 | 200 | 46 |
| 15 | RYCCYYCLTL | LMP2 | 474 | 10 | 400 | 343 |
| 16 | IYVLVMLVLL | LMP2 | 222 | 10 | 300 | 95 |
| 17 | SYAAAQRKLL | LMP2 | 170 | 10 | 200 | 60 |
| 18 | IYLLEMLWRL | LMP1 | 124 | 10 | 300 | 421 |
| 19 | LYLQQNWWTL | LMP1 | 158 | 10 | 300 | 407 |
| 20 | AYQAYAAPQL | BZLF1 | 85 | 10 | 300 | 64 |
| 21 | TYPVLEEMF | BRLF1 | 198 | 9 | 180 | 491 |
| 22 | DYCNVLNKEF | BRLF1 | 28 | 10 | 132 | 84 |
| 23 | SYVKQPLCL | BMLF1 | 347 | 9 | 300 | 437 |
| 24 | DYNFVKQLF | BMLF1 | 320 | 9 | 252 | 221 |
| 25 | SYKTLREFF | BMLF1 | 230 | 9 | 120 | 265 |
| 26 | NYNPGTLSSL | BMLF1 | 387 | 10 | 360 | 332 |
| 27 | SYVKQPLCLL | BMLF1 | 347 | 10 | 300 | 387 |
| 28 | CYDHAQTHL | BMRF1 | 20 | 9 | 200 | 116 |
| 29 | TYTSGEACL | BMRF1 | 199 | 9 | 200 | 109 |
| 30 | FYISLIQGL | BALF2 | 846 | 9 | 432 | 331 |
| 31 | FYPLATYPL | BALF2 | 25 | 9 | 300 | 547 |
| 32 | CYENDNPGL | BALF2 | 636 | 9 | 300 | 78 |
| 33 | TYWQLNQNL | BALF2 | 526 | 9 | 288 | 231 |
| 34 | LYTGLAQAL | BALF2 | 217 | 9 | 288 | 187 |
| 35 | AYAEATSSL | BALF2 | 880 | 9 | 240 | 253 |
| 36 | FYRSLLTIL | BALF2 | 618 | 9 | 240 | 70 |
| 37 | HYQTLCTNF | BALF2 | 655 | 9 | 180 | 322 |
| 38 | EYLLGRFSVL | BALF2 | 1035 | 10 | 360 | 72 |
| 39 | FYMTHGLGTL | BALF2 | 362 | 10 | 300 | 114 |
| 40 | TYPLREVATL | BALF2 | 30 | 10 | 300 | 83 |
| 41 | TYWQINQNLL | BALF2 | 526 | 10 | 240 | 270 |
| 42 | VYAASFSPNL | BALF2 | 407 | 10 | 200 | 244 |
| 43 | RYSIFFDYM | EBNA3A | 246 | 9 | 60 | 450 |
| 44 | TYSAGIVQI | EBNA3B | 217 | 9 | 50 | 481 |
| 45 | TYGPVFMCL | LMP2 | 419 | 9 | 403 | 411 |
| 46 | RYLRDQQLL | HIV envelope | 584 | 9 | 720 | 251 |
| Peptide no. . | Amino acid sequence . | Protein name . | Start position . | Amino acid length . | Score* . | Percentage MFI increase† . |
|---|---|---|---|---|---|---|
| 1 | FYNIPPMPL | EBNA2 | 282 | 9 | 300 | 390 |
| 2 | GYDVGHGPL | EBNA2 | 119 | 9 | 200 | 44 |
| 3 | DYMAIHRSL | EBNA3A | 271 | 9 | 420 | 142 |
| 4 | VYDSMLQSDL | EBNA3B | 114 | 10 | 240 | 66 |
| 5 | RYEDPDAPL | EBNA3C | 721 | 9 | 720 | 41 |
| 6 | GYQEPPAHGL | EBNA3C | 775 | 10 | 360 | 36 |
| 7 | DYSQGAFTPL | EBNA3C | 923 | 10 | 240 | 72 |
| 8 | DYDASTESEL | EBNA3C | 982 | 10 | 220 | 40 |
| 9 | RYAREAEVRF | EBNA3C | 239 | 10 | 200 | 80 |
| 10 | GYRTATLRTL | EBNA3C | 183 | 10 | 200 | 40 |
| 11 | IYVLVMLVL | LMP2 | 222 | 9 | 420 | 86 |
| 12 | SYAAAQRKL | LMP2 | 170 | 9 | 220 | 54 |
| 13 | LYALALLLL | LMP2 | 357 | 9 | 200 | 213 |
| 14 | AYRRRWRRL | LMP2 | 234 | 9 | 200 | 46 |
| 15 | RYCCYYCLTL | LMP2 | 474 | 10 | 400 | 343 |
| 16 | IYVLVMLVLL | LMP2 | 222 | 10 | 300 | 95 |
| 17 | SYAAAQRKLL | LMP2 | 170 | 10 | 200 | 60 |
| 18 | IYLLEMLWRL | LMP1 | 124 | 10 | 300 | 421 |
| 19 | LYLQQNWWTL | LMP1 | 158 | 10 | 300 | 407 |
| 20 | AYQAYAAPQL | BZLF1 | 85 | 10 | 300 | 64 |
| 21 | TYPVLEEMF | BRLF1 | 198 | 9 | 180 | 491 |
| 22 | DYCNVLNKEF | BRLF1 | 28 | 10 | 132 | 84 |
| 23 | SYVKQPLCL | BMLF1 | 347 | 9 | 300 | 437 |
| 24 | DYNFVKQLF | BMLF1 | 320 | 9 | 252 | 221 |
| 25 | SYKTLREFF | BMLF1 | 230 | 9 | 120 | 265 |
| 26 | NYNPGTLSSL | BMLF1 | 387 | 10 | 360 | 332 |
| 27 | SYVKQPLCLL | BMLF1 | 347 | 10 | 300 | 387 |
| 28 | CYDHAQTHL | BMRF1 | 20 | 9 | 200 | 116 |
| 29 | TYTSGEACL | BMRF1 | 199 | 9 | 200 | 109 |
| 30 | FYISLIQGL | BALF2 | 846 | 9 | 432 | 331 |
| 31 | FYPLATYPL | BALF2 | 25 | 9 | 300 | 547 |
| 32 | CYENDNPGL | BALF2 | 636 | 9 | 300 | 78 |
| 33 | TYWQLNQNL | BALF2 | 526 | 9 | 288 | 231 |
| 34 | LYTGLAQAL | BALF2 | 217 | 9 | 288 | 187 |
| 35 | AYAEATSSL | BALF2 | 880 | 9 | 240 | 253 |
| 36 | FYRSLLTIL | BALF2 | 618 | 9 | 240 | 70 |
| 37 | HYQTLCTNF | BALF2 | 655 | 9 | 180 | 322 |
| 38 | EYLLGRFSVL | BALF2 | 1035 | 10 | 360 | 72 |
| 39 | FYMTHGLGTL | BALF2 | 362 | 10 | 300 | 114 |
| 40 | TYPLREVATL | BALF2 | 30 | 10 | 300 | 83 |
| 41 | TYWQINQNLL | BALF2 | 526 | 10 | 240 | 270 |
| 42 | VYAASFSPNL | BALF2 | 407 | 10 | 200 | 244 |
| 43 | RYSIFFDYM | EBNA3A | 246 | 9 | 60 | 450 |
| 44 | TYSAGIVQI | EBNA3B | 217 | 9 | 50 | 481 |
| 45 | TYGPVFMCL | LMP2 | 419 | 9 | 403 | 411 |
| 46 | RYLRDQQLL | HIV envelope | 584 | 9 | 720 | 251 |
Estimated t1/2 of dissociation from the HLA A24 allele (in minutes).
Percentage mean fluorescence intensity (MFI) increase of HLA A∗2402 molecules on T2-A24 cells. Percentage MFI increase = (MFI with sample peptide − MFI without peptide)/(MFI without peptide) × 100. See the MHC stabilization assay section in “Materials and methods.”
MHC stabilization assay
MHC stabilization assays were performed as described earlier.23 33 Briefly, T2-A24 cells were pulsed with each of the peptides (10 μM) at 26°C for 16 hours, followed by incubation at 37°C for 3 hours. Surface HLA A24 molecules were then stained with a specific mAb. Expression was measured by FACSCalibur (Becton Dickinson, San Jose, CA) and mean fluorescence intensity (MFI) was recorded. Percentage MFI increase was calculated as (MFI with the given peptide − MFI without peptide)/(MFI without peptide) × 100.
ELISPOT assays
ELISPOT assays were performed as previously described.37 The following APCs in 100 μL culture medium were seeded into each well: (1) T2-A24 cells (5 × 104cells/well) pulsed with each peptide (the peptide concentrations described in the text and figures are those in the final assay volume); (2) HLA A24–positive or –negative LCLs (1 × 105 cells /well); (3) HLA A24–positive fibroblast cells (1 × 104cells/well) infected 2 hours earlier with recombinant vaccinia virus expressing each of the EBV genes at a multiplicity of infection of 10 (some of the cells had been cultured in the presence of 400 U/mL interferon-γ (IFN-γ) (Strathmann Biotech GmbH, Hamburg, Germany) for 6 days prior to the vaccinia infection); (4) HEK293T or T2-A24 cells (5 × 104 cells/well) transfected 48 hours earlier with plasmids expressing each of the cloned genes.
As responders, polyclonal or monoclonal EBV-specific CD8+ T cells suspended in 100 μL culture medium were seeded into each well. All assays were performed in duplicate.
CTL assays
CTL assays were performed as previously described.33 Briefly, CTLs were suspended in fresh culture medium at the desired cell concentration and seeded in wells of V-bottomed 96-well plates (Costar, Cambridge, MA) containing51Cr-labeled LCLs (2500 cells/well). In some wells, the target cells were pulsed with antigenic peptides to a final concentration of 100 ng/mL. After 5 hours of incubation, the supernatants were harvested and radioactivity counted with a γ-counter. Percentage specific lysis was calculated as previously described.33 Each assay was performed in triplicate.
Tetramer-assisted TCR down-regulation assay
MHC/peptide tetramers were produced as previously described.33,40,41 To quantify engagement-dependent TCR down-regulation,42 43 we set up several stimulation systems preceding the tetramer staining: (1) 1000 to 2000 tetramer-positive cells (analyzed by FACS beforehand) for a given EBV-specific T-cell line were stimulated by incubation with T2-A24 cells (1 × 106) in the presence of the same peptide used for the tetramer construction or a control peptide at a final concentration of 100 ng/mL in 1 mL culture medium supplemented with 20 U/mL IL-2 in 16 × 125–mm culture tubes at 37°C for 16 hours; (2) 1000 to 2000 tetramer-positive cells were incubated with 1 × 106 HLA A24–positive CD40-Bs, or HLA A24–positive or –negative LCLs in 1 mL culture medium supplemented with 20 U/mL IL-2 in the culture tubes at 37°C for 16 hours; (3) 3000 to 10 000 tetramer-positive cells in 2 mL culture medium supplemented with 20 U/mL IL-2 were placed on a full sheet of HLA A24–positive fibroblast cells (approximately 5 × 105/well) infected 2 hours earlier with recombinant vaccinia viruses expressing each of the EBV genes at a multiplicity of infection of 10 in each well of 6-well plates. After the plates were incubated at 37°C for 16 hours, the cells were harvested, washed, resuspended in 1 mL phosphate-buffered saline (PBS) with 0.04% EDTA (ethylenediaminetetraacetic acid) and incubated at room temperature for 10 minutes before staining with the indicated tetramers. For staining, CTLs or PBMCs (2 × 106) were incubated with 40 μg/mL of the tetramer and a TRI-COLOR–labeled anti-CD8 mAb (Caltag, Burlingame, CA) at 37°C for 15 minutes before analysis with FACSCalibur (Becton Dickinson, Mansfield, MA).
Results
Selection of potential HLA A24–binding peptides within EBV proteins
To identify antigenic peptides presented by HLA A*2402 molecules, EBNA2, EBNA3A, EBNA3B, EBNA3C, LMP1, and LMP2 were selected from the EBV latent-cycle proteins because they are reported to be sources of CTL epitopes presented by other HLA class I molecules.2,15,16,23 In addition, BZLF1, BRLF1, BMLF1, BMRF1, and BALF2 were selected from the lytic-cycle proteins because they are targeted by CD8+ CTLs restricted by HLA class I molecules other than A*2402.2,44 To search for potential HLA A24–binding peptides, amino acid sequences of the B95-8 EBV strain35 were analyzed with a computer program.36,37 A total of 42 peptides with estimatedt1/2 dissociation scores of more than 100 were selected (Table 1). All share HLA A24–binding motifs with tyrosine as the second residue and phenylalanine or leucine as the ninth or tenth residues.45-47 Next, MHC stabilization assays were performed to test these peptides for HLA A*2402 binding efficiency using T2-A24 cells. All peptides increased the HLA A24 expression on the cells, indicating that they bound and stabilized the HLA complex on the cell surface (Table 1).
Screening of peptides antigenic for EBV-specific polyclonal CD8+ T-cell lines by ELISPOT assay
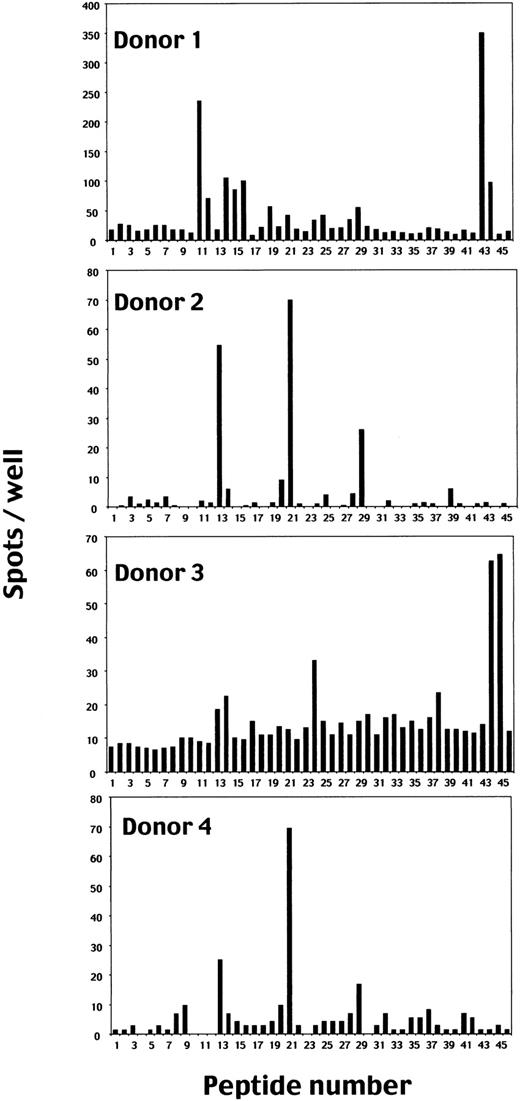
To identify peptides recognized by EBV-specific CD8+ T cells in the context of HLA A*2402 molecules, the ELISPOT assay was performed using T2-A24 cells as APCs. Figure1 summarizes the results using 4 polyclonal EBV-specific CD8+ T cells established from PBMCs of EBV-seropositive and HLA A24–positive donors as responders.
Screening of HLA A*2402–binding candidate peptides from amino acid sequences of EBV proteins for CD8+ T-cell stimulation by ELISPOT assay.
Polyclonal EBV-specific CD8+ T cells (1 × 105) established from PBMCs of 4 EBV-seropositive and HLA A24–positive donors were cocultured with T2-A24 cells (5 × 104) in wells in the presence of each peptide at a final concentration of 10 μM. When the spots were too many to count, 1 × 104 CD8+ T cells were used as responder cells and the numbers of spots were shown after multiplication by 10. We used 3 known EBV epitopes (nos. 43-45) and an HLA A24–restricted HIV-specific CD8+ T-cell epitope as control peptides. Each bar represents the average number of spots in duplicate wells.
Screening of HLA A*2402–binding candidate peptides from amino acid sequences of EBV proteins for CD8+ T-cell stimulation by ELISPOT assay.
Polyclonal EBV-specific CD8+ T cells (1 × 105) established from PBMCs of 4 EBV-seropositive and HLA A24–positive donors were cocultured with T2-A24 cells (5 × 104) in wells in the presence of each peptide at a final concentration of 10 μM. When the spots were too many to count, 1 × 104 CD8+ T cells were used as responder cells and the numbers of spots were shown after multiplication by 10. We used 3 known EBV epitopes (nos. 43-45) and an HLA A24–restricted HIV-specific CD8+ T-cell epitope as control peptides. Each bar represents the average number of spots in duplicate wells.
EBV-specific CD8+ T cells from donors 1 and 3 produced significant numbers of IFN-γ spots with peptide nos. 43, 44, and 45, which have been reported as HLA A*2402–restricted epitopes derived from EBNA3A (peptide no. 43),38 EBNA3B (peptide no. 44),2 and LMP2 (peptide no. 45),16respectively, indicating that the screening system with the ELISPOT assay functions efficiently.
In addition to these reported epitope peptides, CD8+ T cells from donor 1 produced significant numbers of IFN-γ spots when incubated with T2-A24 cells pulsed with peptides 11, 12, 14, 15, 16, 19, 21, 24, 25, and 29; those from donor 2 with peptides 13, 21, and 29; those from donor 3 with peptides 13, 14, 17, 24, and 38; and those from donor 4 with peptides 13, 21, and 29.
Tetramer-assisted TCR down-regulation assay
Peptides at high concentrations such as 10 μM in the ELISPOT assay can stimulate irrelevant T-cell repertoires with low-affinity TCR48 and there seems to be no evidence that these reacting peptides are endogenously processed and presented. We have adopted the novel strategy of confirming that the peptides are endogenously processed and presented with the aid of MHC/tetramer staining. For this purpose, we generated HLA A*2402 tetramers incorporating the 5 candidate peptides 11, 13, 21, 24, and 29, which were relatively commonly recognized in the ELISPOT assay, and 2 known epitopes, RYSIFFDYM (peptide no. 43) derived from EBNA3A,38 and TYSAGIVQI (peptide no. 44) derived from EBNA3B.2 Actually, the nonamer peptide RYSIFFDYM was used throughout this study instead of the reported octamer RYSIFFDY, because the nonamer gave high affinity to HLA A*2402 molecules, demonstrated by the MHC stabilization assay, and was recognized at lower concentrations by specific CD8+ T cells (data not shown).
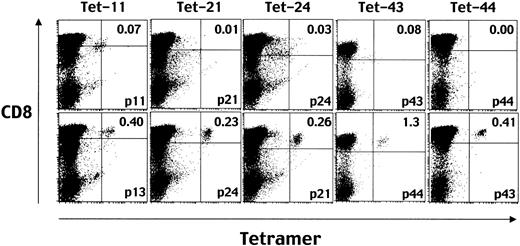
No cells in any EBV-specific CD8+ T cell line were stained with tetramers incorporating peptide no. 13 (Tet-13) and Tet-29 (data not shown). The reasons were not clear, but Tet-13 and Tet-29 were thus excluded from further studies. Tet-11 stained some proportion of CD8+ T cells from donor 1, Tet-21 stained those from all the donors, Tet-24 stained those from donors 1 and 3, Tet-43 stained those from donor 1, and Tet-44 stained those from donor 3 (data not shown). Thus, combinations of the CD8+ T cells from donor 1 and Tet-11, -21, -24, or -43, and those from donor 3 and the Tet-44 were used in further studies to demonstrate ligation-dependent TCR down-regulation.
It has been shown that stimulation with cognate peptides leads to rapid down-regulation of the TCR.42 43 To confirm the phenomenon for the epitopes identified here, CD8+ T cells from donor 1 were stimulated with peptides 11, 21, 24, and 43 for 16 hours and stained with Tet-11, -21, -24, and -43, respectively. CD8+T cells from donor 3 were stimulated with peptide no. 44 and stained with Tet-44. All aliquots of responder T cells were simultaneously stimulated with irrelevant peptides which can bind to HLA A*2402 molecules for control staining. The results are shown in Figure2. The percentage of tetramer-positive cells was specifically decreased in total CD8+ T cells that had been stimulated with the same peptide incorporated in the individual tetramer used for staining. These data not only confirmed the previous observation of rapid stimulation–induced TCR down-regulation but also provided evidence that the down-regulation occurs in an epitope-specific manner.
Down-regulation of TCR after stimulation with cognate peptides demonstrated by decreased levels of tetramer staining.
Before staining with each MHC/peptide tetramer (eg, Tet-11 means one incorporating peptide no. 11), aliquots of EBV-specific CD8+ T cells were incubated with T2-A24 cells in the presence of the same peptide used for making the indicated tetramer (eg, p11 for Tet-11, top row) or control HLA A24–binding peptide (eg, p13 for Tet-11, bottom row). After a 16-hour incubation, the cells were stained with phycoerythrin (PE)–conjugated HLA A*2402-tetramers and TRI-COLOR–labeled anti-CD8 mAb and analyzed by FACSCalibur. Numbers in the top right quadrants represent the percentages of tetramer-positive cells in the total CD8high cells.
Down-regulation of TCR after stimulation with cognate peptides demonstrated by decreased levels of tetramer staining.
Before staining with each MHC/peptide tetramer (eg, Tet-11 means one incorporating peptide no. 11), aliquots of EBV-specific CD8+ T cells were incubated with T2-A24 cells in the presence of the same peptide used for making the indicated tetramer (eg, p11 for Tet-11, top row) or control HLA A24–binding peptide (eg, p13 for Tet-11, bottom row). After a 16-hour incubation, the cells were stained with phycoerythrin (PE)–conjugated HLA A*2402-tetramers and TRI-COLOR–labeled anti-CD8 mAb and analyzed by FACSCalibur. Numbers in the top right quadrants represent the percentages of tetramer-positive cells in the total CD8high cells.
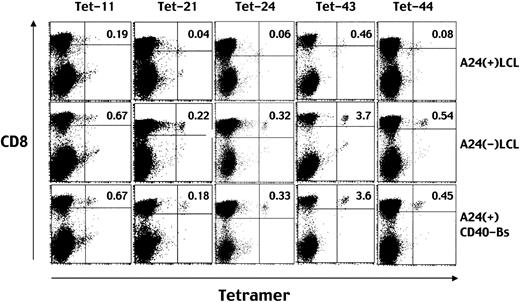
Next we prepared HLA A24–positive or –negative LCLs and CD40-Bs (EBV-negative) to study whether they could induce TCR down-regulation in epitope-specific T cells. As demonstrated in Figure3, the percentage of CD8+ T cells stained with either of the tetramers assembled with the 5 peptides decreased when stimulated with only HLA A*2402–positive LCLs, but not with HLA A24–negative LCLs or HLA A24–positive CD40-Bs. These data suggest specific TCR down-regulation to be caused by ligation of the TCR and HLA A*2402-EBV peptide complexes. In other words, the data suggest all the peptides are presented on the cell surfaces of HLA A24–positive LCLs, recognizable by specific T cells.
Down-regulation of TCR after stimulation with HLA A24–positive LCLs demonstrated by decreased levels of tetramer staining.
Before staining with each MHC/peptide tetramer (eg, Tet-11 means tetramer incorporating peptide no. 11), aliquots of EBV-specific CD8+ T cells were incubated with HLA A24–positive LCLs (top row), HLA A24–negative LCLs (middle row), or HLA A24–positive CD40-activated B cells (bottom row). After a 16-hour incubation, the cells were stained with PE-conjugated HLA A*2402 tetramers and TRI-COLOR–labeled anti-CD8 mAb, and analyzed by FACSCalibur. Numbers in the top right quadrants represent the percentages of tetramer-positive cells in the total CD8highcells.
Down-regulation of TCR after stimulation with HLA A24–positive LCLs demonstrated by decreased levels of tetramer staining.
Before staining with each MHC/peptide tetramer (eg, Tet-11 means tetramer incorporating peptide no. 11), aliquots of EBV-specific CD8+ T cells were incubated with HLA A24–positive LCLs (top row), HLA A24–negative LCLs (middle row), or HLA A24–positive CD40-activated B cells (bottom row). After a 16-hour incubation, the cells were stained with PE-conjugated HLA A*2402 tetramers and TRI-COLOR–labeled anti-CD8 mAb, and analyzed by FACSCalibur. Numbers in the top right quadrants represent the percentages of tetramer-positive cells in the total CD8highcells.
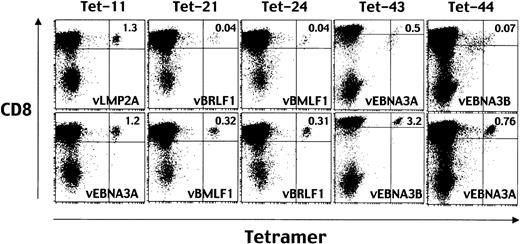
Finally, for the TCR down-regulation assay we prepared HLA A24–positive fibroblast cells infected with recombinant vaccinia viruses expressing each of the EBV genes as APCs. As shown in Figure4, the percentage of CD8+ T cells stained with Tet-21, -24, -43, and -44 decreased when stimulated with HLA A24–positive fibroblast cells infected with vBRLF1, vBMLF1, vEBNA3A, and vEBNA3B, respectively (upper panels). Each of the 4 genes encodes sequences of the epitope peptide assembled in the tetramer used for given staining. Stimulation with APCs expressing irrelevant EBV proteins did not cause TCR down-regulation (lower panels). Thus, peptide ligand–specific TCR down-regulation took place only when stimulated with APCs expressing the separate EBV gene of interest. This strongly indicates that the 4 EBV proteins are endogenously processed and presented for recognition by T cells. Taken together, the data demonstrated in Figures 2, 3, and 4 indicate that the newly identified peptide no. 21, TYPVLEEMF from BRLF1 and peptide no. 24, DYNFVKQLF from BMLF1, in addition to the peptides reported previously, no. 43, RYSIFFDYM, from EBNA3A and no. 44, TYSAGIVQI, from EBNA3B, are endogenously processed and presented by HLA A*2402–positive EBV-infected target cells. In contrast, the TCRs specific to HLA A*2402-peptide no. 11 complexes, identified by staining with Tet-11, were down-regulated when stimulated with T2-A24 cells pulsed with the same peptide (Figure 2) or HLA A24–positive LCLs (Figure 3), but not with the HLA A24–positive fibroblast cells infected with vLMP2 encoding the peptide no. 11 sequence (Figure 4).
Down-regulation of TCR after stimulation with HLA A24–positive fibroblast cells infected with recombinant vaccinia viruses expressing each of the EBV genes.
Before staining with Tet-11, Tet-21, Tet-24, Tet-43, or Tet-44, aliquots of EBV-specific CD8+ T cells were incubated with HLA A24–positive fibroblast cells infected with recombinant vaccinia virus expressing LMP2 (vLMP2), vBRLF1, vBMLF1, vEBNA3A, or vEBNA3B, respectively (top row). For control stimulation, aliquots of EBV-specific CD8+ T cells were incubated with HLA A24–positive fibroblast cells infected with vEBNA3A, vBMLF1, vBRLF1, vEBNA3B, or vEBNA3A, respectively (bottom row). After a 16-hour incubation, the cells were stained with PE-conjugated HLA A*2402 tetramers and TRI-COLOR–labeled anti-CD8 mAb and analyzed by FACSCalibur. Numbers in the top right quadrants represent the percentages of tetramer-positive cells in the total CD8high cells.
Down-regulation of TCR after stimulation with HLA A24–positive fibroblast cells infected with recombinant vaccinia viruses expressing each of the EBV genes.
Before staining with Tet-11, Tet-21, Tet-24, Tet-43, or Tet-44, aliquots of EBV-specific CD8+ T cells were incubated with HLA A24–positive fibroblast cells infected with recombinant vaccinia virus expressing LMP2 (vLMP2), vBRLF1, vBMLF1, vEBNA3A, or vEBNA3B, respectively (top row). For control stimulation, aliquots of EBV-specific CD8+ T cells were incubated with HLA A24–positive fibroblast cells infected with vEBNA3A, vBMLF1, vBRLF1, vEBNA3B, or vEBNA3A, respectively (bottom row). After a 16-hour incubation, the cells were stained with PE-conjugated HLA A*2402 tetramers and TRI-COLOR–labeled anti-CD8 mAb and analyzed by FACSCalibur. Numbers in the top right quadrants represent the percentages of tetramer-positive cells in the total CD8high cells.
Peptide no. 11 is presented on HLA 24–positive LCLs, IFN-γ–treated fibroblast cells, and CD40-Bs infected with vLMP2
Absence of down-regulation of TCR stained with Tet-11 after stimulation by HLA A24–positive fibroblast cells infected with vLMP2 (Figure 4) indicated failure of presentation of the epitope on the cell surface. This could be due to differential processing and/or presentation of peptide no. 11 in different APCs. To elucidate this point, we established a CD8+ T-cell clone specific to peptide no. 11 IYVLVMLVL, designated as IYV-CTL, and one specific to peptide no. 43 RYSIFFDYM as a control clone, designated as RYS-CTL, by limiting dilution of the EBV-specific CD8+ T-cell line from donor 1.
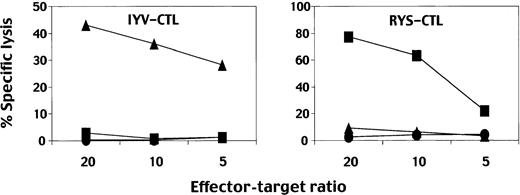
As demonstrated in Figure 5, neither CTL clone efficiently killed autologous LCLs unless cognate peptides were pulsed, which is not uncommon for some EBV-specific CTL clones stimulated with autologous LCLs for growing.49,50 This could be due to the insensitive nature of the 51Cr-release assay if small percentages of the target cells present sufficient peptide/MHC complexes to stimulate cytolysis. On the other hand, assays applying cytokine release could be useful when no less than 1% of the stimulating LCLs display them, especially performed at higher stimulator-to-CTL ratios.49 Thus, further studies were performed applying the ELISPOT assay. Figure6 demonstrates that both IYV-CTL and RYS-CTL produced IFN-γ spots when stimulated with HLA A24–positive LCLs but not with HLA A24–negative LCLs (left boxes). The IYV-CTL did not recognize fibroblast cells infected with vLMP2 (Figure 6, upper right box), which is in accordance with the data shown in Figure 4. In contrast, the RYS-CTL recognized fibroblast cells infected with vEBNA3A (Figure 6, lower right box). The data suggest that peptide no. 11 was inefficiently or not at all processed and/or presented in the fibroblast cells. CD8+ T cells specific to another LMP2 epitope, TYGPVFMCL, produced IFN-γ when stimulated with the same APCs (data not shown), suggesting the defect of presentation to be restricted to peptide no. 11. Then we asked whether IFN-γ treatment, which has been reported to enhance antigen presentation,51could render positive epitope processing and presentation. IYV-CTL produced IFN-γ spots when stimulated with fibroblast cells infected with vLMP2 but not with vEBNA3A, both of which had been incubated with IFN-γ, although the numbers of spots were relatively small (Figure 6, upper right box). RYS-CTL produced more IFN-γ spots when stimulated with fibroblast cells treated with IFN-γ prior to vEBNA3A infection than with infected cells without IFN-γ treatment (Figure 6, lower right box). CD40-Bs proved able to process and present peptide no. 11, suggesting possession of antigen-presenting machinery closer to that in IFN-γ–treated as opposed to untreated fibroblast cells (Figure 6, upper right box).
Results of CTL assays using CD8+ T-cell clones.
CTL assays were performed using 51Cr-labeled autologous LCLs as target cells. IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. The CTLs were incubated with the target cells in the presence of 100 nM peptide, IYVLVMLVL (▴), RYSIFFDYM (▪), or in the absence of peptide (●) at the indicated effector-target ratio.
Results of CTL assays using CD8+ T-cell clones.
CTL assays were performed using 51Cr-labeled autologous LCLs as target cells. IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. The CTLs were incubated with the target cells in the presence of 100 nM peptide, IYVLVMLVL (▴), RYSIFFDYM (▪), or in the absence of peptide (●) at the indicated effector-target ratio.
Results of ELISPOT assays using CD8+ T-cell clones.
IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. LCL1 and 2 were established from PBMCs of HLA A24–positive donors, and LCL3 and 4 from HLA A24–negative donors. Abbreviations used for describing the stimulators are as follows: IYV, T2-A24 cells pulsed with peptide IYVLVMLVL; RYS, T2-A24 cells pulsed with peptide RYSIFFDYM; vL2, cells infected with vLMP2; vE3A, cells infected with vEBNA3A. CD8+ T cells (200 [▪]; 100 [░]; 50 [ ]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.
]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.
Results of ELISPOT assays using CD8+ T-cell clones.
IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. LCL1 and 2 were established from PBMCs of HLA A24–positive donors, and LCL3 and 4 from HLA A24–negative donors. Abbreviations used for describing the stimulators are as follows: IYV, T2-A24 cells pulsed with peptide IYVLVMLVL; RYS, T2-A24 cells pulsed with peptide RYSIFFDYM; vL2, cells infected with vLMP2; vE3A, cells infected with vEBNA3A. CD8+ T cells (200 [▪]; 100 [░]; 50 [ ]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.
]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.
Minigene constructs present peptide no. 11 in TAP-negative T2-A24 cells
As a final set of experiments to characterize processing of peptide no. 11, we constructed plasmids encoding the minimal epitopes, peptide no. 11, IYVLVMLVL (pcIYV), and peptide RYSIFFDYM (pcRYS). Both the minigenes start with methionine, which should be cleaved by aminopeptidase in the endoplasmic reticulum52 and end with a stop codon just after the C-terminus amino acid. HEK293T or T2-A24 cells were independently transfected with pcDNA3 encoding full-length LMP2 (pcLMP2), the EBNA3A (pcEBNA3A) gene, or minimal epitopes. The plasmid expressing HLA A*2402 was cotransfected into HEK293T cells. The results are summarized in Figure 7. IYV-CTL produced IFN-γ spots when stimulated with HEK293T cells transfected with pcIYV but not with pcLMP2 (Figure 7, upper left box), indicating there is a defect in processing but not presentation of the IYVLVMLVL, probably as is the case with fibroblast cells not treated with IFN-γ. In contrast, RYS-CTL produced IFN-γ spots when incubated with HEK293T cells transfected with either pcEBNA3A or pcRYS (Figure 7, lower left box). Next we used TAP-deficient T2-A24 cells as APCs to study whether TAPs are necessary for the epitope presentation. IYV-CTL produced IFN-γ spots when incubated with T2-A24 cells transfected with pcIYV, indicating that IYVLVMLVL can be presented TAP-independently (Figure 7, upper right box). IYV-CTL did not produce IFN-γ spots when incubated with T2-A24 cells transfected with pcLMP2 (Figure 7, upper right box), indicating some defect in processing to yield the peptides in T2-A24 cells, as is the case with HEK293T cells. In addition, IYV-CTL did not produce IFN-γ spots when incubated with T2-A24 cells infected with vLMP2 (data not shown). In contrast, RYS-CTL produced few IFN-γ spots when incubated with T2-A24 cells transfected with either pcRYS or pcEBNA3A, indicating that presentation of RYSIFFDYM is TAP dependent (Figure 7, lower right panel).
Presentation of IYVLVMLVL is TAP independent.
IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. HEK293T or T2-A24 cells were transfected with pcDNA3 expressing LMP2 (pcL2), pcDNA3 expressing EBNA3A (pcE3A), pcDNA3 expressing the minimal IYVLVMLVL epitope (pcIYV), or pcDNA3 expressing the minimal RYSIFFDYM epitope (pcRYS), and used as APCs. Plasmids expressing HLA A*2402 were cotransfected into HEK293T cells. T2-A24 cells pulsed with the peptides IYVLVMLVL (IYV) or RYSIFFDYM (RYS) were also used as stimulators. CD8+ T cells (200 [▪]; 100 [░]; 50 [▨]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.
Presentation of IYVLVMLVL is TAP independent.
IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. HEK293T or T2-A24 cells were transfected with pcDNA3 expressing LMP2 (pcL2), pcDNA3 expressing EBNA3A (pcE3A), pcDNA3 expressing the minimal IYVLVMLVL epitope (pcIYV), or pcDNA3 expressing the minimal RYSIFFDYM epitope (pcRYS), and used as APCs. Plasmids expressing HLA A*2402 were cotransfected into HEK293T cells. T2-A24 cells pulsed with the peptides IYVLVMLVL (IYV) or RYSIFFDYM (RYS) were also used as stimulators. CD8+ T cells (200 [▪]; 100 [░]; 50 [▨]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.
Discussion
Antigenic peptides recognized by virus-specific CTLs are not only useful tools for studying CTL responses but also potential reagents for immunotherapy of those who have presenting MHC class I molecules. For widening the application, efficient strategies to determine such epitopes are a high priority. Meij et al21 recently described an efficient approach using an IFN-γ ELISPOT assay to directly screen PBMCs with 15 mer peptides overlapping by 10 amino acids spanning the LMP2 sequences. However, it would be difficult to identify class I molecules presenting the epitopes and to confirm natural epitope processing with this system. When investigators are interested in particular HLA alleles, prediction of peptide sequences with a specific binding motif could be an option. In our study, potential HLA A24–binding peptides were synthesized after analyzing amino acid sequences of EBV proteins including LMP2 using a computer-assisted program.36,37 Notably, 2 of 3 known A24 epitopes from EBV had estimated t1/2dissociation scores below 100 (Table 1). Although first screening of candidate peptides based on the binding motif is useful,23,33 39 the observation implies that there is a risk of missing epitopes that score lower than the cutoff value applied.
We have employed LCL-stimulated polyclonal EBV-specific CD8+ T cells as effector cells in the ELISPOT assay to ensure a concentration of T cells specific to naturally processed peptides presented by HLA A*2402 molecules on the LCLs used for stimulation. In addition, a novel approach to confirm endogenous processing and presentation was introduced, consisting of (1) production of HLA A*2402 tetramers incorporating each of the candidate peptides; (2) staining the polyclonal EBV-specific CD8+T-cell lines with tetramers; and (3) detecting decline of tetramer staining on the same T-cell lines, which had been stimulated with HLA A*2402–positive cells, whereby EBV genes encoding the epitopes were translated, processed, and presented.
EBV has 2 types of replication cycles, namely, lytic infection, where infectious virions are produced, and latent infection represented by the majority of LCLs.1 This is why the percentages of Tet-21– and Tet-24–staining cells (specific to BRLF1 and BMLF1 proteins, respectively) in polyclonal CTL lines are relatively low and our stimulation protocol is unlikely to be optimal for reactivating and then growing lytic-cycle–specific CTLs. On the other hand, TCRs stained with Tet-21 and Tet-24 were specifically decreased in intensity after stimulation with HLA A24–positive LCLs. Thus, both T cells specific for latent proteins (stained with Tet-43 and Tet-44) and those specific for lytic proteins should be effectively stimulated for TCR down-regulation in our protocol. Most importantly, experiments using recombinant vaccinia viruses demonstrated that TCR down-regulation took place only on CD8+ T cells stimulated with APCs that expressed EBV genes encoding sequences of peptides incorporated in the tetramers used for TCR detection.
Staining patterns using Tet-11 were unique because the TCRs specific to the HLA A*2402-peptide no. 11 complexes were down-regulated with HLA A24–positive LCL stimulation, but not with HLA A24–positive fibroblasts infected with vLMP2 encoding the peptide no. 11 sequence. This observation indicates the peptide to be present on the LCLs but not on vLMP2-infected fibroblasts. The defective presentation of vLMP2-infected fibroblast cells was confirmed using a CD8+T-cell clone specific to peptide no. 11, IYVLVMLVL, designated as IYV-CTL. Defective processing of peptide no. 11 was also demonstrated in the experiment using other APCs, such as HEK293T and T2-A24 cells transfected with pcDNA3 expressing LMP2. Interestingly, IYV-CTL produced IFN-γ spots when incubated with both HEK293T and T2-A24 cells transfected with pcDNA3 encoding the minimal epitope sequence, indicating a defect in processing but not presentation of IYVLVMLVL in the APCs. The results also indicate that IYVLVMLVL is presented in a TAP-independent manner, at least expressed as a minimal construct. Because IFN-γ treatment of fibroblasts partially renders epitope presentation, studies are underway in our laboratory to identify the responsible gene(s) that is (are) inducible by IFN-γ treatment and able to restore the epitope processing and presentation in APCs expressing full-length LMP2 gene products.
EBV LMP2 is a multiple membrane–spanning molecule, within which a number of epitopes recognized by human CD8+ CTLs are included. Recently, Lautscham et al54 reported a clear correlation between hydrophobicity of the LMP2 epitope sequence and TAP independence. Our data for the newly identified HLA A*2402–restricted epitope, IYVLVMLVL, is in accordance with the conclusions they drew. The amino acid sequence has a high hydrophobicity value of 8.7,55 and lies within the fourth membrane-associate domain of LMP2. The hydrophobicity value of the control peptide RYSIFFDYM is 0.67. TAP-independent presentation of IYVLVMLVL was confirmed using a TAP-negative cell line, T2-A24, transfected with plasmid encoding the minimal peptide. In addition, to our knowledge IYVLVMLVL is the first LMP2 epitope that is not presented on HLA-matched fibroblast cells unless treated with IFN-γ prior to infection with vLMP2. To investigate the utility of the peptide for treatment of EBV-positive malignancies, we plan to stain their infiltrating T cells with the tetramer.
Tetramers incorporating peptide no. 29 (Tet-29) were found to be unstable, as disclosed by the final gel filtration profile (data not shown). We could not deny the possibility that peptide no. 29 is an epitope. It is derived from BMRF1, which is produced abundantly in the early lytic cycle of EBV infection.53 Even though peptide no. 29 demonstrated a low affinity for HLA A*2402 molecules, resulting in a short half-life for MHC-peptide complexes on the cell surface, a large amount of peptides might compensate to sustain the ligand density sufficient to elicit CTL responses. To answer the question of whether peptide no. 29 is really processed and presented, CTL clones specific to TYTSGEACL need to be established for testing reactivity to target cells with expression of HLA A*2402 and BMRF1 genes. This has not been achieved so far. On the other hand, there were no problems regarding gel filtration profiles when making tetramers with peptide no. 13 (Tet-13), suggesting stable assembly into MHC molecules. Nevertheless, EBV-specific CD8+ T-cell lines established from PBMCs of donors 2, 3, and 4, which had been shown to produce IFN-γ spots when incubated with the peptide (Figure 1), were not stained with Tet-13 at all (data not shown). Because the screening ELISPOT assays were performed with peptides at a concentration of 10 μM, the high concentration of peptide no. 13 might have triggered T-cell responses with low-affinity TCR48 inefficient for stable binding to Tet-13.
In conclusion, we have identified an LMP2 epitope presented by HLA A*2402 molecules in addition to 2 lytic-cycle EBV-specific CTL epitopes from the amino acid sequences of BRLF1 and BMLF1. The LMP2-derived peptide, IYVLVMLVL, is processed and presented in a manner that appears IFN-γ dependent and TAP independent. The newly identified peptides should prove useful for detection and/or study of EBV-specific CD8+ T-cell responses among populations positive for HLA A*2402.
The authors thank Dr Toshitada Takahashi for critically reading the manuscript.
Prepublished online as Blood First Edition Paper, September 26, 2002; DOI 10.1182/blood-2002-04-1240.
Supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (no. 13218152) and the Japan Society for the Promotion of Science (no. 12670802).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kiyotaka Kuzushima, Division of Immunology, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chigusa-ku, Nagoya 464-8681 Japan; e-mail:kkuzushi@aichi-cc.jp.

![Fig. 6. Results of ELISPOT assays using CD8+ T-cell clones. / IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. LCL1 and 2 were established from PBMCs of HLA A24–positive donors, and LCL3 and 4 from HLA A24–negative donors. Abbreviations used for describing the stimulators are as follows: IYV, T2-A24 cells pulsed with peptide IYVLVMLVL; RYS, T2-A24 cells pulsed with peptide RYSIFFDYM; vL2, cells infected with vLMP2; vE3A, cells infected with vEBNA3A. CD8+ T cells (200 [▪]; 100 [░]; 50 []; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/4/10.1182_blood-2002-04-1240/3/m_h80433810006.jpeg?Expires=1769407863&Signature=byZdIxPqnqO3E13DYUQBgWCIRlVWgEA4NkbXYFEqavnfp4Xn2CFu~1cGyzV655ewyAeZrNQVROb5BL8bdLlODY1IXKs8pd5VDe7Aqqu6uhvacTR-zGVDpSBDzVSA580zYAcMjBABnRVNfO44F8BocUgdnVque0URFclqffywp6tFvy3AuIVecACocH~V2AN24Y~JgmTefTxt9MVRITvz8sr2ixkRS5iBj-Fl1cLXgyLOCCKK~GKDtmYvgEY-6GngxUs6fEA24IcmzEzvS6847m29Pscgq5m4p~lZ~Vew-j9Cl67zvnYX-MPqwH7IQDFsghiaL2LdMxEWXYXYmn-U-g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Presentation of IYVLVMLVL is TAP independent. / IYV-CTL is a clone specific for peptide no. 11, IYVLVMLVL; RYS-CTL, a clone specific for peptide RYSIFFDYM. HEK293T or T2-A24 cells were transfected with pcDNA3 expressing LMP2 (pcL2), pcDNA3 expressing EBNA3A (pcE3A), pcDNA3 expressing the minimal IYVLVMLVL epitope (pcIYV), or pcDNA3 expressing the minimal RYSIFFDYM epitope (pcRYS), and used as APCs. Plasmids expressing HLA A*2402 were cotransfected into HEK293T cells. T2-A24 cells pulsed with the peptides IYVLVMLVL (IYV) or RYSIFFDYM (RYS) were also used as stimulators. CD8+ T cells (200 [▪]; 100 [░]; 50 [▨]; or 25 [■]) were cultured with the indicated APCs in each well. Each bar represents the average number of spots in duplicate wells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/4/10.1182_blood-2002-04-1240/3/m_h80433810007.jpeg?Expires=1769407863&Signature=BBsroaqzKjUS3jjgBh8i23-riVGi7YbMq2Yotjs0Qrsh3XI4VYVTC4pCRetC6iuWN3VgVz5p6P37brDRbMxsgXK4BBI71qhCQzAiTsMS8uSv2~jWFpaFvYLPbIPrlffvavMAk56Y6KUC5q2DRfvkxQ9GzpYoP-UspET3gNvNoRZ4h49EXed~ozT1E9f7DgeOXzDm-PjTze8FAsCtq7oI7fwCwelE-64Cb67b~hfTVBYxAyO5cPbLKJnu9zPf5fx9BNCQwgY9ikZZ8ZRnOugNqbeMl8U3uBD4c8oPxOpsIp0T68wUPcmC9WFaNX0GPmfqz8gchvBbXx~lUsPALAwRXw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal