Previous studies demonstrated that integrin αMβ2 (CD11b/18, Mac-1) forms a physical complex with the urokinase-type plasminogen activator receptor (uPAR/CD87) on leukocytes. In this study, we used human peripheral blood neutrophils and transfected cells expressing αMβ2, uPAR, or both receptors to show that the integrin can directly interact with urokinase (uPA). We demonstrate that αMβ2 supported adhesion and migration of these cells to uPA, and, in each case, blockade of αMβ2 suppressed the response. Within uPA, both the kringle and proteolytic domains are recognized by αMβ2, which are distinct from the growth factor domain that binds to uPAR. Within the αM subunit of the integrin, the I domain interacts with uPA, which is distinct from the region that interacts with uPAR. On cells expressing uPAR and αMβ2, both receptors mediated adhesion and migration. This cooperation was particularly apparent in the responses of neutrophils to uPA, where blockade of αMβ2 reduced uPAR-mediated responses and engagement of uPAR enhanced recognition of uPA by αMβ2. Thus, recognition of uPA by αMβ2 allows for formation of a multicontact trimolecular complex, in which a single uPA ligand may bind simultaneously to both uPAR and αMβ2. This complex may play an important role in the control of inflammatory cell migration and vascular homeostasis.
Introduction
Cell migration is a complex response that requires the coordination and cooperation among multiple cell surface receptors, including sensory receptors that detect migratory stimuli, adhesion receptors that mediate interactions of migrating cells with the extracellular matrix, and protease receptors that facilitate movement of cells through their extracellular environment. Members of the integrin family of receptors recognize many extracellular matrix proteins to which cells adhere and de-adhere as they migrate. Binding partners for proteins of the plasmin(ogen)/fibrinolytic system, such as plasminogen-binding proteins and urokinase-type plasminogen activator receptor (uPAR), focus proteolytic activity, particularly to the leading edge of migrating cells, thus facilitating degradation and remodeling of the extracellular matrix.1-5 Cooperation between these 2 cell surface systems is particularly evident in the recruitment of leukocytes during inflammatory responses, whether outside the vasculature or inside the vessel wall. Mice rendered deficient in members of the β2 subfamily of leukocyte integrins or in components of the plasminogen system exhibit blunted inflammatory responses in a variety of in vivo models.6-11
Evidence for direct interplay between integrins and the fibrinolytic system at cell surfaces has emerged over the past several years. For example, vitronectin, a matrix protein recognized by several integrins, was shown to be a ligand for uPAR,12 and urokinase (uPA) binding to uPAR was shown to change the specificity of several integrins.13,14 Plasminogen activator inhibitor 1 (PAI-1), an inhibitor of uPA,15 bound to vitronectin,16,17 thereby modulating cell migration by inhibiting vitronectin binding to integrins and to uPAR.18,19 An interaction critical to several of these functional relationships between the 2 systems is the direct interaction between uPAR and the integrins.1,20 Several reports of integrin/uPAR interaction have centered on αMβ2 (Mac-1, CD11b/CD18, CR3) of the β2 subfamily of leukocyte integrins. uPAR and αMβ2 copurify in monocyte lysates21 and form a reversible complex on neutrophils as detected by immunolocalization and fluorescence resonance energy transfer studies.22,23 Recently, a peptide sequence from within the αM subunit (M25, residues 424-440)24 was shown to bind uPAR and disrupt its complexes with αMβ2.1,24 The interaction between uPAR and αMβ2influences the functions of each other. Complex formation between uPAR and αMβ2 enhances αMβ2-mediated adhesion to fibrinogen (Fg) and other ligands of this integrin, as well as degradation of Fg by human monocytes.14 uPAR engagement of vitronectin also facilitates αMβ2 function.25On the other hand, αMβ2 activation increases uPAR-dependent cell adhesion to vitronectin.14,25 The importance of the uPA/uPAR system in the regulation of αMβ2-dependent leukocyte functions is underscored by several recent observations. uPAR-deficient mice exhibit reduced neutrophil recruitment to inflammatory stimuli.10 In addition, uPA- and uPAR-deficient mice exhibit impaired T-cell and macrophage recruitment toCryptococcus neoformans, which results in increased mortality of these animals during infection.6
In the present study, we demonstrate an additional and potentially important element to the interrelationship between uPAR and αMβ2. uPA is shown to be a ligand for αMβ2. Moreover, the region of αMβ2 that binds to uPA is different from that which binds to uPAR, and the regions of uPA that bind to αMβ2 are different from those that bind to uPAR. Therefore, a trimolecular complex of uPA/uPAR/αMβ2, in which uPA binds to both receptors, may form on the leukocyte surface and display unique functional properties.
Materials and methods
Reagents, antibodies, and synthetic peptides
Recombinant human high-molecular-weight (HMW)–tc-uPA was a generous gift from Jack Henkin (Abbott Laboratories, Chicago, IL). Neutrophil inhibitory factor (NIF) was provided by Corvas International (San Diego, CA). The recombinant uPA domains growth factor domain (GFD; residues 4-43), kringle domain (KD; residues 47-135), and low-molecular-weight (LMW)–tc-uPA (residues 136-411) were purchased from Calbiochem (San Diego, CA). Recombinant human intercellular adhesion molecule 1 (ICAM-1) was from R & D Systems (Minneapolis, MN) and human Fg from Enzyme Research Labs (South Bend, IN). The Cyquant-Cell Proliferation Kit, phosphatidylinositol-specific phospholipase C (PI-PLC), and Alexa 488–labeled goat antimouse IgG were purchased from Molecular Probes (Eugene, OR). The αMI domain was expressed as glutathione-S-transferase (GST)–fusion protein and purified on glutathione-Sepharose 4B (Pharmacia Biotech, Piscataway, NJ) as previously described.26Monoclonal antibody (mAb) CBRM1/527 was kindly provided by Dr T. Springer (Harvard Medical School, Boston, MA). The mAbs 44a, IB4, and W6/32 were from American Type Culture Collection (Rockville, MD); mAb P4H9 was from Chemicon International (Temecula, CA); and uPAR mAbs 62022.11 and 3936 were from R & D Systems and American Diagnostica (Greenwich, CT), respectively. M25 (PRYQHIGLVAMFRQNTG) and control scrambled M25 (M25 SCR; HQIPGAYRGVNQRFTLM)13 24 peptides were synthesized on an Applied Biosystems model 430A peptide synthesizer (Foster City, CA) using N-(9-fluorenyl)methoxycarbonyl chemistry.
Cell lines and neutrophil preparations
Granulocytes were isolated from human peripheral blood of healthy volunteers drawn into sterile acid-citrate-dextrose (1:7 volume 145 mM sodium citrate, pH 4.6, and 2% dextrose). Isolation was performed by density gradient centrifugation onto Ficoll-Hypaque (Pharmacia, Uppsala, Sweden), followed by dextran sedimentation of erythrocytes and hypotonic lysis of residual erythrocytes. The remaining cells were 98% granulocytes, of which more than 96% were neutrophils and 2% were eosinophils. Contaminating lymphocytes and monocytes were less than 2% as determined by Wright staining.
Human epithelial kidney 293 cells transfected with αMβ2 were maintained as described previously.28 Nontransfected and αΜβ2 293 cells were stably transfected in the absence of serum using the LipofectAMINE Plus reagent (Invitrogen, Carlsbad, CA) with 0.5 to 5 μg pcDNA 3.1 containing the cDNA for uPAR or vector alone. Cell clones were selected using neomycin sulfate (Invitrogen), and cells expressing uPAR were detected and isolated by fluorescence-activated cell sorting (FACS).
Soluble HMW-tc-uPA binding
uPA was labeled with Alexafluor-488 according to manufacturer's protocol (Molecular Probes). Unstimulated, phorbol myristate acetate (PMA)–treated human neutrophils or HEK293 cells were suspended in Dulbecco modified Eagle medium (DMEM)/F-12 medium, 1 mM Mg2+, 0.1% bovine serum albumin (BSA), and 100 nM Alexa 488–HMW-tc-uPA and incubated at 37°C for 0 to 4 hours. Cells were centrifuged through a cushion of fetal calf serum (FCS) twice and resuspended in 1% paraformaldehyde/phosphate-buffered saline (PBS). Cell-bound HMW-tc-uPA was detected by FACS.
Adhesion assays
The 96-well nontissue culture–treated plates (Falcon, Becton Dickinson, San Diego, CA) were coated with HMW-tc-uPA, GFD, KD, LMW-tc-uPA, or BSA (200 nM in PBS) for 3 hours at 37°C and then blocked with 0.5% polyvinylpyrrolidone (PVP) for 1 hour at room temperature. The 293 cells and neutrophils were resuspended in the serum-free DMEM/F-12 medium. Neutrophils were stimulated with 20 nM PMA (Sigma Chemical, St Louis, MO) for 20 minutes at 37°C. Only about 5% to 8% of neutrophils were apoptotic as determined by fluorescein isothiocyanate (FITC)–labeled annexin V binding (R & D Systems). In inhibition experiments, the cells were pretreated with respective antibodies or reagents for 30 minutes at 37°C, then seeded at 1 to 2 × 105 cells/well onto the coated plates and incubated at 37°C for 30 minutes. The plates were washed with PBS, and the number of adherent cells in each well was quantified using the Cyquant Cell Proliferation Assay Kit (Molecular Probes), according to the manufacturer's instructions. The data from cell adhesion and migration assays are presented as the total number of adherent or migrated cells, determined from standard curves developed with a known number of Cyquant-labeled cells.
Migration assays
Cell migration assays were performed in serum-free DMEM/F-12 medium using Costar 24-transwell plates with 3 μm (for neutrophils) or 8 μm (for 293 cells) pore polycarbonate filters (Corning, Corning, NY). HMW-tc-uPA and its domains (0-100 nM) were added to the lower chambers in total volume of 600 μL medium, whereas the upper wells contained a final volume of 200 μL after addition of the cells. To commence the assay, 50 μL cell suspension (2 × 105cells/well) was added to the upper chambers, and the plates were placed in a humidified incubator at 37°C and 5% CO2. For inhibition experiments, the function-blocking mAbs were added to the upper chambers at 20 μg/mL. Assays were stopped after 6 hours by removing the upper wells and wiping the inside of upper wells with a cotton swab to remove nonmigrated cells. The migrated cells were quantitated using the Cyquant Cell Proliferation Kit.
Results
Neutrophil responses to uPA are mediated by both uPAR and αMβ2
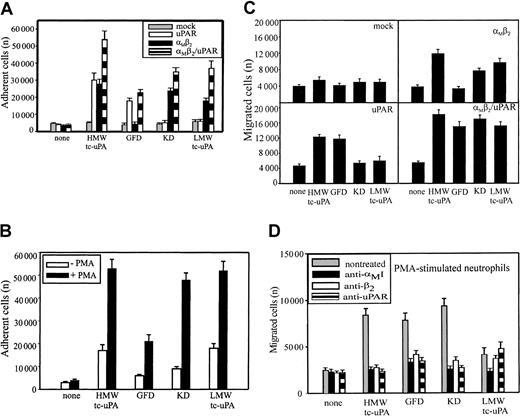
Most studies of uPAR/integrin interactions have emphasized the influence of uPAR on integrin-mediated responses. In view of the physical association between αMβ2 and uPAR on the surface of leukocytes,13,21-23 we hypothesized that the integrin might influence uPAR-dependent responses. Because the uPA/uPAR system is implicated in cell migration, a process indispensable in inflammation, we first assessed the influence of αMβ2 on neutrophil migration toward uPA. Consistent with previous reports,29 neutrophils migrated toward HMW-tc-uPA; the increase in migration to uPA was 3-fold greater than to buffer (Figure 1A). The function-blocking mAb (clone 62022.11) to uPAR inhibited this migration to background levels (Figure 1A). When 2 different mAbs to αMβ2 were added, they were as effective as anti-uPAR in suppressing the migratory response. One of these mAbs (44a) was directed to the αM and the other (IB4) to the β2 subunit. Each completely blocked neutrophil migration to uPA, whereas a control mAb, W6/32, to an unrelated surface antigen (major histocompatability complex class I [MHC-I]), had no effect.
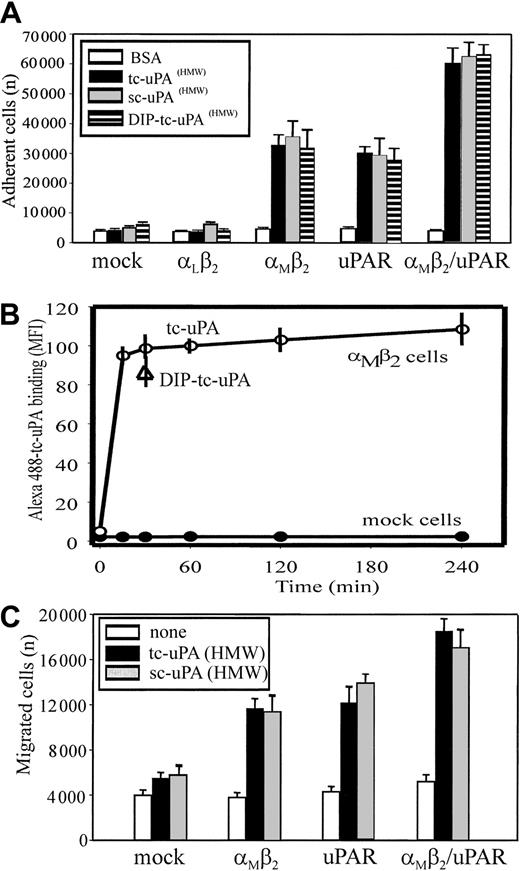
Recognition of uPA by neutrophils is αMβ2 integrin- and uPAR-dependent.
(A) Migration to uPA. Human polymorphonuclear cells (PMNs) were resuspended in a serum-free DMEM/F-12 medium containing PMA (20 nM) and added (2 × 105 cells/well) to the upper chambers of transwells. HMW-tc-uPA was added to the lower chambers, and selected function-blocking mAbs (20 μg/mL) were added to both chambers. PMNs were allowed to migrate to the lower chamber for 6 hours in a humidified incubator at 37°C and 5% CO2. The number of migrated cells was determined as described in “Materials and methods.” The data are expressed as means ± SEM of duplicate wells from 3 independent experiments. (B) Adhesion to uPA. PMA-stimulated PMNs (1 × 105 cells/well) were seeded onto wells of microtiter plates coated with HMW-tc-uPA or BSA and allowed to adhere for 30 minutes at 37°C. Nonadherent PMNs were removed by 3 washings with PBS, and representative photomicrographs of the wells were taken (Original magnification, × 200). (C) Direct binding of uPA. Unstimulated or PMA-stimulated PMNs (20 nM, 20 minutes at 37°C) were incubated in DMEM/F-12 medium/0.1% BSA with 100 nM Alexa 488–HMW-tc-uPA in the absence or presence of the indicated blocking mAbs for 30 minutes at 37°C. Cells were centrifuged through a cushion of FCS twice and resuspended in 1% paraformaldehyde/PBS. Cell-bound HMW-tc-uPA was detected by FACS analysis.
Recognition of uPA by neutrophils is αMβ2 integrin- and uPAR-dependent.
(A) Migration to uPA. Human polymorphonuclear cells (PMNs) were resuspended in a serum-free DMEM/F-12 medium containing PMA (20 nM) and added (2 × 105 cells/well) to the upper chambers of transwells. HMW-tc-uPA was added to the lower chambers, and selected function-blocking mAbs (20 μg/mL) were added to both chambers. PMNs were allowed to migrate to the lower chamber for 6 hours in a humidified incubator at 37°C and 5% CO2. The number of migrated cells was determined as described in “Materials and methods.” The data are expressed as means ± SEM of duplicate wells from 3 independent experiments. (B) Adhesion to uPA. PMA-stimulated PMNs (1 × 105 cells/well) were seeded onto wells of microtiter plates coated with HMW-tc-uPA or BSA and allowed to adhere for 30 minutes at 37°C. Nonadherent PMNs were removed by 3 washings with PBS, and representative photomicrographs of the wells were taken (Original magnification, × 200). (C) Direct binding of uPA. Unstimulated or PMA-stimulated PMNs (20 nM, 20 minutes at 37°C) were incubated in DMEM/F-12 medium/0.1% BSA with 100 nM Alexa 488–HMW-tc-uPA in the absence or presence of the indicated blocking mAbs for 30 minutes at 37°C. Cells were centrifuged through a cushion of FCS twice and resuspended in 1% paraformaldehyde/PBS. Cell-bound HMW-tc-uPA was detected by FACS analysis.
Soluble complexes of uPA and uPAR can be sequestered by vitronectin within the extracellular matrix30 and uPA can bind to uPAR on the surfaces of cells (for reviews, see Chapman and Wei1 and Preissner et al4). Such interactions would present uPA as an immobilized substrate, which could support adhesion of αMβ2-bearing cells. To test this possibility, neutrophils were added to plates coated with BSA or HMW-tc-uPA, and adhesion was measured after 30 minutes at 37°C. As shown in Figure 1B, when neutrophils were stimulated with PMA, they attached and spread on HMW-tc-uPA but not BSA. Nonstimulated neutrophils showed little adhesion to either substratum. The mAbs not only to uPAR but also to αMβ2 reduced adhesion of the stimulated neutrophils onto HMW-tc-uPA (Figure 1B). Control mAb W6/32 or nonimmune mouse IgG decreased adhesion by less than 10% under these conditions (not shown). Taken together, these data suggest a significant role for both uPAR and αMβ2 in the adhesive and migratory response of human neutrophils to uPA.
Migration and adhesion are complex responses, and we sought to more directly examine the role of the 2 receptors in uPA recognition by measuring the binding of Alexa 488–labeled HMW-tc-uPA to neutrophils by FACS (Figure 1C). Unstimulated neutrophils showed little binding of soluble Alexa 488–labeled HMW-tc-uPA (mean fluorescence intensity [MFI] = 10.9); however, on stimulation of the cells with PMA, binding increased by 8-fold (MFI = 92.3). The Alexa 488–HMW-tc-uPA binding was completely inhibited by a 50-fold molar excess of unlabeled HMW-tc-uPA, verifying specificity (data not shown). Binding of labeled HMW-tc-uPA to PMA-stimulated cells was significantly abrogated by blocking mAbs to uPAR (MFI = 32.3) and to the αM(MFI = 12.1) and β2 subunits (MFI = 16.8) of αMβ2. Control mAb W6/32 or nonimmune mouse IgG did not inhibit uPA binding to neutrophils (data not shown). In addition, NIF, a high-affinity ligand of the αMI domain of αMβ2,31 32 also blocked binding of the Alexa 488–labeled HMW-tc-uPA to the cells. Thus, recognition of uPA by neutrophils and the functional responses of the cells to uPA clearly are influenced by αMβ2as well as by uPAR.
αMβ2 recognizes HMW-tc-uPA and uPAR enhances the interaction
A likely explanation for these observations is that αMβ2 may directly recognize uPA. This possibility was pursued using transfected HEK293 cells expressing αMβ2, uPAR, or both αMβ2/uPAR. FACS analyses were performed to estimate the expression levels of the various receptors, and the results are summarized in Table 1. These data indicate that the expression level of uPAR or αMβ2 was not influenced by the presence or absence of the other receptor. The nontransfected HEK293 cells exhibited no detectable expression of αMβ2and very low expression (∼2-fold above background) of uPAR. The level of uPAR expression in the transfected cells was 8- to 9-fold higher than that of the endogenous receptor and was similar in the presence or absence of αMβ2. The αMβ2 expression levels were similar in the presence or absence of uPAR. Thus, functional differences between αMβ2 in the αMβ2 cells and the αMβ2/uPAR cells or between uPAR in the uPAR cells and αMβ2/uPAR cells could not be attributed to the expression levels of the receptors.
Expression levels of αMβ2 and uPAR on wild-type and transfected HEK293 cells
| HEK293 cell lines . | Normal mouse IgG, MFI . | Anti-αM mAb, MFI . | Anti-β2 mAb, MFI . | Anti-uPAR mAb, MFI . |
|---|---|---|---|---|
| αMβ2 | 38.0 | 558.9 | 550.1 | 48.0 |
| αMβ2/uPAR | 26.3 | 541.2 | 600.1 | 390.9 |
| uPAR | 32.1 | 25.2 | 30.5 | 425.5 |
| Mock | 24.0 | 22.4 | 29.1 | 59.7 |
| HEK293 cell lines . | Normal mouse IgG, MFI . | Anti-αM mAb, MFI . | Anti-β2 mAb, MFI . | Anti-uPAR mAb, MFI . |
|---|---|---|---|---|
| αMβ2 | 38.0 | 558.9 | 550.1 | 48.0 |
| αMβ2/uPAR | 26.3 | 541.2 | 600.1 | 390.9 |
| uPAR | 32.1 | 25.2 | 30.5 | 425.5 |
| Mock | 24.0 | 22.4 | 29.1 | 59.7 |
Expression of αMβ2 and uPAR on the surface of HEK293 cells was determined by FACS analysis using mAbs 3936 and 44a, IB4 to quantitate uPAR and αMβ2, respectively. The data are expressed as mean fluorescence intensity (MFI) and are representative of 3 independent analyses.
Adhesion of the various cell lines to different forms of uPA, enzymatically inactive, single-chain uPA (HMW-sc-uPA), enzymatically active 2-chain uPA (HMW-tc-uPA), and diisoproprylfluorophosphate-inactivated (DIP)–HMW-tc-uPA, was measured. All 3 forms of uPA supported adhesion of the αMβ2 cells, even though uPAR expression was minimal on these cells (Figure 2A), indicating that recognition of uPA by αMβ2does not require proteolytically active uPA. The extent of adhesion to these 3 uPA forms was similar and was similar to that observed with the cells expressing uPAR alone. In contrast, mock-transfected cells or cells expressing a different β2 integrin, αLβ2, which expressed similarly low levels of endogenous uPAR as the αMβ2 cells, failed to adhere to uPA. Additionally, uPA recognition by αMβ2/uPAR cells was enhanced 2-fold compared with the cells expressing αMβ2 or uPAR alone, suggesting additive contributions of the 2 receptors to recognition of the ligand. Adhesion of all cell lines to a control protein, BSA, was negligible.
Integrin αMβ2 binds HMW-tc-uPA and uPAR enhances the interaction.
(A) Adhesion to uPA. HEK293 cells (2 × 105cells/well) expressing uPAR or β2-integrin receptors or both were seeded onto wells of microtiter plates coated with HMW-sc-uPA, HMW-tc-uPA, DIP–HMW-tc-uPA (HMW-tc-uPA was treated with 5 mM DFP for 2 hours at room temperature and extensively dialyzed against PBS), or BSA. Cells were allowed to adhere for 30 minutes at 37°C. After several washings, the number of adherent cells was quantified using the Cyquant Cell Proliferation Kit as described in “Materials and methods.” The data are means ± SEM of quadruple measurements from 3 independent experiments. (B) Direct binding of tc-uPA. HEK αMβ2 and mock cells were incubated in DMEM/F-12 medium/1 mM Mg2+/0.1% BSA with 200 nM Alexa 488–HMW-tc-uPA for 0 to 4 hours at 37°C. Cell-bound uPA was detected by FACS. (C) Migration to uPA. HEK293 cells were added (2 × 105 cells/well) to the upper chamber of transwells in a serum-free DMEM/F-12 medium. HMW-sc-uPA or HMW-tc-uPA was added to the lower chamber (100 nM). The cells were allowed to migrate for 6 hours in a humidified incubator at 37°C and 5% CO2. The number of migrated cells was determined as described in “Materials and methods.” The data are expressed as means ± SEM of duplicate wells from 3 independent experiments.
Integrin αMβ2 binds HMW-tc-uPA and uPAR enhances the interaction.
(A) Adhesion to uPA. HEK293 cells (2 × 105cells/well) expressing uPAR or β2-integrin receptors or both were seeded onto wells of microtiter plates coated with HMW-sc-uPA, HMW-tc-uPA, DIP–HMW-tc-uPA (HMW-tc-uPA was treated with 5 mM DFP for 2 hours at room temperature and extensively dialyzed against PBS), or BSA. Cells were allowed to adhere for 30 minutes at 37°C. After several washings, the number of adherent cells was quantified using the Cyquant Cell Proliferation Kit as described in “Materials and methods.” The data are means ± SEM of quadruple measurements from 3 independent experiments. (B) Direct binding of tc-uPA. HEK αMβ2 and mock cells were incubated in DMEM/F-12 medium/1 mM Mg2+/0.1% BSA with 200 nM Alexa 488–HMW-tc-uPA for 0 to 4 hours at 37°C. Cell-bound uPA was detected by FACS. (C) Migration to uPA. HEK293 cells were added (2 × 105 cells/well) to the upper chamber of transwells in a serum-free DMEM/F-12 medium. HMW-sc-uPA or HMW-tc-uPA was added to the lower chamber (100 nM). The cells were allowed to migrate for 6 hours in a humidified incubator at 37°C and 5% CO2. The number of migrated cells was determined as described in “Materials and methods.” The data are expressed as means ± SEM of duplicate wells from 3 independent experiments.
Interaction of uPA as a soluble ligand with the αMβ2-transfected cells also was demonstrated. Binding of the Alexa-labeled HMW-tc-uPA with the αMβ2-expressing HEK cells was detectable by FACS (Figure 2B). The interaction was time dependent and reached a plateau within 30 minutes and remained constant for up to 4 hours. Inactivation of the HMW-tc-uPA with diisopropyl-fluorophosphate (DFP) did not affect its binding to the αMβ2 cells, and no uPA binding to mock cells was detected (Figure 2B).
Analyses were also undertaken to determine whether uPA could support αMβ2-mediated cell migration of the transfected cells and whether uPAR could influence this response. HMW-sc-uPA and HMW-tc-uPA supported migration not only of uPAR cells but also cells expressing αMβ2, whereas mock-transfected cells failed to migrate to both forms of uPA (Figure2C). The migration of the αMβ2-transfected cells to HMW-tc-uPA was blocked by mAbs 44a and IB4 to the αM and β2 subunits, respectively. Consistent with the data of adhesion experiments, the migration of cells expressing both receptors was higher than that observed with transfectants expressing the individual receptors.
αMβ2 and uPAR recognize different domains of uPA
HMW-tc-uPA is composed of an N-terminal growth factor domain (GFD), a kringle domain (KD), and LMW-tc-uPA, which contains the protease domain. These various domains of uPA were used to compare the recognition specificity of αMβ2 and uPAR. The analyses were first performed in adhesion assays with the various HEK293 cell lines. uPAR binds to the GFD of uPA33,34 and, as expected, the HEK293 cells expressing uPAR adhered to the GFD and to HMW-tc-uPA, which contains the GFD, but not to the KD and LMW-tc-uPA (Figure 3A). The αMβ2 cells adhered to HMW-tc-uPA, KD, and LMW-tc-uPA, but not to the GFD. Thus, αMβ2recognizes a site in HMW-uPA distinct from the GFD recognized by uPAR. Although coexpression of uPAR with αMβ2 on αMβ2/uPAR cells increased adhesion by 50% to 100%, αMβ2 did not appear to enhance HMW-uPA binding mediated by uPAR; that is, adhesion of αMβ2/uPAR cells to the GFD was similar to that of the cells expressing uPAR alone. However, coexpression of the 2 receptors did enhance αMβ2 recognition of the KD and LMW-tc-uPA (Figure 3A); the adhesion of the cells expressing both receptors was enhanced to the uPA domains recognized selectively by αMβ2. Mock-transfected cells did not adhere to uPA or to any of its fragments. As shown on Figure 3B, PMA stimulation enhanced adhesion of neutrophils to HMW-tc-uPA by 2.5-fold compared with nonstimulated cells. The PMA-stimulated neutrophils bound weakly to the GFD, indicating that uPAR can support a low level of adhesion under the conditions of the assay. However, the adhesion of the stimulated neutrophils to KD and LMW-tc-uPA was much more extensive (Figure 3B), consistent with a prominent role of αMβ2 in the interaction. The adhesion of neutrophils to KD, as well as the binding of Alexa 488–labeled HMW-tc-uPA to αMβ2-expressing HEK293 cells detected by FACS (Figure 2B), was not inhibited by the carboxy-terminal lysine analog, 6-aminohexanoic acid (5 mM), or by another kringle-containing molecule, plasminogen (2 μM). These results are consistent with previous data indicating that the KD of uPA lacks a lysine-binding capacity35 and indicate specificity for recognition of the uPA kringle by αMβ2.
Recognition of uPA domains by αMβ2 and uPAR.
HEK293 cell lines (A) and unstimulated or PMA-stimulated (20 nM) PMNs (B) were allowed to adhere to microtiter plates coated with HMW-tc-uPA or its domains for 30 minutes at 37°C. Nonadherent cells were removed by washing with PBS, and the number of adherent cells was quantitated as described in “Materials and methods.” HEK293 cells expressing uPAR or αMβ2 or both (C) and PMA-stimulated PMNs (D) were added to the transwells (2 × 105cells/well) and allowed to migrate for 6 hours at 37°C in 5% CO2. HMW-tc-uPA or its domains were added to the lower chamber of the transwell at 100 nM (C) and 10 nM (D) concentrations. The data are means ± SEM of triple measurements from 3 independent experiments.
Recognition of uPA domains by αMβ2 and uPAR.
HEK293 cell lines (A) and unstimulated or PMA-stimulated (20 nM) PMNs (B) were allowed to adhere to microtiter plates coated with HMW-tc-uPA or its domains for 30 minutes at 37°C. Nonadherent cells were removed by washing with PBS, and the number of adherent cells was quantitated as described in “Materials and methods.” HEK293 cells expressing uPAR or αMβ2 or both (C) and PMA-stimulated PMNs (D) were added to the transwells (2 × 105cells/well) and allowed to migrate for 6 hours at 37°C in 5% CO2. HMW-tc-uPA or its domains were added to the lower chamber of the transwell at 100 nM (C) and 10 nM (D) concentrations. The data are means ± SEM of triple measurements from 3 independent experiments.
Migration of HEK293 cells and neutrophils was assessed to determine which uPA domains supported the response. As shown on Figure 3C, mock-transfected HEK293 cells migrated neither to HMW-tc-uPA nor its domains. The background migration of these cells was 4000 to 5000 cells migrated per transwell in the presence and the absence of all uPA derivatives. HMW-tc-uPA, KD, and LMW-tc-uPA induced migration of the αMβ2 cells by 2- to 3-fold compared with buffer alone, but the GFD failed to support migration of these cells. In contrast, for the uPAR cells (Figure 3C), it was HMW-tc-uPA and the GFD that supported migration, whereas the KD and LMW-tc-uPA did not. This response pattern is consistent with the recognition of the GFD by uPAR and the KD and LMW-tc-uPA by αMβ2. Finally, in the cells transfected with both αMβ2 and uPAR, robust migration was observed with HWM-tc-uPA, GFD, KD, and LMW-tc-uPA (Figure 3C). The migration of the αMβ2/uPAR to the uPA derivatives was more extensive than observed with the single transfectants, again suggesting a combined contribution of the 2 receptors in mounting the response.
As shown in Figure 3D, not only HMW-tc-uPA and its GFD, the uPAR recognition sites, supported migration of PMA-stimulated neutrophils, but also the KD and weakly LMW-tc-uPA elicited a response. The αMβ2 function blocking mAbs 44a and IB4 decreased neutrophil migration to all the uPA domains to background levels, suggesting that this integrin not only directly mediates neutrophil migration toward KD and LMW-tc-uPA but also influences the function of uPAR in mediating cell migration toward the GFD. The function-blocking mAb directed against uPAR reduced neutrophil migration toward HMW-tc-uPA and GFD as anticipated, but also fully suppressed migration toward the KD and partially toward LMW-tc-uPA. Neither normal mouse IgG nor control mAb W6/32 influenced neutrophil migration toward uPA or any of its domains (not shown). Thus, on PMA-stimulated neutrophils, uPAR and αMβ2appear to be strongly linked in regulating the migratory response to uPA.
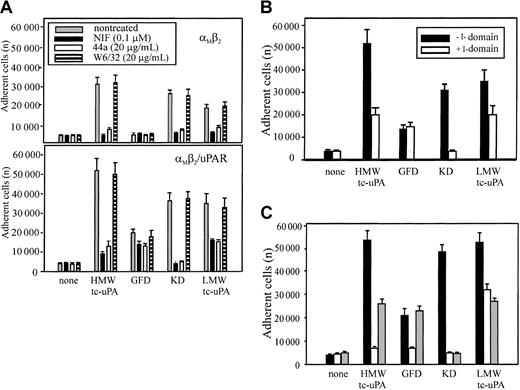
αMβ2 recognizes uPA via its I domain
The mAb 44a and NIF, which blocked HMW-tc-uPA recognition by αMβ2 on neutrophils and HEK293 transfectants, bind to the αMI-(A) domain, the inserted domain of about 200 amino acids in the αMsubunit.36,37 The αMI domain is involved in the binding of many protein ligands to αMβ2.38-40 To determine if uPA also is an αMI domain ligand, αMβ2 and αMβ2/uPAR cells were allowed to adhere to HMW-tc-uPA or its domains in the presence or absence of mAb 44a or NIF. As shown in Figure 4A, both NIF and mAb 44a, but not mAb W6/32, blocked adhesion of the αMβ2 (upper panel) and αMβ2/uPAR (lower panel) cells to intact HMW-tc-uPA and the KD almost completely. Adhesion of these cells to LMW-tc-uPA was reduced by about 50% to 80% by these reagents. The αMβ2 cells did not adhere to the GFD, and the adhesion of αMβ2/uPAR cells to this domain was reduced slightly (20%) by the αMI domain–blocking reagents.
Effect of αMI domain–blocking reagents on the adhesion of αMβ2 and αMβ2/uPAR-expressing cells to uPA and its domains.
(A) Effect of αMI domain reagents. The αMβ2 (upper) or αMβ2/uPAR (lower) HEK293 cells were pretreated with NIF (0.1 μM), mAb 44a, or control mAb W6/32 to MHC-I (20 μg/mL) for 30 minutes at 37°C, and then seeded (2 × 105 cells) onto HMW-tc-uPA or its domains. (B) Effect of αMI domain. Wells, coated with HMW-tc-uPA or its derivatives, were preincubated with or without recombinant αMI domain (100 nM) for 1 hour at 37°C, washed once, and the αMβ2/uPAR cells added. (C) Effect of αMI domain reagents on neutrophil adhesion. The influence of NIF (0.1 μM; white bars) and αMI domain (100 nM; gray bars), or no treatment (black bars) on the adhesion of PMA-stimulated neutrophils to HMW-tc-uPA or its domains. In each case, adhesion was measured after 30 minutes. The data are means ± SEM of triple measurements from 3 independent experiments.
Effect of αMI domain–blocking reagents on the adhesion of αMβ2 and αMβ2/uPAR-expressing cells to uPA and its domains.
(A) Effect of αMI domain reagents. The αMβ2 (upper) or αMβ2/uPAR (lower) HEK293 cells were pretreated with NIF (0.1 μM), mAb 44a, or control mAb W6/32 to MHC-I (20 μg/mL) for 30 minutes at 37°C, and then seeded (2 × 105 cells) onto HMW-tc-uPA or its domains. (B) Effect of αMI domain. Wells, coated with HMW-tc-uPA or its derivatives, were preincubated with or without recombinant αMI domain (100 nM) for 1 hour at 37°C, washed once, and the αMβ2/uPAR cells added. (C) Effect of αMI domain reagents on neutrophil adhesion. The influence of NIF (0.1 μM; white bars) and αMI domain (100 nM; gray bars), or no treatment (black bars) on the adhesion of PMA-stimulated neutrophils to HMW-tc-uPA or its domains. In each case, adhesion was measured after 30 minutes. The data are means ± SEM of triple measurements from 3 independent experiments.
In a separate approach to evaluate the role of the αMI domain in uPA recognition, αMβ2/uPAR cells were allowed to adhere to HMW-tc-uPA or its fragments, which had been preincubated with recombinant αMI domain. As shown in Figure 4B, this preincubation reduced cell binding to intact HMW-tc-uPA by 60%, to LMW-tc-uPA by 40%, and completely blocked adhesion of the cells to the KD. However, preincubation of αMI domain had no effect on cell adhesion to the GFD, which is mediated by uPAR. These results are entirely consistent with the data in Figure 4A and suggest that recognition of KD is mediated by the αMI domain. Recognition of LMW-tc-uPA is at least partially dependent on the αMI domain. Similar results were obtained with PMA-stimulated neutrophils (Figure 4C). The presence of the αMI domain (gray bars) reduced adhesion of the stimulated neutrophils to intact HMW-tc-uPA and LMW-tc-uPA by 50% and reduced adhesion to KD to background levels. One distinguishing feature with neutrophils was the effect of NIF (clear bars) on adhesion to the GFD (Figure 4C). In contrast to the HEK293 αMβ2/uPAR cells, where NIF had a marginal effect on uPAR-mediated adhesion to the GFD, it produced substantial (∼80%) inhibition of neutrophil adhesion to this domain. Thus, αMβ2 may exert significant control on uPAR function on the blood cells.
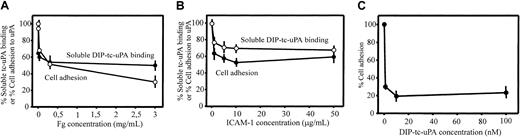
To further examine the involvement of the I domain in uPA binding to αMβ2, we assessed the effects of 2 other αMβ2 ligands, Fg and ICAM-1, which interact with the I domain of the integrin.41 Varying concentrations of these ligands were tested for their effects on the binding of soluble, Alexa 488–labeled HMW-tc-uPA (DFP-inactivated) to αMβ2 cells by FACS and on adhesion of these cells to immobilized HMW-tc-uPA. As shown on Figure 5A, Fg decreased soluble uPA binding and adhesion of the αMβ2 cells in a dose-dependent manner; at the highest concentration tested (3.0 mg/mL), the inhibition of these 2 functions was 50% and 70%, respectively. With ICAM-1, inhibition of soluble HMW-tc-uPA binding and adhesion to the ligand was also observed although the inhibition was less extensive than with Fg. These responses were inhibited by 40% to 50% by ICAM-1 (Figure 5B). In the inverse experiment, wells were coated with ICAM-1, and the effect of varying concentrations of DFP-inactivated HMW-tc-uPA on the adhesion of the αMβ2cells to this immobilized ligand was assessed. As shown on Figure 5C, as little as 1 nM DIP–HMW-tc-uPA reduced αMβ2 cell adhesion to immobilized ICAM-1 by 70%. These results indicate that the binding sites for these αMI domain ligands are overlapping but probably not identical and their interrelationships are complex.28
Relationship between uPA-, fibrinogen-, and ICAM-1–binding sites in αMβ2.
The αMβ2 cells were preincubated with increasing concentrations of Fg (A) or ICAM-1 (B) in DMEM/F-12/1 mM Mg2+, washed once and allowed to bind soluble (DFP-inactivated) Alexa 488–HMW-tc-uPA or the mixture was allowed to adhere to immobilized DIP–HMW-tc-uPA for 30 minutes at 37°C. Cell-bound uPA was detected by FACS, and the number of adherent cells was counted as described in “Materials and methods.” (C) αMβ2 cells were preincubated with DIP–HMW-tc-uPA for 30 minutes at 37°C, washed once, and then seeded onto plates coated with 10 μg/mL ICAM-1. After 30 minutes at 37°C, plates were washed and adherent cells were counted.
Relationship between uPA-, fibrinogen-, and ICAM-1–binding sites in αMβ2.
The αMβ2 cells were preincubated with increasing concentrations of Fg (A) or ICAM-1 (B) in DMEM/F-12/1 mM Mg2+, washed once and allowed to bind soluble (DFP-inactivated) Alexa 488–HMW-tc-uPA or the mixture was allowed to adhere to immobilized DIP–HMW-tc-uPA for 30 minutes at 37°C. Cell-bound uPA was detected by FACS, and the number of adherent cells was counted as described in “Materials and methods.” (C) αMβ2 cells were preincubated with DIP–HMW-tc-uPA for 30 minutes at 37°C, washed once, and then seeded onto plates coated with 10 μg/mL ICAM-1. After 30 minutes at 37°C, plates were washed and adherent cells were counted.
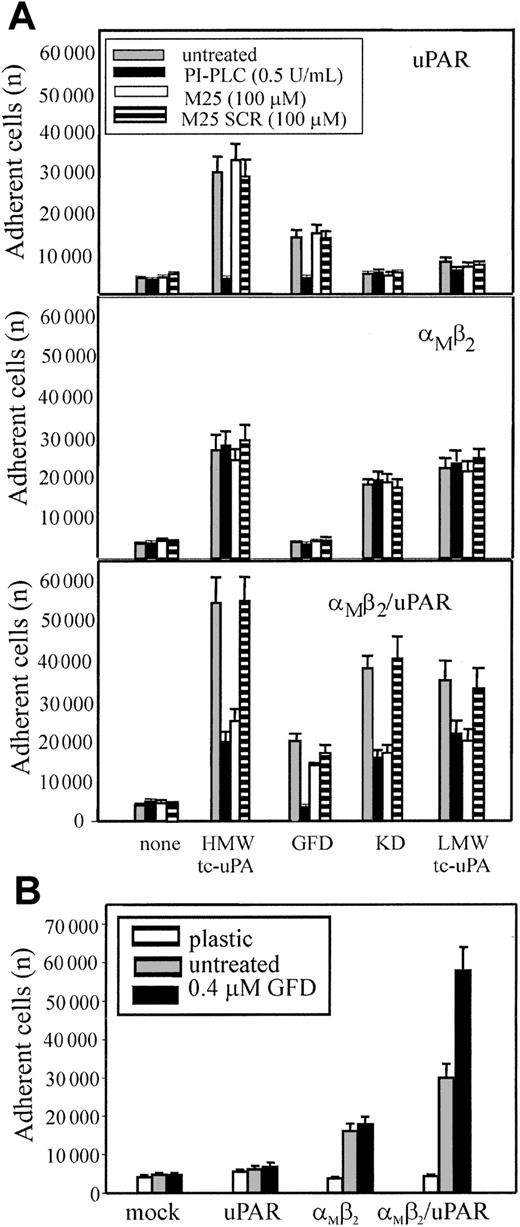
αMβ2 binds HMW-tc-uPA independently of uPAR but uPAR enhances the interaction
To exclude that low levels of uPAR intrinsic to the HEK293 cells might be necessary for αMβ2 to bind uPA, 2 reagents were used: phosphatidylinositol-specific phospholipase C (PI-PLC), which releases uPAR and other glycosylphosphatidylinositol (GPI)–anchored proteins from cell surfaces42 and the M25 peptide, corresponding to the uPAR-binding sequence in the αM subunit, which disrupts αMβ2/uPAR complexes.13 24HEK293-transfected cells were pretreated with PI-PLC, M25 peptide, or a sequenced scrambled peptide (M25 SCR) and then tested for adhesion to the various tc-uPA fragments (Figure 6A). Adhesion of uPAR cells (upper panel) to HMW-tc-uPA and the GFD was completely blocked by PI-PLC treatment, verifying that the reagent was functional. As expected, the M25 peptide or its scrambled (SCR) control did not affect uPAR-mediated adhesion to HMW-tc-uPA of the GFD. PI-PLC treatment and the M25 peptide did not affect adhesion of αMβ2 cells (middle panel) to HMW-tc-uPA, KD, and LMW-tc-uPA, verifying that the αMβ2-mediated adhesion of these cells to these derivatives was uPAR independent. Treatment of αMβ2/uPAR cells (lower panel) with PI-PLC or M25 peptide, but not with SCR, reduced cell adhesion to HMW-tc-uPA, KD, and LMW-tc-uPA by 70%, 60%, and 40%, respectively, to levels similar to that observed for cells expressing only αMβ2. Recognition of GFD was abolished by PI-PLC, whereas M25 and SCR had a negligible effect on adhesion, consistent with the exclusive role of uPAR in recognition of the GFD. With PMA-stimulated neutrophils (not shown), the M25 peptide, but not M25 SCR, reduced their adhesion to KD (as well as to HMW-tc-uPA and LMW-tc-uPA) but not to GFD. PI-PLC reduced their adhesion by more than 80% not only to the GFD but also to the KD, confirming the extensive involvement of uPAR in αMβ2-mediated recognition of uPA.
Influence of uPAR on αMβ2-mediated adhesion to uPA.
(A) uPAR (upper panel), αMβ2 (middle panel), or αMβ2/uPAR (lower panel) transfected HEK293 cells were treated with 0.5 U/mL PI-PLC (black bars), 100 μM M25 peptide (white bars), the control-scrambled M25 peptide SCR (striped bars), or untreated (gray bars) for 30 minutes at 37°C, and then allowed to adhere HMW-tc-uPA or its domains for 30 minutes at 37°C. (B) The indicated transfected cells were preincubated with GFD (0.4 μM) for 30 minutes and then seeded onto kringle domain (KD)–coated wells for 30 minutes at 37°C. After 4 washings with PBS, the adherent cells were quantitated. The results are expressed as means ± SEM of quadruplets from 3 independent experiments.
Influence of uPAR on αMβ2-mediated adhesion to uPA.
(A) uPAR (upper panel), αMβ2 (middle panel), or αMβ2/uPAR (lower panel) transfected HEK293 cells were treated with 0.5 U/mL PI-PLC (black bars), 100 μM M25 peptide (white bars), the control-scrambled M25 peptide SCR (striped bars), or untreated (gray bars) for 30 minutes at 37°C, and then allowed to adhere HMW-tc-uPA or its domains for 30 minutes at 37°C. (B) The indicated transfected cells were preincubated with GFD (0.4 μM) for 30 minutes and then seeded onto kringle domain (KD)–coated wells for 30 minutes at 37°C. After 4 washings with PBS, the adherent cells were quantitated. The results are expressed as means ± SEM of quadruplets from 3 independent experiments.
Based on the observation that considerably more (2- to 3-fold) αMβ2/uPAR cells adhered and migrated to HMW-tc-uPA than cells expressing either αMβ2 or uPAR alone, we hypothesized that uPAR may enhance αMβ2-mediated recognition of uPA. To test this possibility, the various HEK293-transfected cells were preincubated with soluble GFD to allow occupancy of uPAR and then seeded onto KD (Figure 6B). Pretreatment of the mock-transfected 293 and uPAR cells with the GFD did not induce adhesion to KD and did not affect adhesion of the αMβ2 cells to the KD. However, the presence of GFD increased the binding of αMβ2/uPAR cells to KD by 2-fold. In the inverse experiment (data not shown), pretreatment of the αMβ2/uPAR cells with KD did not enhance their adhesion to the GFD. These results suggest that binding of GFD to uPAR may alter the function of αMβ2 to enhance its recognition of KD; on the other hand, recognition of KD of uPA by αMβ2 does not appear to stimulate uPAR recognition of the GFD of uPA.
Discussion
In this study, we have examined the relationship between 2 cell surface receptors, uPAR and αMβ2, and the proteolytic enzyme, uPA. Although these 3 molecules and their interrelationship have been the subject of numerous analyses over the past decade (for reviews, see Chapman and Wei,1 Preissner et al,4 and Chapman et al20), our studies have led to a novel data set. The underpinning of these findings is that uPA is a ligand for αMβ2. This interaction was demonstrated with transfected cells expressing αMβ2 and with cells that express this integrin naturally, peripheral blood neutrophils. The interaction of uPA with αMβ2 supported soluble binding of the ligand and both the adhesion and migration of cells bearing this integrin. The regions of uPA that are recognized by αMβ2 are the kringle and the protease domains. These are distinct from the GFD, which interacts with uPAR. Thus, in addition to the already established interaction between uPAR and αMβ2, uPA has the potential to bind to both receptors simultaneously. Consequently, our data suggest the possible formation of a trimolecular complex involving multiple contacts between uPA/uPAR/αMβ2, which may act as a unique functional unit on the leukocyte surface.
Although the direct binding of uPA to αMβ2and other integrins has not been documented, several recent reports are consistent with uPAR-independent interactions of uPA with cells that may be mediated by integrin. The mitogenic effects of uPA on human lung epithelial and melanoma cells are independent of high-affinity binding of uPA to uPAR43,44 and are consistent with the role of integrins in proliferation. Cell-associated plasminogen activation catalyzed by uPA on the surface of cells lacking uPAR has also been demonstrated.45 Olivier et al documented that uPA inhibits integrin α4β1-mediated T-lymphocyte adhesion and migration to fibronectin.46 This suppression was independent of uPAR and is consistent with a competition between fibronectin and uPA for α4β1 binding. This hypothetical mechanism is particularly interesting because recently 2 other proteinases were shown to reduce integrin-mediated adhesion: matrix metalloproteinase 2 (MMP2) binds to αvβ3 and inhibits ligand binding, leading to decreased cell adhesion,47 and leukocyte elastase binds to αMβ2 and competes with C3bi binding, which results in the detachment of leukocytes from this ligand.48 The latter observations are analogous to those of Sitrin et al,25 Simon et al,14 and May et al9 showing that uPA significantly reduces αMβ2-mediated adhesion of leukocytes to fibrinogen and endothelium as well as fibrin degradation. Our observation that uPA interacts with αMβ2and effectively inhibits (Figure 5C) recognition of ICAM-1 suggests that the interference of uPA with αMβ2-dependent leukocyte adhesion to endothelium9 may be due to direct competition for receptor rather than to disruption of αMβ2-uPAR interaction as previously interpreted.
We have shown that both αMβ2 transfectants and neutrophils migrate toward uPA. The KD and LMW-uPA induced αMβ2-mediated migration, whereas the GFD supported uPAR-dependent cell migration. The capacity of mAbs directed against the αMI domain, the β2 subunit, and uPAR to significantly reduce neutrophil migration toward intact uPA and the various uPA domains supports the model of the multicontact trimolecular complex shown in Figure 7. It is known that uPAR binds to the αM subunit via the M25 sequence located in the W4 repeat, which resides outside but close to the αMI domain (A in Figure 7),13,24 and that uPAR recognizes the GFD of uPA.49 Because GFD enhanced adhesion of αMβ2/uPAR cells to KD, it appears that occupancy of uPAR by uPA influences the functional activity of αMβ2. Such “activation” may be similar to an integrin activator that induces an inside-out signal from within the cell to the ligand binding domain50 or may be the direct result of the interaction between the extracellular domains of the 2 receptors. The consequence of such activation is the enhanced binding of the KD and proteolytic domains of uPA to the αMI domain of αMβ2 (C in Figure 7). Simon et al24 have shown that uPA is required for the interaction of uPAR with αMβ2, and, thus, in the reaction sequence shown in Figure 7, it would be necessary for uPA to bind to uPAR for optimal presentation to αMβ2. Accordingly, uPAR and αMβ2, when in complex, can share uPA as a common ligand to form a trimolecular complex (C in Figure 7). The trimolecular complex may not only be a physical but also a function entity. Cao et al51 observed that intact, but not fragmented uPA, increases cytosolic calcium levels in human neutrophils, which express both αMβ2 and uPAR, but does not do so in neutrophils from patients with leukocyte adhesion deficiency, which lack the β2 integrins including αMβ2. This uPA-mediated calcium rise was inhibited by mAbs to uPAR and to αMβ2 as predicted from the model. In our own cell adhesion and migration studies, the responses of cells to a ligand containing the recognition sites for both uPAR and αMβ2, HMW-tc-uPA, were greater when both receptors could be occupied than when only a single receptor or a ligand that could occupy only a single receptor was presented. On HEK293 cells expressing both uPAR and αMβ2, the 2 receptors appeared to behave relatively independently; that is, the responses were additive compared with the single transfectants. With neutrophils, there appeared to be communication between the 2 receptors; for example, when uPA binding to αMβ2 was blocked, uPAR-mediated responses to uPA were substantially suppressed. Although our model considers only the trimolecular complex of uPA, αMβ2, and uPAR, we do not exclude additional regulatory roles of still higher order multimers induced by uPAR dimerization,52αMβ2 clustering,53 or uPA-mediated bridging of αMβ2 to uPAR on adjacent cells.
Schematic model for uPA/αMβ2/uPAR interactions.
uPAR binds to the GFD of HMW-uPA (A), and this interaction enhances complex formation between uPAR and αMβ2 (B) on the cell surface. In this complex (B), uPAR binds the αM subunit of αMβ2, via M25 sequence, adjacent to the αMI domain. When uPA/uPAR is in complex with αMβ2, this facilitates recognition of the kringle domain (KD) and proteolytic domain (LMW-uPA) of HMW-uPA to the αMI domain of αMβ2 (C). However, αMβ2 is capable of binding HMW-uPA independently from uPAR. (D) Formation of the trimolecular complex (C) enhances adhesion and migration of cells cotransfected with both αMβ2 and uPAR and on cells naturally expressing both receptors, such as neutrophils.
Schematic model for uPA/αMβ2/uPAR interactions.
uPAR binds to the GFD of HMW-uPA (A), and this interaction enhances complex formation between uPAR and αMβ2 (B) on the cell surface. In this complex (B), uPAR binds the αM subunit of αMβ2, via M25 sequence, adjacent to the αMI domain. When uPA/uPAR is in complex with αMβ2, this facilitates recognition of the kringle domain (KD) and proteolytic domain (LMW-uPA) of HMW-uPA to the αMI domain of αMβ2 (C). However, αMβ2 is capable of binding HMW-uPA independently from uPAR. (D) Formation of the trimolecular complex (C) enhances adhesion and migration of cells cotransfected with both αMβ2 and uPAR and on cells naturally expressing both receptors, such as neutrophils.
Although the ordered formation of the trimolecular complex may be the favored reaction sequence, our data indicate that αMβ2 can bind uPA in the absence of uPAR; that is, cells expressing only αMβ2 bound to uPA. Thus, uPA binding to uPAR is not a prerequisite for recognition by αMβ2 (D in Figure 7). uPA resembles certain other αMβ2 ligands, such as Fg, ICAM-1, and C3bi. All are recognized by αMβ2; they interact primarily with the αMI domain, but their binding to αMβ2 is also inhibited by β2mAbs.41 Involvement of the αMI domain in uPA recognition is based on inhibition by mAbs that bind to the domain, NIF, a high-affinity αMI domain ligand, and recombinant αMI domain. In addition, uPA is again similar in being an activation-dependent ligand of αMβ254,55; PMA stimulation of the neutrophil is required for αMβ2 to recognize uPA. However, uPA could bind to the αMβ2 transfectants without an activating stimulus, and its binding did not induce integrin activation as assessed by induction of the activation-dependent epitope of αMβ2, recognized by mAb CBRM1/5,27 although this epitope can be induced on these cells by PMA or Mn2+ (not shown).
In summary, we described a novel interaction between uPA and αMβ2, which would allow for formation of a multicontact trimolecular complex with uPA contacting both uPAR and αMβ2. The interaction of uPA with each receptor is shown to influence the cell adhesion and migration, and uPA allows for cross-talk between uPAR and αMβ2on cells bearing both receptors, including peripheral blood neutrophils. The interactions of uPA with these 2 receptors are likely to influence leukocyte inflammatory and thrombolytic functions.
We thank Dr Timothy A. Springer, Center for Blood Research, Harvard Medical School, Boston, MA, and Drs Li Zhang and Dudley Strickland of the Holland Laboratories, American Red Cross, Rockville, MD, for provision of reagents, and Jane Rein for secretarial assistance.
Prepublished online as Blood First Edition Paper, September 26, 2002; DOI 10.1182/blood-2002-06-1842.
Supported in part by National Institutes of Health grants HL66197 and HL17964.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Edward F. Plow, Joseph J. Jacobs Center for Thrombosis and Vascular Biology, Department of Molecular Cardiology, NB50, Lerner Research Institute, Cleveland Clinic Foundation, 9500 Euclid Ave, Cleveland, OH 44195; e-mail:plowe@ccf.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal