There is evidence that neutrophil production is a balance between the proliferative action of granulocyte–colony-stimulating factor (G-CSF) and a negative feedback from mature neutrophils (the chalone). Two neutrophil serine proteases have been implicated in granulopoietic regulation: pro–proteinase 3 inhibits granulocyte macrophage–colony-forming unit (CFU-GM) growth, and elastase mutations cause cyclic and congenital neutropenia. We further studied the action of the neutrophil serine proteases (proteinase 3, elastase, azurocidin, and cathepsin G) on granulopoiesis in vitro. Elastase inhibited CFU-GM in methylcellulose culture. In serum-free suspension cultures of CD34+ cells, elastase completely abrogated the proliferation induced by G-CSF but not that of GM-CSF or stem cell factor (SCF). The blocking effect of elastase was prevented by inhibition of its enzymatic activity with phenylmethylsulfonyl fluoride (PMSF) or heat treatment. When exposed to enzymatically active elastase, G-CSF, but not GM-CSF or SCF, was rapidly cleaved and rendered inactive. These results support a role for neutrophil elastase in providing negative feedback to granulopoiesis by direct antagonism of G-CSF.
Introduction
The generation of neutrophils from pluripotent progenitors is regulated primarily by granulocyte macrophage–colony-stimulating factor (GM-CSF) and granulocyte–colony-stimulating factor (G-CSF).1 However, at the stage of committed progenitor of neutrophils and monocytes (granulocyte macrophage–colony-forming unit [CFU-GM]), G-CSF is the only growth factor required for neutrophil granulopoiesis. G-CSF stimulates proliferation and differentiation of CFU-GM and is required for cell survival. Mice deficient for the G-CSF growth factor show a deficit in absolute neutrophil number,2 and neutropenia is seen in a canine model by inducing antibody against the G-CSF protein.3 For many years, it has been debated whether the positive G-CSF–mediated regulation of granulopoiesis requires a compensatory negative-feedback arm. Various regulatory models involving the monocyte4 and negative feedback from a granulocyte chalone have been proposed. The granulocyte chalone (never completely identified) present in serum and granulocytes was believed to specifically, but reversibly, inhibit granulopoiesis.5Although no definitive candidate molecule causing negative feedback of granulopoiesis has been found, there is evidence that some neutrophil granule proteins inhibit granulopoiesis. Lactoferrin inhibits hematopoiesis by blocking the production of growth factors by monocytes,6 and pro–proteinase 3 inhibits granulopoiesis by reducing the number of CFU-GMs in S phase.7,8 In 1991 Shellard et al9 showed that a membrane extract from acute myelogenous leukemia (AML) cells (“CAMAL”) could inhibit mouse and human hematopoiesis. Sequencing revealed that CAMAL contained elastase, together with cathepsin G, azurocidin, and proteinase 3. Proteolytic inhibitors were able to reverse this inhibition.9-11 These findings suggested that some of the neutrophil serine proteases might act like a chalone. Finally, recent studies12 13 have shown that mutations in the elastase gene are responsible for congenital and cyclic neutropenia. These results are of interest because they raise the possibility that some neutrophil primary granule proteases may negatively regulate granulopoiesis.
Here we further explore the effect of purified neutrophil serine protease on neutrophil granulopoiesis in culture. Of the proteases tested, elastase alone rapidly digested G-CSF (but not other growth factors) and could directly antagonize proliferation and maturation-inducing effects of G-CSF in culture. Our results suggest a role for elastase as a negative regulator of G-CSF–stimulated neutrophil granulopoiesis.
Materials and methods
Materials
Elastase, proteinase 3 (Elastin Product, Owensville, MO), cathepsin G, and azurocidin (Athens Research Technology, GA) were resuspended in 50% glycerol/50% 0.02 M sodium acetate (NaOAc) and were stored at −70°C until use. In all experiments, 50% glycerol/50% 0.02 M NaOAc solution without neutrophil serine protease was used as a control. Recombinant human (rh) stem cell factor (SCF), rhG-CSF, and rhGM-CSF were purchased from Peprotech (Rocky Hill, NJ). Human fibronectin was purchased from Biomedical Technologies (Stoughton, MA). Iscove medium was from Cellgro (Herndon, VA). Methocult H4230 and StemSpan serum-free media were purchased from StemCell Technologies (Vancouver, BC, Canada). Fluorescein isothiocyanate (FITC)–conjugated anti-CD66b mouse antibody was from Beckman Coulter (Miami, FL), and the phycoerythrin (PE)–conjugated anti-CD114 (G-CSF receptor) was from Becton Dickinson Biosciences (San Jose, CA). The electrophoresis NuPAGE Bis-Tris (4%-12%) gel and the MES buffer were purchased from Invitrogen (Carlsbad, CA), and the gel was stained using the Gelcode Blue Stain Reagent (Pierce, Rockford, IL). Phenylmethylsulfonyl fluoride (PMSF) was from Sigma (St Louis, MO), and Camco Stain Pak was from Cambridge Diagnostic Products (Fort Lauderdale, FL). Apoptosis was determined using the Annexin V–PE apoptosis detection kit I (Becton Dickinson). Flow cytometry acquisition and analysis were performed using FACScalibur (Becton Dickinson). The cell number was determined using a cell counter (Coulter Counter ZM; Beckman Coulter, Miami, FL).
Cells
All samples were obtained from healthy persons after informed consent. CD34+ cells were obtained from leukapheresis donations from healthy volunteers undergoing G-CSF stimulation and stem cell mobilization under National Institutes of Health Institutional Review Board (NIH IRB)–approved protocol 96-H-0049. Healthy bone marrow was obtained from donors under NIH IRB-approved protocol 99-H-0046. Healthy blood samples were obtained from healthy volunteers after informed consent. Bone marrow mononuclear cells (BMMCs) were separated by Ficoll-Hypaque density separation, counted, and stored in liquid nitrogen until use. In some experiments, positive selection of CD34+ cells was made before counting and freezing. Healthy neutrophils were obtained from fresh blood after Ficoll-Hypaque separation. Neutrophils were recovered from the bottom pellet, after lysing the red cells, and were used immediately.
Progenitor cell (CFU-GM) assays in methylcellulose
BMMCs (3 × 105) were incubated in serum-free Iscove medium with varying concentration of elastase, proteinase 3, cathepsin G, or azurocidin (10 ng/mL-10 μg/mL) for 2 hours.10 Cells were washed once, resuspended in 1 mL serum-free Iscove medium, and added to 4.5 mL Methocult H4230. GM-CSF (5 ng/mL) was then added, and 1-mL cultures were plated in triplicate in 6-well plates and were incubated at 37°C, 5% CO2 for up to 14 days, at which time colonies of more than 20 cells were scored using an inverted microscope. Percentage CFU-GM inhibition was calculated as 100 − (mean colonies in elastase-stimulated wells/mean of colonies in unstimulated cultures × 100).
Suspension cultures of CD34 cells
CD34+ cells (105) were plated in triplicate in 24-well plates in 1 mL StemSpan serum-free medium with varying additions of growth factors, with or without neutrophil serine proteases at the concentrations indicated. The plates were then incubated at 37°C, 5% CO2 for 10 days, at which time cells were counted using a Coulter particle counter ZM as follows: 200 μL each well were diluted to 20 mL with isotone, and cells larger than 5.5 μm were counted. To inhibit elastase enzymatic activity, PMSF was preincubated with elastase (20 μg/mL) for 1 hour at 5 mM at 37°C before use. Alternatively elastase (20 μg/mL) was heat inactivated at 95°C for 15 minutes.
Proliferation assay
CD34+ cells (2.5 × 104) were plated in triplicate in 96-well round-bottom plates in 200 μL StemSpan; serum-free medium was supplemented with various growth factor combinations with or without neutrophil elastase at the concentrations indicated in the figures. Plates were then incubated at 37°C in 5% CO2. Proliferation was assessed by 3H-thymidine incorporation (final concentration 1 μCi (0.037 MBq)/well), added for the final 16 hours of culture. All assays, with or without protein, were carried out in triplicate. Cultures were harvested on fiberglass paper, and incorporation of 3H-thymidine was determined by scintillation counting.
Measurement of cell death
CD34+ cells (106) were plated in 12-well plates in 2 mL StemSpan serum-free medium with 25 ng/mL of G-CSF, GM-CSF, and SCF, with or without 1 μg/mL neutrophil elastase. Plates were then incubated at 37°C, 5% CO2. Apoptosis was measured daily by flow cytometry using the Annexin V–PE apoptosis detection kit. Daily samples were also examined morphologically: maturation of the myeloid series and apoptotic cell numbers were evaluated on Camco Stain Pak-stained slides. Maturation index was determined by the following formula: (Stab cell number + segmented nuclear cell number)/total number of all myeloid series. Apoptotic cells were morphologically assessed by shrinkage of the cells, condensation and fragmentation of the nuclei, and presence of apoptotic bodies, and the apoptotic cell ratio to total cell number was calculated. Data were expressed as the mean of 3 counts of 1000 cells each in 3 areas of the slide. On day 9, remaining cells were harvested, labeled with monoclonal anti-CD66b–fluorescein isothiocyanate (FITC), and analyzed by flow cytometry.
Flow cytometry
CD34+ (105) cells were plated in 24-well plates in 1 mL StemSpan serum-free medium with 10 ng/mL G-CSF, GM-CSF, and SCF with or without 1 μg/mL neutrophil elastase. Plates were incubated at 37°C in 5% CO2. On day 7, cells were harvested, labeled with FITC-conjugated anti-CD66b or phycoerythrin (PE)–conjugated anti-CD114 (G-CSF receptor) mouse antibodies for 30 minutes on ice, washed twice, fixed in 1% paraformaldehyde, and analyzed by flow cytometry. Neutrophils (106) were incubated overnight at 37°C, 5% CO2 in 2 mL StemSpan serum-free medium with or without 1 μg/mL neutrophil elastase, directly stained with FITC-conjugated anti-CD66b or PE-conjugated anti-CD114, and analyzed by flow cytometry.
Cleavage of G-CSF by elastase
Proteolytic digestion assays were performed as previously described14 with the following modification. Lyophilized rhG-CSF, rhGM-CSF, rhSCF, and fibronectin were resuspended in phosphate-buffered saline (PBS) at 100 μg/mL. Aliquots of 20 μL were mixed with 2 μg/mL neutrophil elastase, cathepsin G, 25 μg/mL proteinase 3, or no enzyme and were incubated at 37°C. After 15 minutes the reaction was stopped by the addition of NuPAGE sample buffer (106 mM Tris HCl, 10% glycerol, and 2% lithium dodecyl sulfate) and incubated for 10 minutes at 70°C. Samples were separated by electrophoresis on NuPAGE Bis-Tris gels (4%-12%). Gels were stained using the Gelcode Blue Stain Reagent.
Statistical analysis
Values shown are the mean ± standard error of the mean (SEM). Significant differences between groups were evaluated using the Student 2-tailed t test.
Results
Effect of serine proteases on CFU-GM
CFU-GMs were cultured with varying concentrations of elastase, proteinase 3, azurocidin, and cathepsin G. In 4 persons tested, colony numbers in the presence of elastase were reduced in a dose-dependent fashion, with the maximum colony inhibition of 32% observed at 10 μg/mL. Colonies grown in the presence of elastase also appeared smaller, and the cells were less refractile. In contrast, no consistent CFU-GM inhibition was observed with other serine proteases (Table1).
Effect of neutrophil serine proteases on CFU-GM growth
| . | No. colonies/5 × 104 BMMCc ± SD (inhibition %) . | ||||
|---|---|---|---|---|---|
| Control . | Elastase (%) . | Proteinase 3 (%) . | Cathepsin G (%) . | Azurocidin (%) . | |
| 10 μg/mL | |||||
| Donor 1 | 140 ± 22 | 82 ± 50 (41) | 136 ± 20 (9) | 122 ± 38 (13) | 134 ± 50 (5) |
| Donor 2 | 130 ± 2 | 90 ± 26 (31) | 98 ± 14 (24) | 126 ± 10 (4) | 136 ± 2 (−4) |
| Donor 3 | 107 ± 7 | 80 ± 6 (26) | 108 ± 5 (−1) | ND | ND |
| Donor 4 | 228 ± 6 | 161 ± 4 (29) | 228 ± 3 (0) | ND | ND |
| Inhibition, mean % | 32 | 8 | 9 | 1 | |
| 1 μg/mL | |||||
| Donor 1 | — | 116 ± 50 (17) | 158 ± 36 (−12) | 134 ± 34 (4) | 136 ± 12 (3) |
| Donor 2 | — | 98 ± 12 (25) | 84 ± 24 (36) | 126 ± 8 (4) | 128 ± 8 (2) |
| Donor 3 | — | 74 ± 9 (31) | 109 ± 5 (−2) | ND | ND |
| Donor 4 | — | 148 ± 17 (35) | 225 ± 8 (1) | ND | ND |
| Inhibition, mean % | 27 | 6 | 4 | 3 | |
| 0.1 μg/mL | |||||
| Donor 1 | — | 158 ± 22 (−13) | 148 ± 24 (−5) | 146 ± 16 (−4) | 150 ± 26 (−7) |
| Donor 2 | — | 106 ± 6 (19) | 88 ± 26 (31) | 120 ± 14 (8) | 136 ± 6 (−4) |
| Donor 3 | — | 72 ± 3 (33) | 107 ± 10 (0) | ND | ND |
| Donor 4 | — | 183 ± 7 (20) | 225 ± 10 (1) | ND | ND |
| Inhibition, mean % | 15 | 7 | 2 | −6 | |
| 0.01 μg/mL | |||||
| Donor 1 | — | 128 ± 92 (9) | 144 ± 48 (−3) | 136 ± 54 (2) | 122 ± 50 (13) |
| Donor 2 | — | 94 ± 2 (28) | 94 ± 18 (28) | 128 ± 12 (2) | 126 ± 14 (4) |
| Donor 3 | — | 103 ± 11 (4) | 100 ± 6 (7) | ND | ND |
| Donor 4 | — | 228 ± 3 (0) | 230 ± 2 (−1) | ND | ND |
| Inhibition, mean % | 10 | 8 | 2 | 9 | |
| . | No. colonies/5 × 104 BMMCc ± SD (inhibition %) . | ||||
|---|---|---|---|---|---|
| Control . | Elastase (%) . | Proteinase 3 (%) . | Cathepsin G (%) . | Azurocidin (%) . | |
| 10 μg/mL | |||||
| Donor 1 | 140 ± 22 | 82 ± 50 (41) | 136 ± 20 (9) | 122 ± 38 (13) | 134 ± 50 (5) |
| Donor 2 | 130 ± 2 | 90 ± 26 (31) | 98 ± 14 (24) | 126 ± 10 (4) | 136 ± 2 (−4) |
| Donor 3 | 107 ± 7 | 80 ± 6 (26) | 108 ± 5 (−1) | ND | ND |
| Donor 4 | 228 ± 6 | 161 ± 4 (29) | 228 ± 3 (0) | ND | ND |
| Inhibition, mean % | 32 | 8 | 9 | 1 | |
| 1 μg/mL | |||||
| Donor 1 | — | 116 ± 50 (17) | 158 ± 36 (−12) | 134 ± 34 (4) | 136 ± 12 (3) |
| Donor 2 | — | 98 ± 12 (25) | 84 ± 24 (36) | 126 ± 8 (4) | 128 ± 8 (2) |
| Donor 3 | — | 74 ± 9 (31) | 109 ± 5 (−2) | ND | ND |
| Donor 4 | — | 148 ± 17 (35) | 225 ± 8 (1) | ND | ND |
| Inhibition, mean % | 27 | 6 | 4 | 3 | |
| 0.1 μg/mL | |||||
| Donor 1 | — | 158 ± 22 (−13) | 148 ± 24 (−5) | 146 ± 16 (−4) | 150 ± 26 (−7) |
| Donor 2 | — | 106 ± 6 (19) | 88 ± 26 (31) | 120 ± 14 (8) | 136 ± 6 (−4) |
| Donor 3 | — | 72 ± 3 (33) | 107 ± 10 (0) | ND | ND |
| Donor 4 | — | 183 ± 7 (20) | 225 ± 10 (1) | ND | ND |
| Inhibition, mean % | 15 | 7 | 2 | −6 | |
| 0.01 μg/mL | |||||
| Donor 1 | — | 128 ± 92 (9) | 144 ± 48 (−3) | 136 ± 54 (2) | 122 ± 50 (13) |
| Donor 2 | — | 94 ± 2 (28) | 94 ± 18 (28) | 128 ± 12 (2) | 126 ± 14 (4) |
| Donor 3 | — | 103 ± 11 (4) | 100 ± 6 (7) | ND | ND |
| Donor 4 | — | 228 ± 3 (0) | 230 ± 2 (−1) | ND | ND |
| Inhibition, mean % | 10 | 8 | 2 | 9 | |
ND indicates not determined; —, not applicable.
Suspension cultures
To further examine the effect of elastase on granulopoiesis, suspension cultures of CD34+ cells stimulated with G-CSF, SCF, and GM-CSF were sequentially analyzed over a 10-day period. Proliferation was measured by nucleated cell count and tritiated thymidine uptake. Apoptosis and maturation index were quantitated morphologically. In control cultures, cell numbers increased from 1 × 105 to 2.76 × 106 ± 0.15 (28-fold increase). Cell counts in the presence of 1000 ng/mL and 500 ng/mL elastase increased only 2.5 ± 0.4- and 9.5 ± 2-fold, respectively. There was a concordantly lower tritiated thymidine uptake in cells cultured with elastase (Figure1). Although a decrease in counts and proliferation relative to the controls was observed by day 1 of culture, no morphologic differences between control or elastase cultures were observed in the first 3 days. However, on day 4, an increased proportion of apoptotic cells was observed, reaching 58% ± 12% by day 6. Correspondingly, the maturation index ([Stab cell number + segmented nuclear cell number]/total number) of all myeloid series fell dramatically after day 6 (Figures2-3). On day 9, cells were harvested and examined by flow cytometry. In the untreated culture, more than 35% of the cells were positive for the neutrophil-specific marker CD66b, compared with only 4% of cells cultured in elastase (Figure 2), confirming the elastase-induced neutrophil maturation arrest observed morphologically. Apoptosis was confirmed using annexin V staining; CD34+ cells were cultured with the same cocktail of cytokine in the presence or absence of elastase (1000 ng/mL) and was analyzed daily by flow cytometry. In elastase-treated cultures, apoptosis was already higher by day 2 of culture and reached a maximum on day 5, showing that, in the presence of elastase, neutrophil precursors switched from proliferation and maturation to apoptosis. To determine whether elastase had a nonspecific antiproliferative effect, cultures of B-lymphoblastoid cell lines, K-562 cells, and IL-2–stimulated T cells were incubated for 5 days with or without 1 μg/mL elastase, at which time cells were counted. No effect on cell proliferation was observed in these cultures (data not shown).
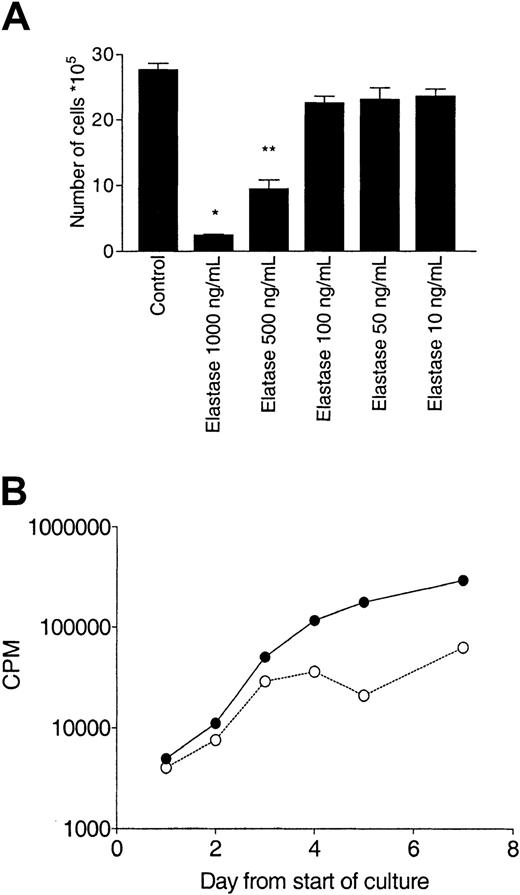
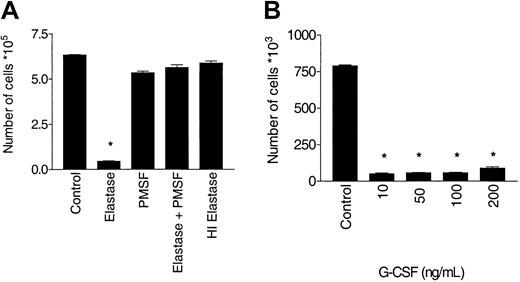
Effect of elastase on CD34+ growth.
CD34+ cells (105/mL) were cultured in the presence of 10 ng/mL G-CSF, GM-CSF, and SCF with different concentrations of elastase. (A) After 10 days of culture, nucleated cells were counted. Asterisks indicate significant differences from control values (*P < .005; **P < .01). Data are representative of 3 independent experiments. (B) Daily measurement of proliferation by tritiated thymidine uptake. Data are presented as mean (± SEM) of the triplicates. CPM indicates counts per minute; ●, control; and ○, elastase.
Effect of elastase on CD34+ growth.
CD34+ cells (105/mL) were cultured in the presence of 10 ng/mL G-CSF, GM-CSF, and SCF with different concentrations of elastase. (A) After 10 days of culture, nucleated cells were counted. Asterisks indicate significant differences from control values (*P < .005; **P < .01). Data are representative of 3 independent experiments. (B) Daily measurement of proliferation by tritiated thymidine uptake. Data are presented as mean (± SEM) of the triplicates. CPM indicates counts per minute; ●, control; and ○, elastase.
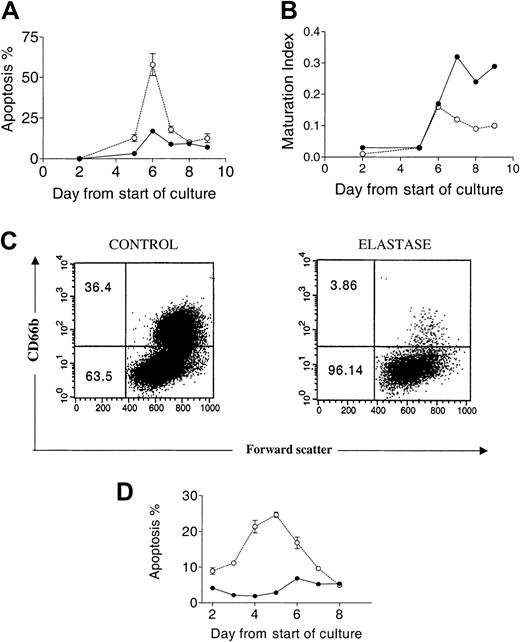
Apoptosis and maturation arrest induced by elastase.
CD34+ cells (5 × 105/mL) were cultured in the presence of 25 ng/mL G-CSF, GM-CSF, and SCF with or without 1 μg/mL elastase. (A) Percentage apoptotic cells determined by morphologic analysis. (B) Maturation index. (C) CD66b+cells day 9. (D) Percentage apoptotic cells determined by annexin V analysis. Data are presented as mean ± SEM of 2 independent experiments. ● indicates control; and ○, elastase.
Apoptosis and maturation arrest induced by elastase.
CD34+ cells (5 × 105/mL) were cultured in the presence of 25 ng/mL G-CSF, GM-CSF, and SCF with or without 1 μg/mL elastase. (A) Percentage apoptotic cells determined by morphologic analysis. (B) Maturation index. (C) CD66b+cells day 9. (D) Percentage apoptotic cells determined by annexin V analysis. Data are presented as mean ± SEM of 2 independent experiments. ● indicates control; and ○, elastase.
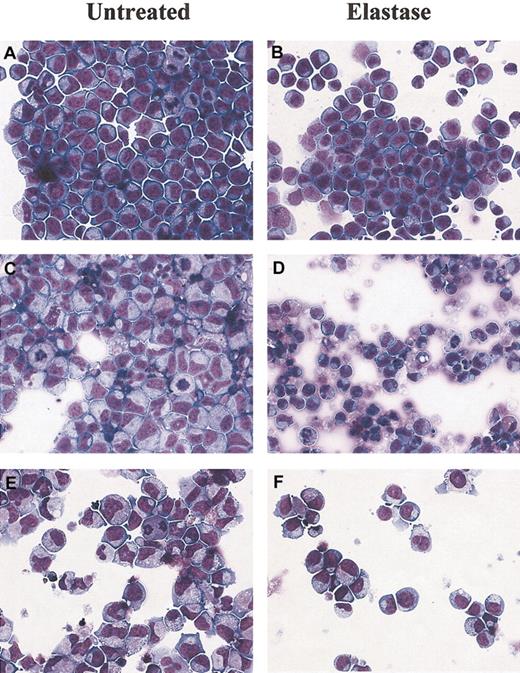
Morphologic changes in culture induced by elastase.
Cytospin preparations of CD34+ cells cultured with 25 ng/mL G-CSF, GM-CSF, and SCF and with or without 1 μg/mL elastase on days 3, 6, and 8. (A-B) Day 3: no differences between control and elastase-treated cells. (C-D) Day 6: widespread apoptosis in differentiating elastase-treated CD34+ cells. (E-F) Day 8: mature neutrophils observed almost exclusively in control cultures. Original magnification, × 400.
Morphologic changes in culture induced by elastase.
Cytospin preparations of CD34+ cells cultured with 25 ng/mL G-CSF, GM-CSF, and SCF and with or without 1 μg/mL elastase on days 3, 6, and 8. (A-B) Day 3: no differences between control and elastase-treated cells. (C-D) Day 6: widespread apoptosis in differentiating elastase-treated CD34+ cells. (E-F) Day 8: mature neutrophils observed almost exclusively in control cultures. Original magnification, × 400.
Interactions between elastase and growth factors
The effect of elastase in cultures stimulated with single growth factors or combinations of growth factors was further explored. SCF and GM-CSF, but not G-CSF, induced proliferation in CD34+ elastase-treated cultures (Figure4). Untreated cultures combining G-CSF and GM-CSF showed enhanced proliferation over cultures in the presence of GM-CSF alone. Elastase weakly affected the proliferation of GM-CSF–stimulated cultures but reduced the proliferation of G-CSF plus GM-CSF cultures to the level observed with GM-CSF alone. Finally, elastase completely blocked proliferation of cultures grown in the presence of G-CSF alone. The abrogation of G-CSF–stimulated proliferation by elastase was dose dependent and required at least 500 ng/mL elastase (Figure 4; Table2).
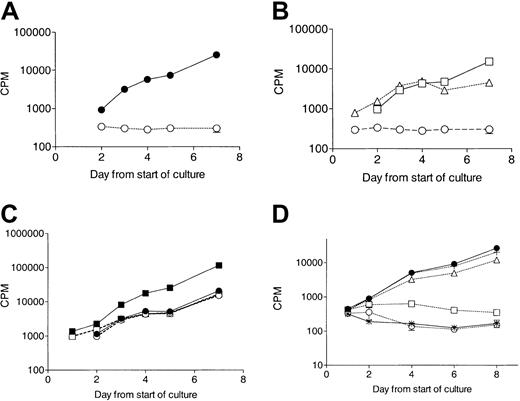
No response to G-CSF in elastase-treated CD34+ cells.
CD34+cells (105/mL) were cultured in the presence of 10 ng/mL cytokine with or without 1 μg/mL elastase. Proliferation was measured daily by tritiated thymidine uptake. (A) No proliferation in response to G-CSF in elastase-treated culture. ● indicates control; and ○, elastase. Data are representative of 3 independent experiments. (B) Proliferation in response to GM-CSF or SCF but not G-CSF in the presence of elastase (1 μg/mL). ○ indicates G-CSF; ■, GM-CSF; and ▵, SCF. Data are representative of 3 independent experiments. (C) Loss of synergy between GM-CSF and G-CSF in elastase-treated cultures. Data presented are the mean (± SEM) of triplicates. ● indicates control GM-CSF; ▪, control GM-CSF + G-CSF; ○, elastase GM-CSF; and ■, elastase GM-CSF + G-CSF. (D) Proliferation response to G-CSF incubated with increasing doses of elastase (mean ± SEM of triplicates). ● indicates control; *, no G-CSF/no elastase; ○, 1 μg/mL elastase; ■, 500 ng/mL elastase; ▵, 250 ng/mL elastase; and +, 62.5 ng/mL elastase).
No response to G-CSF in elastase-treated CD34+ cells.
CD34+cells (105/mL) were cultured in the presence of 10 ng/mL cytokine with or without 1 μg/mL elastase. Proliferation was measured daily by tritiated thymidine uptake. (A) No proliferation in response to G-CSF in elastase-treated culture. ● indicates control; and ○, elastase. Data are representative of 3 independent experiments. (B) Proliferation in response to GM-CSF or SCF but not G-CSF in the presence of elastase (1 μg/mL). ○ indicates G-CSF; ■, GM-CSF; and ▵, SCF. Data are representative of 3 independent experiments. (C) Loss of synergy between GM-CSF and G-CSF in elastase-treated cultures. Data presented are the mean (± SEM) of triplicates. ● indicates control GM-CSF; ▪, control GM-CSF + G-CSF; ○, elastase GM-CSF; and ■, elastase GM-CSF + G-CSF. (D) Proliferation response to G-CSF incubated with increasing doses of elastase (mean ± SEM of triplicates). ● indicates control; *, no G-CSF/no elastase; ○, 1 μg/mL elastase; ■, 500 ng/mL elastase; ▵, 250 ng/mL elastase; and +, 62.5 ng/mL elastase).
Effect of elastase on CD 34+ cell growth in 10 ng/mL G-CSF
| . | Cell count, ×105/mL ± SD . | Proliferation index . |
|---|---|---|
| Donor 1 | ||
| Control | 7.89 ± 0.22 | 8 |
| Elastase, 1 μg/mL | 0.51 ± 0.10* | 0.5 |
| Donor 2 | ||
| Control | 6.34 ± 0.05 | 6 |
| Elastase, 1 μg/mL | 0.45 ± 0.05* | 0.5 |
| Donor 3 | ||
| Control | 8.04 ± 2.66 | 8 |
| Elastase, 1 μg/mL | 0.43 ± 0.03† | 0.5 |
| . | Cell count, ×105/mL ± SD . | Proliferation index . |
|---|---|---|
| Donor 1 | ||
| Control | 7.89 ± 0.22 | 8 |
| Elastase, 1 μg/mL | 0.51 ± 0.10* | 0.5 |
| Donor 2 | ||
| Control | 6.34 ± 0.05 | 6 |
| Elastase, 1 μg/mL | 0.45 ± 0.05* | 0.5 |
| Donor 3 | ||
| Control | 8.04 ± 2.66 | 8 |
| Elastase, 1 μg/mL | 0.43 ± 0.03† | 0.5 |
P < .001.
P < .05.
Inhibition of G-CSF–stimulated growth requires enzymatic activity of elastase
The inhibitory effect of elastase on G-CSF was potent and could only be modestly overcome by increasing the dose of G-CSF to 200 ng/mL. To determine whether the inhibitory effect of elastase on G-CSF was mediated by enzymatic activity, the effect of blocking enzymatic activity was studied using PMSF, which irreversibly inhibits the serine proteases, or using heat inactivation to 95°C for 15 minutes. PMSF treatment and heat inactivation completely blocked the inhibitory effect of elastase on G-CSF–stimulated proliferation by cell count and also by tritiated thymidine uptake (data not shown), indicating that enzymatically active elastase was required to inhibit G-CSF (Figure5).
Antagonism of G-CSF stimulation requires enzymatically active elastase.
CD34+ cells (105/mL) were cultured in the presence of 10 ng/mL G-CSF, with or without 1 μg/mL elastase. After 10 days of culture, nucleated cells were counted. (A) Abrogation of the blocking effect of elastase by PMSF or heat inactivation. Data are representative of 3 independent experiments. (B) Increased dose of G-CSF does not override inhibition by elastase (mean ± SEM of triplicates) (*P < .001).
Antagonism of G-CSF stimulation requires enzymatically active elastase.
CD34+ cells (105/mL) were cultured in the presence of 10 ng/mL G-CSF, with or without 1 μg/mL elastase. After 10 days of culture, nucleated cells were counted. (A) Abrogation of the blocking effect of elastase by PMSF or heat inactivation. Data are representative of 3 independent experiments. (B) Increased dose of G-CSF does not override inhibition by elastase (mean ± SEM of triplicates) (*P < .001).
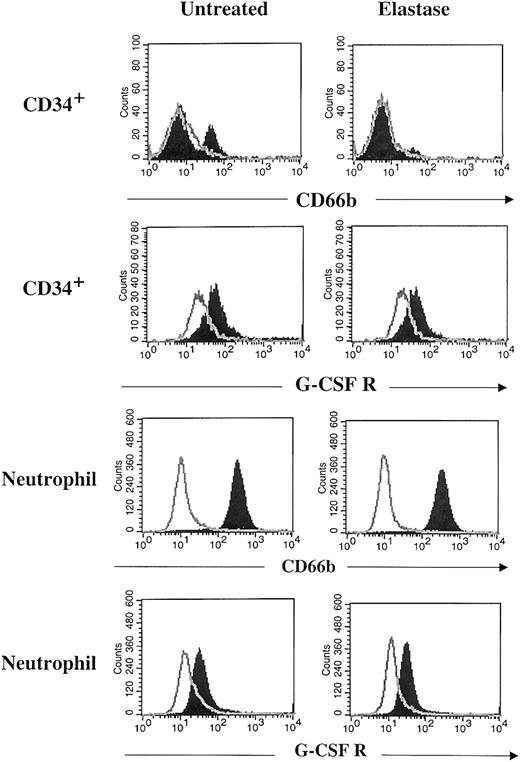
Interaction of elastase with G-CSF and its receptor
To determine whether elastase blocked the effect of G-CSF by its receptor or by direct interaction with the growth factor, G-CSF receptor expression on neutrophils and CD34+ cultivated cells was measured by flow cytometry in the presence and absence of elastase. Although CD66b expression was lost on elastase-exposed CD34+ cultures, no reduction in the surface expression of the G-CSF receptor was seen, nor was it seen on CD66b+blood neutrophils exposed to elastase (Figure6). To study the enzymatic inactivation of G-CSF by elastase, G-CSF, SCF, and GM-CSF were incubated for 15 minutes with 2μg/mL elastase and were analyzed by gel electrophoresis. G-CSF alone was rapidly cleaved by elastase. When the 3 growth factors were first mixed and treated with elastase, only G-CSF was cleaved by elastase. In contrast, proteinase 3 did not cleave G-CSF, and cathepsin G only induced minimal cleavage of G-CSF. Fibronectin, used as a positive control, was cut by the 3 proteases. However, because it has lower affinity for fibronectin, the concentration of proteinase 3 was increased to 25 μg/mL (Figure7).
No cleavage of G-CSF receptor by elastase.
After 7 days of culture with 10 ng/mL G-CSF, GM-CSF, SCF with or without 1 μg/mL elastase, CD34+ cells were harvested and stained with anti–CD66b–FITC, anti–G-CSFR–PE antibodies (filled histogram), or appropriate isotype control (open histogram). Peripheral blood neutrophils were incubated overnight with 1 μg/mL elastase and then stained with the same antibodies.
No cleavage of G-CSF receptor by elastase.
After 7 days of culture with 10 ng/mL G-CSF, GM-CSF, SCF with or without 1 μg/mL elastase, CD34+ cells were harvested and stained with anti–CD66b–FITC, anti–G-CSFR–PE antibodies (filled histogram), or appropriate isotype control (open histogram). Peripheral blood neutrophils were incubated overnight with 1 μg/mL elastase and then stained with the same antibodies.
Digestion of G-CSF by elastase.
(A) Two micrograms cytokine incubated with or without 40 ng elastase for 15 minutes and analyzed by gel electrophoresis. Data are representative of 3 independent experiments. (B) Two micrograms G-CSF or fibronectin incubated with 40 ng elastase, cathepsin G, 500 ng proteinase 3, or without protease for 15 minutes and analyzed by gel electrophoresis. Lanes 4, 9, and 10 show 3 bands corresponding to proteinase 3 in addition to the degradation product of G-CSF (lane 4) and fibronectin (lane 9). Data are representative of 3 independent experiments.
Digestion of G-CSF by elastase.
(A) Two micrograms cytokine incubated with or without 40 ng elastase for 15 minutes and analyzed by gel electrophoresis. Data are representative of 3 independent experiments. (B) Two micrograms G-CSF or fibronectin incubated with 40 ng elastase, cathepsin G, 500 ng proteinase 3, or without protease for 15 minutes and analyzed by gel electrophoresis. Lanes 4, 9, and 10 show 3 bands corresponding to proteinase 3 in addition to the degradation product of G-CSF (lane 4) and fibronectin (lane 9). Data are representative of 3 independent experiments.
Discussion
Here we show that neutrophil elastase blocks the ability of G-CSF to provide proliferative, maturational, and survival signals to neutrophils and their precursors. First, elastase inhibited normal CFU-GM growth, with the result that colonies that form do not attain the size of control colonies and appear to senesce before day 10. The mechanism of colony inhibition is unclear. It is possible that elastase digests G-CSF produced by BMMCs during preincubation in the absence of growth factor or in the culture in methylcellulose bound to the membrane of the BMMCs.15 To further study the effect of elastase on granulopoiesis, suspension cultures of CD34 cells in the presence of abundant growth factors were studied. Elastase blocked the generation of mature CD66b+ neutrophils, increasing the apoptotic score of maturing neutrophil progenitors but only minimally affecting undifferentiated cells. Apoptosis of developing neutrophils was confirmed by annexin V flow cytometric analysis. Because annexin V is an early marker of apoptosis, the maximum apoptosis was seen on day 5, preceding the morphologic changes seen on day 6. To determine which growth factor was antagonized by elastase, we cultured CD34 cells in the presence of G-CSF, GM-CSF, or SCF. CD34 cells proliferated with GM-CSF or SCF alone, but proliferation under the influence of G-CSF was almost completely blocked by elastase. The elastase preparation used was more than 95% pure, but it is derived from neutrophil extracts and could contain other granule proteins. However, the other neutrophil serine protease granules (proteinase 3, cathepsin G, and azurocidin) did not inhibit colony growth and showed no comparable effect on proliferation in suspension culture. The blocking effect on G-CSF–stimulated proliferation was thus attributable to elastase. The possibility that elastase had a nonspecific effect on proliferating cells was also considered. However, elastase did not inhibit proliferation in leukemic cell lines, B cells, or T cells. Furthermore, identical results in repeated experiments using another commercial source of elastase (Calbiochem, La Jolla, CA) were found (data not shown). Finally, elastase appeared to counteract primarily the proliferative effect of G-CSF but not GM-CSF or SCF.
Inactivation of elastase with PMSF or heat treatment showed that the blocking effect on G-CSF required enzymatic activity. This raised the possibility that elastase might act by cleaving G-CSF or its receptor. The G-CSF receptor contains a fibronectin 3 domain in the extracellular region near the cytoplasmic junction.16 Elastase is known to cleave fibronectin.17 The possibility was considered that the G-CSF receptor could be enzymatically clipped from the cell surface by elastase, thus depriving the cell of a mechanism for G-CSF signaling. This did not appear to be the case; using antibodies specific for the G-CSF receptor, neither elastase-treated CD34 cells nor neutrophils lost surface G-CSF receptors. Because the epitope recognized by the antibody is unknown, the possibility that elastase cuts above this epitope cannot be excluded. Rather, the main antagonistic action of elastase appeared to be caused by enzymatic digestion of G-CSF (but not SCF or GM-CSF). By extrapolation from the in vitro experiment, a single molecule of elastase appears to be capable of rapidly digesting more than 70 molecules of G-CSF. The rapid digestion of G-CSF was not observed with the other serine proteases tested, suggesting a specific elastase–G-CSF interaction.
How might these in vitro observations relate to the action of G-CSF in vivo? Normally serum neutrophil elastase levels are in the order of 200 ng/mL, but much of it is complexed with serpins and therefore is inactive. However, elastase levels correlate directly with the number of peripheral blood neutrophils,18 and 106 neutrophils can release up to 2 μg/mL elastase.19 Furthermore, changes in elastase accompany the administration of G-CSF. After a single subcutaneous dose of G-CSF, elastase levels rise, both in the blood20 and in the bone marrow.14 Thus the concentration of 500 ng/mL, effective in our cultures, could easily be reached in G-CSF–stimulated bone marrow. The potent inactivation of G-CSF by elastase could explain its rapid disappearance from the circulation seen after parenteral administration. Interestingly, a large body of data defines an inverse relationship between neutrophil count and G-CSF level.21-23 It has been suggested that the presence of G-CSF receptors on neutrophils is responsible for this relationship, but because neutrophil counts and elastase levels also correlate, this correlation could alternatively be reinterpreted as a direct effect of elastase on G-CSF. It is therefore possible that elastase release by neutrophils represents a negative feedback loop of granulopoietic regulation.
The fact that mice deficient in the elastase gene have normal neutrophil counts24,25 could be explained by redundancy in the system controlling granulopoiesis. Recent studies have shown that granulopoiesis is negatively regulated by pro–proteinase 3 and pro-azurocidin (but not pro-elastase, pro–cathepsin G, mature proteinase 3, or azurocidin).7 8 Because these neutrophil serine protease proforms are only produced in primary granules at the promyelocyte stage, they may represent a primary granule feedback path, distinct from an elastase–G-CSF feedback from mature neutrophils.
Elastase is normally retained within the neutrophil granule until the cell degranulates or dies in the tissue compartment. Although large amounts of elastase must be generated daily from the normal process of neutrophil senescence, the events occur in tissues peripheral to the marrow. Furthermore, elastase is normally inactivated by circulating α-1 antitrypsin and α-2 macroglobulin, thus limiting its impact on G-CSF–regulated events in the marrow. Nevertheless, elastase released in the tissues could affect neutrophil survival by locally destroying G-CSF and promoting apoptosis. During pus formation, the release of elastase into the environment in which neutrophils accumulate and degranulate might trigger a chain reaction of elastase release and death of neutrophils deprived of the antiapoptotic effect of G-CSF.
Recently, neutrophil serine proteases have been implicated in the process of neutrophil mobilization.14 In mice, exogenously administered G-CSF increases intramedullary elastase and cathepsin G, which in turn cleaves stromal cell vascular cellular adhesion molecule-1 (VCAM-1), reducing cellular adhesiveness and mobilizing marrow cells into the circulation. Again, by inactivating G-CSF, elastase may modulate mobilization.
Recent findings implicate elastase mutations in the pathophysiology of cyclic neutropenia and congenital neutropenia. However, the pattern of mutation in the neutrophil elastase gene differs in the 2 disorders,26 suggesting that the mutated enzyme acts by a different mechanism in each disease. Mutated elastase complexes poorly with α-1 antitrypsin27 and may be less sensitive to other serpins. This raises the possibility that neutropenia could be a consequence of increased destruction of G-CSF by more abundant elastase. Our findings support this hypothesis: mutated elastase, uninhibited by serpin family members, would increase with the generation of mature neutrophils to accumulate in sufficient concentrations to block G-CSF function. The subsequent neutropenia would persist until the concentration of elastase falls to a level permitting G-CSF stimulation of a further burst of neutrophil production. This hypothesis is consistent with mathematical models of cycling in cyclic neutropenia.26 Conversely, our findings do not readily implicate elastase in severe congenital neutropenia. Mosaicism for the elastase mutation in the healthy father of a child with severe congenital neutropenia (SCN) suggests that the mutant enzyme mediates its effect within the cell in which it is produced rather than by a paracrine effect.28 Furthermore G-CSF–deficient mice, although neutropenic, do not show a block in myeloid maturation in the marrow, making it unlikely that the pathogenesis of congenital neutropenia is solely caused by enhanced degradation of G-CSF. Additional studies are needed to clarify whether digestion of G-CSF by elastase may play a role in triggering cyclic or congenital neutropenia.
Finally, elastase levels are very high in certain myeloid leukemias.18,29 30 The question arises whether an inhibitory effect of elastase on normal granulopoiesis contributes to the dominance of a G-CSF–independent leukemic clone or, alternatively, whether elastase release by leukemic cells could contribute to the maturation arrest of a G-CSF–sensitive leukemic blast cell.
In conclusion, our study points toward an important role for elastase in antagonizing G-CSF, which blocks granulopoiesis in vitro and may modulate the actions of the growth factor in vivo. Further studies will be necessary to define the significance of elastase in the regulation of neutrophil production and G-CSF function in vivo in normal and pathologic states.
We thank Dr Magnusson Magnus for critical reading of this manuscript. We also thank Dr Philippe Martiat, Dr Philippe Lewalle, and Dr Redouane Rouas for their help.
Prepublished online as Blood First Edition Paper, October 17, 2002; DOI 10.1182/blood-2002-06-1734.
Supported by grants from MEDIC Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John Barrett, Hematology Branch, National Heart, Lung and Blood Institute, National Institutes of Health, Bldg 10, Rm 7C103, 9000 Rockville Pike, Bethesda, MD 20892; e-mail:barrettj@nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal